Abstract
AIM:
The main purpose of the present study was to test the β-tubulin and rodletA genes for rapid identification of Aspergillus fumigatus.
MATERIALS AND METHODS:
Fifty-one A. fumigatus strains including environmental, clinical and reference isolates were tested in this research. PCR was carried out based on βtub and rodA partial gene sequences.
RESULTS:
A 198 bp DNA fragment was obtained using βtub gene. PCR amplification of the rodA gene resulted in a 313 bp band. The βtub and rodA genes PCR products exhibited a 100% homology with the associated sequences in the GenBank.
CONCLUSION:
In the present study, we used a PCR approach that was able to discriminate A. fumigatus from other related species within the section Fumigati.
Keywords: β-tubulin, RodletA, Aspergillus fumigatus
Introduction
Aspergillus fumigatus is an environmental filamentous fungus and is the main causal agent of the aspergillosis. Increasing the use of immunosuppressive therapy for treating many human diseases, the incidence of mortality rate of invasive aspergillosis (IA) is raised between 30–95% [1].
Teleomorphic species of the Aspergillus section Fumigati (AsF) belong to the Neosartorya genus. Aspergillus section Fumigati (AsF) is an economically important fungus and teleomorphic species of this section belong to the genus Neosartorya. Seventeen species of Neosartorya and 8 strictly mitotic species are typically recognized [2].
Aspergillus section Fumigati and also its teleomorph Neosartorya are significantly important in clinic because they are able to pathogenic or allergenic to human [3-5], responsible for food spoilage and producing mycotoxins [6, 7].
The clinical isolates of Aspergillus species are not necessary morphological the same and wrong recognitions of species with morphological characteristics have frequently happened. With the intention of development the diagnostic method, including DNA detection it is important to elucidate intra and interspecies variety in A. fumigatus and closely related species.
Misrecognition of species within the section Fumigati has been frequently reported by clinical laboratories. Species, for example, Aspergillus viridinutans, Aspergillus lentulus, Aspergillus fumisynnematus, Aspergillus fumigatiaffinis, Neosartorya pseudofischeri, Neosartorya udagawae and Neosartorya hiratsukae, are commonly reported as A. fumigatus [8, 9].
A number of biochemical and molecular approaches have been performed for identification of A. fumigstus and related species. Sequencing of genes, for example, ITS, calmodulin, actin, βtubulin (βtub) and rodlet A (rodA), has been used to discrimination A. fumigatus from related species [10, 11]. In the current study, the βtub and rodA gene were tested for identification of Aspergillus section Fumigati.
Materials and Methods
Microorganisms
A total of 51 A. fumigatus strains including environmental, clinical and reference isolates were used in this study. The following strains of A. fumigatus were used as a reference: IBRC-M 30033, IBRC-M 30040, IBRC-M 30048. Eight clinical isolates were kindly provided by Dr Mojtaba Taghizadeh (Mazandaran University of Medical Sciences, Mazandaran, Iran). The environmental isolates were recovered from soil or air. The strains were incubated on Sabouraud dextrose agar (Merck KGaA, Darmstadt, Germany) at 37°C. All A. fumigatus isolates were recognized by morphology. For getting a pure culture, the isolates were subcultured three times and then stained with lactophenol aniline blue. Phenotypic techniques including colony morphology, conidial arrangement, philiades, vesicles and conidiophores were considerated for identification.
DNA extraction
One ml thick spore suspension from each isolate was transferred to an Erlenmeyer flask containig 50 ml yeast extract peptone dextrose medium (Merck KGaA).
The flasks were incubated at 200 rpm under agitation at 27°C for 72 h to allow for mycelium growth.
The harvested mycelia were washed with 0.5 M ethylenediamine tetraacetic acid (EDTA) and sterile distilled water (dH2O) and freeze-dried at -70°C for DNA extraction. Then, the mycelia were ground into a fine powder with a pestle and mortar. The DNA was extracted using the GF-1 Plant DNA Extraction Kit (vivantis, Malysia).
