Abstract
The long noncoding RNA (lncRNA) H19 has been proven to be overexpressed in human pancreatic ductal adenocarcinoma (PDAC). H19-induced PDAC cell proliferation is cell cycle-dependent by modulating E2F-1. However, the mechanism of how H19 regulates E2F-1 remains unclear. In this study, we investigated the expression of miR-675 in PDAC tumours and cells, the biological function of miR-675 in PDAC cell proliferation and the possible relationship among H19, miR-675 and E2F-1. As a transcript of the first exon of H19, the level of miR-675 was negatively correlated with H19 expression in microdissected PDAC tissues (r=-0.0646, P=0.001). The serum miR-675 expression was significantly down-regulated in patients with PDAC compared to those in healthy individuals. Moreover, an evaluation of five PDAC cases showed that there was a remarkable increase of serum miR-675 levels after resection of the primary tumours. Ectopic overexpression of miR-675 in AsPC-1 and PANC-1 cells decreased cell viability, the colony-forming ability and the percentage of cells in S phase; contrarily, miR-675 knockdown resulted in enhanced cell proliferation. Furthermore, the suppressed cell proliferation caused by H19 knockdown could be rescued by inhibiting miR-675 expression. Additionally, intratumoural injection of either miR-675 agomir or antagomir could significantly affect tumour growth in vivo. Both the bioinformatic prediction and luciferase activity assay confirmed that E2F-1 was a direct target of miR-675. And the decrease of E2F-1 protein expression caused by siH19 could be partially reversed by miR-675 knockdown. We concluded that there might be a H19/miR-675/E2F-1 regulatory loop in cell cycle modulation. Serum miR-675 might serve as a potential biomarker for not only early diagnosis but also outcome prediction in PDAC.
Keywords: long noncoding RNA H19, miR-675, E2F-1, pancreatic ductal adenocarcinoma, cell proliferation, cell cycle.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) ranks as the fourth-leading cause of cancer-related death. Lack of obvious early symptoms and high metastatic rate make most patients miss the optimal period for operation upon diagnosis. Although an increasing number of treatments, including chemotherapy, have greatly improved, the mortality rate of PDAC is still on the growth 1. In that case, exploring novel biomarkers and potential therapeutic targets for the early diagnosis and treatment of pancreatic cancer is essential.
H19 is a classic imprinting lncRNA discovered in 1984. It is located on human chromosome 11p15.5 in close proximity to insulin-like growth factor 2 (IGF2) 2. The abundance of H19 is extremely high in fetus, but has a drastic reduction after birth and it is only expressed in few specific tissues and organs 3. Differential expression of H19 has been observed in many cancers. In most malignancies, H19 could function as an oncogene involved in cell proliferation 4, 5, cell cycle regulation 6, apoptosis 7, migration, invasion 8-10, stemness 11 as well as in the promotion of tumourigenesis to a certain extent.
Although loss of H19/IGF2 cluster imprinting could drive tumourigenesis 12, 13, a large share of the cancer-promoting effects of H19 is delivered via microRNA-675 (miR-675), a short noncoding RNA transcribed from the first exon of H19. MiR-675 has been demonstrated to be differentially expressed in many tumours. The levels of miR-675 are significantly increased in several cancer tissues such as glioma 14, gastric cancer 10, 15, colorectal cancer 4, non-small cell lung cancer 16, hepatocellular cancer 17, breast cancer 18, 19 and esophageal squamous cell carcinoma 20. On the other hand, low miR-675 expression has also been observed in bile duct cancer tissue 21. Liu et al. revealed that miR-675 could inhibit cardiomyocyte hypertrophy 22. The functional roles of miRNAs in tumourigenesis depend on the types of target genes. Liu et al. suggested that miR-675 overexpression promoted gastric cancer cell proliferation by targeting the tumour suppressor RUNX1 at the mRNA and protein levels 15, whereas Wang et al. found that miR-675 could decrease the protein expression of ZEB1, which promoted the EMT process in pancreatic cancer 23. To date, there are few studies on the correlation between miR-675 and PDAC. After examining formalin-fixed paraffin-embedded PDAC tissue samples from 225 patients, Schultz et al. discovered five differentially expressed miRNAs, including increased level of miR-675 24. Wang et al. reported that decreased miR-675-5p expression was associated with enhanced cell proliferation and patient survival in PDAC 23.