PCR amplification
In our study, PCR was performed based on βtub and rodA partial gene sequences. The primer sets, βtub-F (5’- TGACGGGTGATTGGGATCTC-3’) and βtub-R (5’- CGTCCGCTTCTTCCTTGTTT-3’) was used to amplify a 198bp DNA fragment of the βtub gene. The primer sets, rodA -F (5’- ACATTGACGAGGGCATCCTT -3’) and rodA -R (5’- ATGAGGGAACCGCTCTGATG -3’) was used to amplify a 313bp DNA fragment of the βtub gene. The PCR reactions were prepared to a final volume of 30 μl, comprised of 3 μl 10X reaction buffer, 2.2 mM MgCl2, 200 μM of each dNTP, 2.5 unit of Taq DNA polymerase (CinnaGen, Karaj, Iran), a 30 ng DNA template and 50 pmol of each primer.
An initial denaturation for 5 min at 94°C was followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 68°C for 1 min and extension at 68°C for 2 min. The PCR products were electrophoresed on 2% agarose gel in TAE buffer at 50 V for 2 h and stained with ethidium bromide.
Sequencing
A number of βtub and rodA genes amplicons were submitted for sequencing (Bioneer Corporation, Daejeon, South Korea). Searching in the NCBI database (http://www.ncbi.nlm.nih.gov/) showed that sequences had 100% identity with A. fumigatus βtub and rodA genes. The MEGA5 software package (http://www.megasoftware.net) was employed for alignment of sequences.
Results
Fifty moulds, all formerly recognized as A. fumigatus by morphology, were screened by the PCR method to identify A. fumigatus isolates. The βtub and rodA genes were considered as genes markers.
PCR amplification of the rodA gene for all 51 isolates with primers rodA -F and rodA -R resulted in a 313 bp band (Fig. 1). PCR amplification of the rodA gene for all 51 isolates with primers βtub-F and βtub-R resulted in a 198 bp band (Fig. 2).
Figure 1.
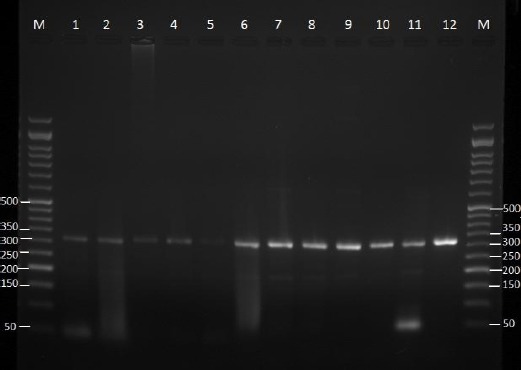
Agarose gel electrophoresis of rodA gene products (313 bp) of the Aspergillus fumigatus (lane 1, 2 references strains; lanes 3-5, clinical isolates; lane 6-12, environmental isolates. Lane M, 50-bp ladder; lane 1, IBRC-M 30040 lane 2, IBRC-M 30048; lane 3, MF6; lane 4, MF30; lane 5, MF34; lane 6, MF13; lane 7, MF17; lane 8, MF35; lane 9, MF39; line 10, MF42; line 11, MF46; line 12, MF53
Figure 2.

Agarose gel electrophoresis of βtub gene products (198 bp) of the Aspergillus fumigatus (lane 1, 2 references strains; lanes 3-5, clinical isolates; lane 6-12, environmental isolates. Lane M, 50-bp ladder; lane 1, IBRC-M 30040 lane 2, IBRC-M 30033; lane 3, MF6; lane 4, MF30; lane 5, MF34; lane 6, MF13; lane 7, MF17; lane 8, MF35; lane 9, MF39; line 10, MF42; line 11, MF46; line 12, MF53
The βtub and rodA genes products were sequenced for several isolates, including the reference strains. A Basic Local Alignment Search Tool (BLAST) search demonstrated that the βtub and rodA genes PCR products exhibited a 100% homology with the associated sequences in the GenBank.
Discussion
Identification of filamentous fungi dissimilar to bacteria, rely mostly on morphological characterizations, however, limitations of phenotypic typing of pathogenic fungi are being progressively more recognized. The new fungal species have recently been recognized within the section Fumigati, Some of them have been associated in severe cases of aspergillosis including pulmonary, cerebral, liver, cutaneous and trabecular bone invasions.
In regard to the A. fumigatus may shows a significant part of all aspergillosis clinical cases, molecular description is important for the accurate detection of species within the section of Fumigati.
Hong et al (2005) reported the variability within A. fumigatus section in Korea by morphology, growth temperature, extrolite patterns and DNA analyses of the partial β-tubulin, actin and calmidulin gene and they suggested two new species which were A. fumigatiaffinis, A. novofumigatus [12].
The phylogenetic associations between A. fumigatus and related species has also been analysed by partial sequencing of cytochrome b gene [13].
Identification of A. fumigatus is vital because this fungus is one of the most significant fungal pathogens. The recognition of Aspergillus spp. isolated from clinical samples depends mostly on morphological features. However, morphology is not enough for the detection of some clinical isolates because of the occurrence of polymorphism and the deprived development of reproductive structure. Consequently, several additional techniques have been used performed in the study of A. fumigatus [10, 14-16]. Burnie et al. employed restriction fragment length polymorphism analysis (RFLP) to discriminate the clinical isolates of A. fumigatus. They were succeeded classify 21 isolates into six types with XbaI digestion [17].
In the present study, we used a PCR approach that was able to discriminate A. fumigatus from other related species within the section Fumigati.
Sequence analysis of PCR products the benA and rodA genes of several isolates revealed that this approach accurately differentiated the non-A.fumigatus from the A. fumigatus isolates.
Acknowledgments
The present study was supported by the Health Research Institute (Infectious and Tropical Diseases Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran) (grant no. 94110).
Footnotes
Funding: The present study was supported by the Health Research Institute (Infectious and Tropical Diseases Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran) (grant no. 94110).
Competing Interests: The authors have declared that no competing interests exist.
References
- 1.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers:human fungal infections. Sci Transl Med. 2012;4(165):165rv13. doi: 10.1126/scitranslmed.3004404. https://doi.org/10.1126/scitranslmed.3004404 PMCid:PMC3627207. [DOI] [PubMed] [Google Scholar]
- 2.Samson RA, Hong SB, Frisvad JC. 2006. Old and new concepts of species differentiation in Aspergillus. Med Mycol. 2006;44:S133–48. doi: 10.1080/13693780600913224. https://doi.org/10.1080/13693780600913224. [DOI] [PubMed] [Google Scholar]
- 3.Brakhage AA, Langfelder K. Menacing mold:the molecular biology of Aspergillus fumigatus. Annu Rev Microbiol. 2002;56:533–455. doi: 10.1146/annurev.micro.56.012302.160625. https://doi.org/10.1146/annurev.micro.56.012302.160625 PMid:12142473. [DOI] [PubMed] [Google Scholar]
- 4.Frisvad JC, Larsen TO. Extrolites of Aspergillus fumigatus and Other Pathogenic Species in Aspergillus Section Fumigati. Front Microbiol. 2016;6:1485. doi: 10.3389/fmicb.2015.01485. https://doi.org/10.3389/fmicb.2015.01485 PMid:26779142 PMCid:PMC4703822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamoth F. Aspergillus fumigatus-Related Species in Clinical Practice. Front Microbiol. 2016;7:683. doi: 10.3389/fmicb.2016.00683. https://doi.org/10.3389/fmicb.2016.00683 PMid:27242710 PMCid:PMC4868848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole RJ, Cox RH. Handbook of toxic fungal metabolites. New York: Academic; 1981. [Google Scholar]