Although H19 encodes the primary miRNA precursor for miR-675, the relationship between the two ncRNAs remains controversial. A positive correlation between H19 and miR-675 expression was identified in the tissues of gastric cancer 10, 25 and colorectal cancer 26. However, Matouk et al. questioned how H19 and miR-675 were both up-regulated in many types of cancers and both were up-regulated by common triggers given that miR-675 was processed at the expense of H19 27. Aside from those reports, the serum miR-675 expression in individuals with cancer has not been described. Thus, it is worth exploring the levels of serum miR-675 in patients with PDAC.
Our previous study revealed that H19 was highly expressed in microdissected PDAC tissues and pancreatic cancer cell lines. Forced diffferential expression of H19 in PDAC cells significantly affected cell proliferation via cell cycle modulation and contributed to consistent changes in the E2F-1 protein levels 28. As a transcription factor associated with cell cycle regulation, E2F-1 was considered to be the potential molecular target of H19. However, the underlying mechanism of H19's regulation on E2F-1 remains unclear. Based on the fact that down-regulated H19 expression was associated with a decreased protein level of E2F-1, but not mRNA level 28, we assumed that the modulation of E2F-1 expression resulted from the post-transcriptional regulation instead of transcriptional repression by H19. MiR-675 is an important functional executor of H19, and the microRNA target database FindTAR3 indicated that the 3' untranslated region (3'UTR) of E2F-1 contained two putative miR-675 binding sites. Hence, we designed this study to demonstrate if the post-transcriptional regulation of E2F-1 is induced by miR-675. Additionally, the correlation between H19 and miR-675 in PDAC is also a major concern of this study. Furthermore, a full investigation of miR-675's expression and function would clarify whether this miRNA could be a candidate for the diagnosis and potential treatment target of PDAC.
Materials and Methods
Human tissue and serum specimens
Thirty pairs of tumours and adjacent normal tissues were collected from patients who were pathologically diagnosed with PDAC at Peking University First Hospital. All the samples were immediately frozen in liquid nitrogen and stored at -80°C until further use.
Thirty-five serum samples were obtained from patients with PDAC before surgery and neoadjuvant chemoradiatherapy. Thirty-eight normal serum samples were collected from healthy individuals aged from 20 to 50 and served as a control group. Moreover, the preoperative and postoperative serum samples from five patients with PDAC were also collected for miR-675 detection.
All the patients and control subjects provided written informed consent for the use of their tissues and sera. This study was approved by the Ethics Committee of Peking University First Hospital.
Laser captured microdissection
All the thirty pairs of frozen PDAC tumour and adjacent pancreatic tissue samples were processed into frozen sections followed by laser captured microdissection (LCM) using a Leica LMD7000 instrument (Germany) according to the manufacturer's protocol. Pure PDAC and normal ductal tissues were isolated from the frozen sections using the LCM technique.
Cell culture
The human pancreatic cancer cell lines COLO357, CAPAN-1, MIA PaCa-2, BxPC-3, AsPC-1, PANC-1, and T3M4 were kindly provided by Dr. Marko Kornmann at the University Hospital of Ulm in Germany, SW 1990 cells and the human pancreatic normal epithelial cell line hTERT-HPNE were purchased from American Type Culture Collection (Manassas, USA). COLO357, MIA PaCa-2 and PANC-1 cells were cultured in DMEM (Gibco, USA), and CAPAN-1, BxPC-3, T3M4 and AsPC-1 cells were cultured in RPMI 1640 (Gibco, USA) medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin. The cells were maintained at 37°C with 5% CO2. SW 1990 cells were cultured in Leibovitz's L-15 Medium in 100% atmospheric air. hTERT-HPNE cells were cultured in 75% glucose-free DMEM (Gibco, USA) and 25% Medium M3 Base (INCell Corp., USA) supplemented with 5% fetal bovine serum, 10 ng/ml human recombinant EGF, 5.5 mM D-glucose, 750 ng/ml puromycin and maintained at 37°C with 5% CO2. The cell culture supernatant from all nine cell lines were collected for miR-675 detection.
RNA isolation and real-time quantitative PCR
In the previous study, total RNA was successfully extracted from twenty-five of thirty microdissected PDAC tissues, all nine cultured cell lines and the cell culture supernatant 28. Real-time quantitative PCR (qPCR) was performed using SYBR Green PCR Master Mix (TOYOBO, Japan) and the ABI7500 system (Applied Biosystems, USA) according to the manufacturer's instruments. U6 and GAPDH were used as endogenous controls for the tissues and cell lines, respectively. Cel-miR-39 was adopted as an exogenous control for the sera and supernatant and was put into the samples after TRIzol reagent and chloroform were added. The specific primers for hsa-miR-675-5p and cel-miR-39 were purchased from RiboBio (Guangzhou, China). The other qPCR primer sequences were as follows: 5'-ACGTGACGTGTCAGGACCT-3' (sense) and 5'-GATCGGGCCTTGTTTGCTCTT-3' (antisense) for E2F-1, 5'-GTATTGGGCGCCTGGTCACC-3' (sense) and 5'-CGCTCCTGGAAGATGGTGATGG-3' (antisense) for GAPDH. Each sample was examined in triplicate, and all the data were analysed using the comparative threshold cycle (CT) (2-ΔΔCT) method.
siRNA transfection
Mimics and inhibitors for hsa-miR-675-5p (as well as respective negative control siRNAs) were purchased from RiboBio (Guangzhou, China). AsPC-1 and PANC-1 cells were transfected with either 50 nM hsa-miR-675-5p mimic or 100 nM hsa-miR-675-5p inhibitor according to the manufacturer's protocol. Either a mixture of three siRNAs targeting H19 (siH19) or negative control siRNAs (NC) were used as previously described 28. Lipofectamine 2000 was adopted as a transfection reagent, and the transfection efficiency was confirmed using qPCR.
Cell viability assay (CCK-8 assay)
The cell viability was determined by Cell Counting kit-8 (CCK-8) (Dojindo Laboratories, Japan). The transfected cells were plated into 96-well plates at a density of 3000-5000 cells/well. At 24 h, 48 h and 72 h, the target cells were incubated with CCK-8 reagent (10 μl/well) for 2 h at 37°C, after which the absorbance at 450 nm was measured with a microplate reader (Bio-Rad Laboratories, USA). Three independent experiments were performed in quadruplicate.
Colony formation assay
The colony-forming ability was determined by colony formation assay. The transfected cells were seeded into 6-well plates at a density of 200 cells/well. Approximately 2-3 weeks later, the cells were stained by crystal violet. The number of macroscopic colonies was counted, and images of the representative colonies were obtained.
EdU incorporation assay
The percentage of cells in the DNA synthesis phase (S phase) was evaluated using an EdU Apollo®567 In Vitro Imaging Kit (RiboBio, Guangzhou, China). Transfected cells were plated into 96-well plates at a density of 3000 cells/well. After 24 h, the EdU reagent was added to the medium at concentration of 50 μM, and the plates were incubated at 37°C for 2 h. After the cells took up the EdU reagent, they were fixed with 4% formaldehyde for 15 min and permeabilized with 0.5% Triton X-100 for 20 min at room temperature. The cells were then washed with PBS three times, reacted with 100 μl of 1×Apollo® reaction cocktail per well for 30 min and counterstained with DAPI for 30 min. Fluorescence microscopy was used to observe the staining and obtain images. Ten randomly chosen visual fields were observed, and the percentage of cells that incorporated EdU was calculated. Three independent experiments were performed in triplicate.
Cell cycle analysis
Transfected cells were seeded into 6-well plates and cultured to 80% confluence, after which they were released using trypsin and harvested. The suspensions were centrifuged, and the cell pellets were washed twice with pre-cooled PBS and fixed with 70% ethanol. Then, the cells were resuspended in PBS, filtered through a 400-mesh sieve, and stained with 50 μg/mL propidium iodide and 100 μg/mL RNase in PBS at 37°C for 30 min. The cell cycle distribution was then determined using flow cytometry (FACSCalibur, BD, USA).
Western blot analysis
Total proteins were extracted from transfected AsPC-1 and PANC-1 cells, and the protein concentration was measured using the BCA assay (Thermo Scientific, USA). Equal amounts of proteins (20 μg/well) were separated on 10% SDS polyacrylamide gels and transferred to PVDF membranes. After the membranes were blocked with 5% fat-free milk in TBST buffer (0.1% Tween-20) for 2 h at room temperature, they were incubated with antibodies against E2F-1 (1:2000, CST, USA) and β-actin (1:40000, MBL, USA) at 4℃ overnight. The membranes were then incubated with HRP-conjugated secondary antibody (Jackson Lab, USA) at 4℃ for 2 h. The protein bands were detected using chemiluminescence (Millipore, USA) and exposed to X-ray films.