- 7.Larsen TO, Smedsgaard J, Nielsen KF, Hansen MA, Samson RA, Frisvad JC. Production of mycotoxins by Aspergillus lentulus and other medically important and closely related species in section Fumigati. Med Mycol. 2007;45(3):225–32. doi: 10.1080/13693780601185939. https://doi.org/10.1080/13693780601185939 PMid:17464844. [DOI] [PubMed] [Google Scholar]
- 8.Balajee SA, Nickle D, Varga J, Marr KA. Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryot Cell. 2006;5(10):1705–12. doi: 10.1128/EC.00162-06. https://doi.org/10.1128/EC.00162-06 PMid:17030996 PMCid:PMC1595351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong SB, Shin HD, Hong J, Frisvad JC, Nielsen PV, Varga J, Samson RA. New taxa of Neosartorya and Aspergillus in Aspergillus section Fumigati. Antonie Van Leeuwenhoek. 2008;93(1-2):87–98. doi: 10.1007/s10482-007-9183-1. https://doi.org/10.1007/s10482-007-9183-1 PMid:17610141 PMCid:PMC2140094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samson R A, Hong S, Peterson S W, Frisvad JC, Varga Polyphasic taxonomy of Aspergillus section Fumigati and its teleomorph Neosartorya. Stud Mycol. 2007;59:147–203. doi: 10.3114/sim.2007.59.14. https://doi.org/10.3114/sim.2007.59.14 PMid:18490953 PMCid:PMC2275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balajee SA, Houbraken J, Verweij PE, Hong SB, Yaghuchi T, Varga J, Samson RA. Aspergillus species identification in the clinical setting. Stud Mycol. 2007;59:39–46. doi: 10.3114/sim.2007.59.05. https://doi.org/10.3114/sim.2007.59.05 PMid:18490954 PMCid:PMC2275201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong SB, Go SJ, Shin HD, Frisvad JC, Samson RA. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97(6):1316–29. doi: 10.3852/mycologia.97.6.1316. https://doi.org/10.1080/15572536.2006.11832738 PMid:16722222. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Yokoyama K, Miyaji M, Nishimura K. Mitochondrial cytochrome b gene analysis of Aspergillus fumigatus and related species. J Clin Microbiol. 2000;38(4):1352–8. doi: 10.1128/jcm.38.4.1352-1358.2000. PMid:10747106 PMCid:PMC86444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ricardo Araujo, António Amorim, Leonor Gusmão. Diversity and specificity of microsatellites within Aspergillus section Fumigati. BMC Microbiol. 2012;12:154. doi: 10.1186/1471-2180-12-154. https://doi.org/10.1186/1471-2180-12-154 PMid:22838495 PMCid:PMC3438126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etienne KA, Gade L, Lockhart SR, Diekema DJ, Messer SA, Pfaller MA, Balajee SA. Screening of a Large Global Aspergillus fumigatus Species Complex Collection by Using a Species-Specific Microsphere-Based Luminex Assay. J Clin Microbiol. 2009;47(12):4171–4172. doi: 10.1128/JCM.01415-09. https://doi.org/10.1128/JCM.01415-09 PMid:19794043 PMCid:PMC2786642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Molina JV, Abad-Diaz-de-Cerio A, Sueiro-Olivares M, Pellon A, Ramirez-Garcia A, Garaizar J, Pemán J, Hernando FL, Rementeria A. Rapid and specific detection of section Fumigati and Aspergillus fumigatus in human samples using a new multiplex real-time PCR. Diagn Microbiol Infect Dis. 2014;80(2):111–8. doi: 10.1016/j.diagmicrobio.2014.06.003. https://doi.org/10.1016/j.diagmicrobio.2014.06.003 PMid:25063549. [DOI] [PubMed] [Google Scholar]
- 17.Burnie JP, Coke A, Matthews RC. Restriction endonuclease analysis of Aspergillus fumigatus DNA. J Clin Pathol. 1992;45:324–327. doi: 10.1136/jcp.45.4.324. https://doi.org/10.1136/jcp.45.4.324 PMid:1349614 PMCid:PMC495273. [DOI] [PMC free article] [PubMed] [Google Scholar]