Xenograft study
Wild type AsPC-1 (2×106) and PANC-1 cells (1×107) were subcutaneously injected into the axillary fossa of five-week-old male Balb/c (nu/nu) nude mice. After 2 weeks when the tumour reached a certain size (0.1 cm3), 1 nM hsa-miR-675-5p agomir (mimic), antagomir (inhibitor) or negative control was intratumourally injected into the tumour twice a week. The tumour volumes were measured (0.5×length×width2) every 2 days. After 2 weeks the mice were sacrificed and the tumours were precisely excised and weighted. The miR-675 and H19 expression levels were determined using qPCR.
Luciferase activity assay
Based on the results of bioinformatics prediction, two 3'UTR fragments from the E2F-1 mRNA containing binding sites for miR-675-5p (wt1 and wt2) as well as two 3'UTR fragments with corresponding mutations (mut1 and mut2) were designed and amplified. The PCR products were then subcloned into the renilla and firefly luciferase reporter vector psiCHECK-2 (Promega, USA).
AsPC-1 cells were cultured in a 24-well plate at a density of 30000 cells/well and then co-transfected with 50 nM of either miR-675 mimic or negative control and 300 ng of either the E2F-1 3'UTR wt or E2F-1 3'UTR mut plasmids using Lipofectamine 2000 (Invitrogen, USA) as the transfection reagent. After 48 hours, the renilla and firefly luciferase activities were measured according to the Dual-Luciferase Reporter Assay's protocol (Promega, USA). Relative renilla luciferase activity was normalized to firefly luciferase activity. Three independent experiments were performed.
Statistical analysis
All the data were presented as the mean values ± standard deviation (s.d.). Statistical analysis of differences between different groups was determined using either Student's t-test or paired t-test. The correlation between miR-675 and H19 expression was analysed using Pearson correlation. All statistical analyses were performed using SPSS 13.0 software. P<0.05 was considered statistically significant.
Results
miR-675 was negatively correlated with H19 expression in PDAC tissues and down-regulated in the sera of patients with PDAC
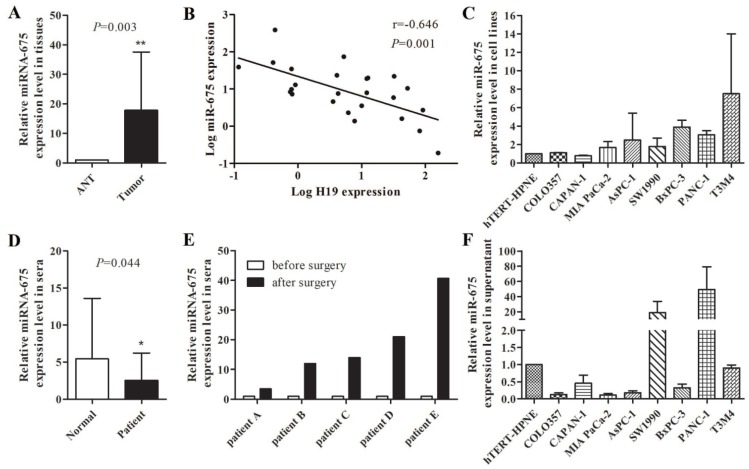
Increased expression level of miR-675 was observed in microdissected PDAC tissues compared with those in adjacent normal tissues (P=0.003) (Fig. 1A), which was consistent with the high level of H19 expression in PDAC tumours in our previous study 28. However, a negative correlation between miR-675 and H19 expression levels in PDAC tissues was identified (r=-0.0646, P=0.001) (Fig. 1B). To explore the role of serum H19 and miR-675 in PDAC, we further examined the expression of H19 and miR-675 in sera. The serum H19 expression was barely detected because of the length of its transcript. Interestingly, the serum miR-675 levels were significantly reduced in patients with PDAC compared to those of healthy individuals (P=0.044) (Fig. 1D). Moreover, comparison of the preoperative and postoperative sera from five PDAC cases indicated a remarkable increase in serum miR-675 after resection of the primary tumour (Fig. 1E). We already noted that H19 gradually up-regulated in COLO357, CAPAN-1, MIA PaCa-2, AsPC-1, SW 1990, BxPC-3, PANC-1 and T3M4 cells by hundred- to thousand-folds when compared with hTERT-HPNE cells 28; however, miR-675 expression did not exhibit the similar trends within these cell lines. The levels of miR-675 were decreased in CAPAN-1 cells but were increased in most cell lines only by two- to three-folds (Fig. 1C). qPCR analysis indicated that miR-675 was lowly expressed in the cell culture supernatant from six pancreatic cancer cell lines when compared with those in hTERT-HPNE cells, except for SW 1990 and PANC-1 cells (Fig. 1F). This result was consistent with the decreased level of miR-675 expression in the PDAC patients' sera, which suggests that the transcription and extracellular transportation of miR-675 might be influenced by the tumourigenesis nature of PDAC cells.
Figure 1.
The levels of miR-675 in PDAC and the correlation between miR-675 and H19 expression. (A) Expression of miR-675 in microdissected PDAC tissues (Tumor) and adjacent normal tissues (ANT). (B) The correlation between miR-675 and H19 expression in microdissected PDAC tissues. The data regarding the H19 level were provided in our previous study 28. (C) Expression of miR-675 in eight PDAC cell lines and hTERT-HPNE cells. (D) Levels of serum miR-675 in PDAC patients (Patient) and healthy individuals (Normal). (E) Comparison of preoperative and postoperative levels of serum miR-675 in five PDAC cases. (F) Expression of miR-675 in the cell culture supernatant from nine cell lines. The results were presented as the mean ± s.d., n=3, *P<0.05, **P<0.01.
Modulated miR-675 expression significantly regulated pancreatic cancer cell proliferation, and suppression of cell proliferation caused by H19 knockdown can be rescued by inhibiting miR-675
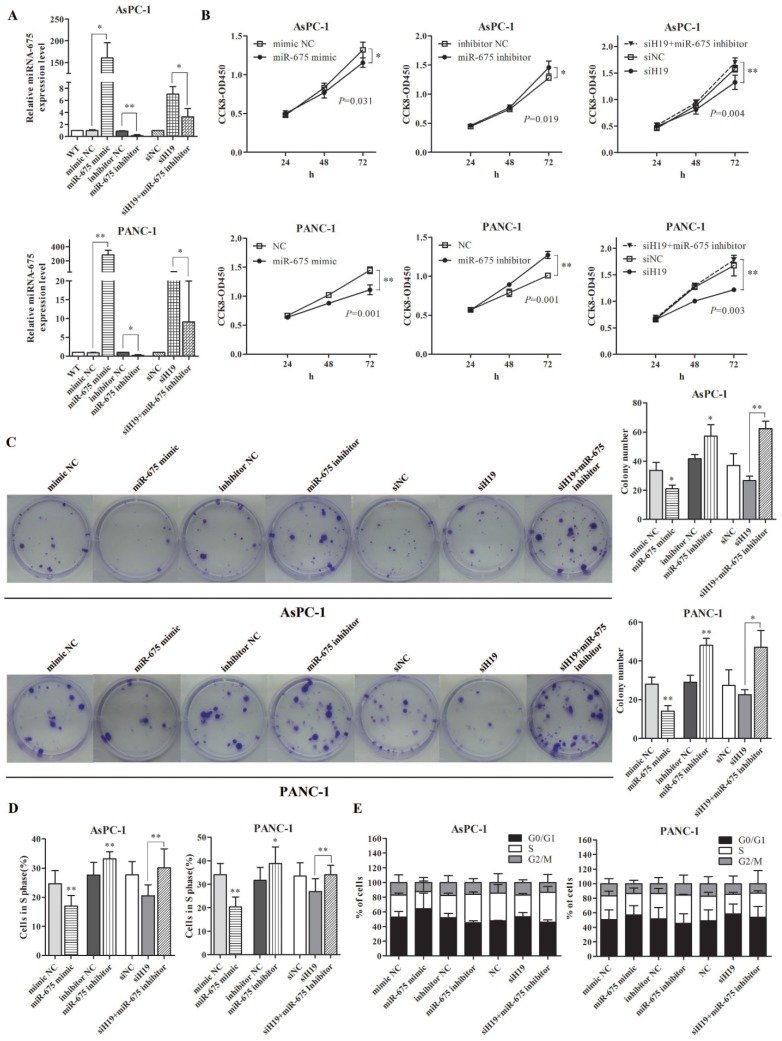
The biological functions of miR-675 in PDAC were examined in vitro. The expression levels of miR-675 in AsPC-1 and PANC-1 cells were successfully altered by miRNA mimic and inhibitor. Based on the negative correlation between miR-675 and H19 expression, we then co-transfected pancreatic cancer cells with siH19 and miR-675 inhibitor. qPCR revealed that miR-675 expression was increased by H19 knockdown. However, the up-regulation of miR-675 level induced by siH19 could be partially reversed by miR-675 inhibitor (Fig. 2A). Ectopic overexpression of miR-675 remarkably inhibited cell viability (Fig. 2B) and the colony-forming ability (Fig. 2C). EdU incorporation assay showed that there was a decrease of the percentage of cells in the S phase in cells with high levels of miR-675 (Fig. 2D). Contrarily, miR-675 knockdown led to obvious increases in cell viability and the colony-forming ability, and an increased cell proportion of S phase (Fig. 2B-2D). The flow cytometry results indicated a G0/G1 to S phase arrest induced by miR-675 overexpression and a re-entry of cells into G2/M phase by miR-675 inhibition (Fig. 2E). Moreover, when the up-regulated miR-675 induced by siH19 transfection was reversed by miR-675 inhibitor, the rescue effects on the cell proliferation and cell cycle distribution were observed. The suppression of cell proliferation and cell cycle arrest caused by H19 knockdown could be reversed by miR-675 inhibition confirmed by CCK-8 assay, colony formation assay, EdU incorporation and cell cycle analysis (Fig. 2B-2E).
Figure 2.
The effect of miR-675 on the proliferation and cell cycle distribution of PDAC cells, and the suppression of cell proliferation caused by H19 knockdown could be rescued by inhibiting miR-675. AsPC-1 and PANC-1 cells were transfected with miR-675 mimic, miR-675 inhibitor, siH19, siH19+miR-675 inhibitor or corresponding negative controls respectively. (A) The transfection efficiency for each group was verified by qPCR. (B) Cell viability was determined by the CCK-8 assay. The cell proliferation curves were drawn based on the measured absorbance at 450 nm every 24 h after transfection. (C) The colony-forming ability of the transfected cells was examined by colony formation assay. The numbers of macroscopic colonies were counted after 2-3 weeks. (D) Cells in S phase (which could incorporate the EdU reagent) were detected by fluorescence microscopy and counted. (E) The distribution of cell cycle progression was determined by flow cytometry. The results were expressed as the mean ± s.d., n=3, *P<0.05, **P<0.01.
miR-675 regulated the growth of PDAC cells in vivo
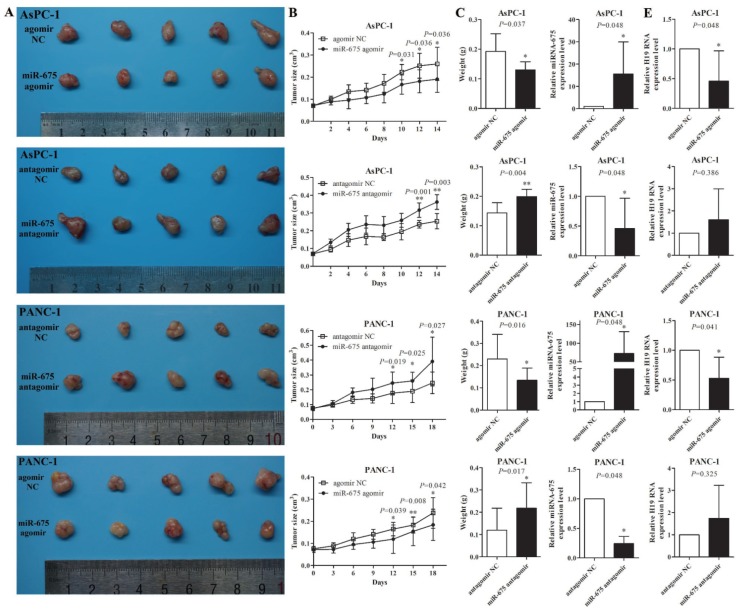
To further demonstrate the role of miR-675 in xenograft tumour growth, after the subcutaneous tumours created by wild type AsPC-1 or PANC-1 cells reached approximately 0.1cm3, 1 nM of miR-675 agomir, antagomir or negative control siRNAs were intratumourally injected into the xenografts twice a week for two weeks. qPCR analysis showed that in vivo injections of miR-675 agomir or antagomir could significantly change the levels of miR-675 in tumours (Fig. 3D). Overexpression of miR-675 decreased the tumour growth rate, tumour size and tumour weight, whereas knocking down of miR-675 remarkably promoted tumour growth (Fig. 3A-3C). Moreover, we examined H19 expression in tumour xenografts and observed that the H19 levels exhibited an inverse trend to the miR-675 levels (Fig. 3E). This result was consistent with our findings in microdissected PDAC tissues.
Figure 3.
The effect of miR-675 on PDAC xenografts in vivo. (A) Photographs of tumours at two weeks after intratumoural injection of miR-675 agomir, antangomir, or respective negative control siRNAs (NC). (B) The volume of the xenografts was measured every two days to construct tumour growth curves. (C) Tumour weight was calculated and incorporated into the statistical analyses. The miR-675 expression (D) and H19 expression (E) in the tumour xenografts was detected by qPCR. The results were expressed as the mean ± s.d., n=5, *P<0.05, **P<0.01.
E2F-1 was a direct target of miR-675 in pancreatic cancer cell proliferation regulation
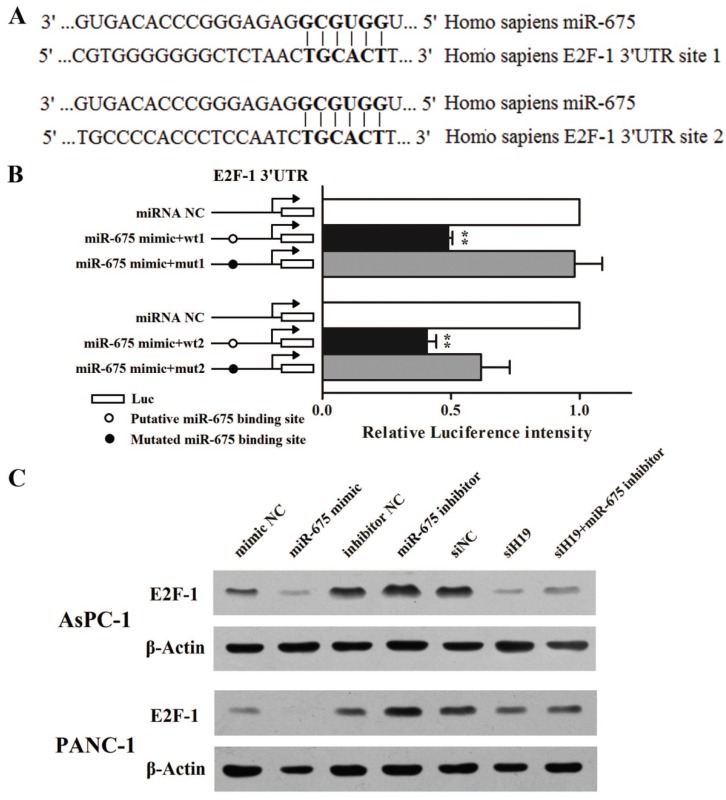
Cytology experiments had proved that the regulatory mechanism of miR-675 on pancreatic cancer cell proliferation was associated with G1-S transition. As observed in our previous study, H19 knockdown evidently decreased the protein level of E2F-1 as well as the mRNA levels of its downstream targets involved in cell cycle, such as Cyclin A1 and MCM3. Meanwhile, H19 overexpression led to an opposing effect 28. Based on those data, we assumed that miR-675 might be a bridge between H19 and E2F-1. Coincidentally, the miRNA target database FindTAR3 suggested that the 3'UTR of E2F-1 contains two putative miR-675 binding sites (Fig. 4A). As shown in Fig. 4B, when miR-675 was overexpressed in AsPC-1 cells, the levels of luciferase activity were significantly diminished when compared with those in the NC group. Moreover, the decrease in the intensity of the luciferase signal was reversed in cells transfected with the plasmid containing the mut1 3'UTR of E2F-1. However, the mut2 plasmid failed to restore the renila luciferase intensity. Taken together, we could confirm that there was at least one credible miR-675 binding site located in the 3'UTR region of the E2F-1 transcript that, when bound to miR-675, could significantly repress E2F-1 protein translation. Western blot analysis further verified that miR-675 could reversely regulated E2F-1 protein expression (Fig. 4C). Furthermore, the repression of E2F-1 protein level caused by H19 knockdown was partially rescued by miR-675 inhibition.
Figure 4.
Regulation of miR-675 on PDAC cell cycle was mediated via direct inhibition of E2F-1 translation. (A) Bioinformatic miRNA target prediction software indicated that there were two putative miR-675 binding sites in the 3'UTR of the E2F-1 mRNA. (B) Luciferase reporter plasmids containing the wild-type or mutated 3'UTR fragments were co-transfected with either miR-675 mimic or negative control. The intensity of luciferase activity was measured using luminescence. (C) The protein levels of E2F-1 in the cells transfected with miR-675 mimic, miR-675 inhibitor, siH19, siH19+miR-675 inhibitor or corresponding negative controls were determined by western blot analysis. The results were expressed as the mean ± s.d., n=5, *P<0.05, **P<0.01.
Discussion
In our previous study, we elucidated the functional role of H19 in PDAC cell proliferation and the possible correlation between H19 and E2F-1 28. However, the specific mechanism of how E2F-1 was regulated by H19 remains unclear. As miR-675 was a transcript of H19, we speculated that miR-675 might be an intermediary molecule that conduct the regulatory effect of H19 on E2F-1.
Significant up-regulation of H19 expression in PDAC tumours and PDAC cell lines was demonstrated to enhance cell proliferation previously. Although the levels of miR-675 in the same microdissected PDAC tissues were slightly increased, our data indicated that the level of miR-675 was negatively correlated with H19 expression. We failed to identify a correlation between serum miR-675 and serum H19 because the length of H19. Although some earlier studies have reported a positive correlation between miR-675 and H19 expression in gastric cancer 15, 25, colorectal cancer 4, 29, glioma 14, esophageal squamous cell carcinoma 20 and osteoarthritis 30, this is the first time to reveal the relationship of H19 and miR-675 expression in human PDAC. It seems that in some kinds of tumours, miR-675 might have some other transcriptional regulatory mechanisms independent of H19. Considering the fact that H19 could encode the primary miRNA precursor for miR-675, which means miR-675 might be processed at the expense of H19, our results seemed quite reasonable. These findings confirmed the concept proposed by Matouk et al., that H19 and miR-675 could not be both increased in cancer tissues or both up-regulated by common triggers 27.
Interestingly, we noticed the inconsistent miR-675 expression levels between the PDAC tissues and sera. The miR-675 expression observed in both the PDAC tissues and PDAC cell lines were slightly increased when compared with the non-tumour ductal tissue or normal epithelial cells. However, the levels of serum miR-675 exhibited a significant decrease in PDAC patients. Moreover, after the primary tumour has been removed, the serum miR-675 expression regained significantly. These data suggest that miR-675 might serve as s sensitive serum biomarker corresponding to human PDAC. Further assessment for its clinical significance on the early diagnosis, tumour recurrence, and even the prognosis of PDAC patients, would be necessary and promising. Although the exact mechanism is still unclarified, this contradictory expression profiles of miRNAs between the intracellular and the sera has been reported previously. Ohshima et al. revealed that in patients with gastric cancer, the let-7 miRNA family was expressed at low levels in serum while overexpressed in the tumours 31. Tanaka et al. demonstrated that the levels of miR-92a decreased in the blood plasma of acute leukaemia patients, whereas strong expression was observed in the tissue samples 32. To explore the preliminary mechanism of this contradictory miRNA expression profile, we detected the miR-675 expression levels in cultured PDAC cell lines and their supernatant. Similar to the expression profiles in PDAC tissue and the sera, most of the cultured PDAC cell lines showed a decreased level of miR-675 in their supernatant, in spite of the relatively high expression levels of the tumour cells. These results indicate that the transcription and extracellular transportation of miR-675 might be affected by the tumourigenesis of PDAC. Although the PDAC cells express high levels of miR-675, it cannot be transported outside the cells, so the levels of miR-675 in PDAC sera and supernatant of the cultured cells remain low. For healthy individuals or after removal of the primary tumours for PDAC cases, miR-675 can be transported extracellularly via some unknown mechanisms, which makes the serum level of miR-675 relatively high.
Consistent with the fact that miR-675 was negatively correlated with H19 expression in PDAC tissues and cells, the functional investigation of miR-675 showed an anti-proliferative effect both in vitro and in vivo, via regulating the distribution of cell cycle. This means that miR-675 might act as a tumour suppressor in PDAC, which was contrary to H19. Taken together, our data indicated that during the tumourigenesis of PDAC, the imprinted H19 gene was overexpressed and act as an oncogene in cancer cells. Meanwhile, the high level of H19 transcription might encode more miR-675 precursors, which makes the level of miR-675 in the PDAC tissues and cells also increased to some degree. Since the expression of miR-675 is processed at the expense of H19, miR-675 is negatively correlated with H19 expression and serves as a tumour suppressor in PDAC. These hypotheses require confirmation in the future studies.
Our previous research has disclosed that E2F-1 is a key transcription factor in H19-mediated regulation of proliferation and cell cycle distribution 28. In this study, we performed the bioinformatic prediction and luciferase activity assay to verify that miR-675 could affect E2F-1 protein expression by binding to the 3'UTR on E2F-1 mRNA. This post-transcriptional regulation might be a part of a negative regulatory loop among H19, miR-675 and E2F-1. Berteaux et al. discovered two putative E2F consensus sites within the minimal promoter of H19 and demonstrated that endogenous E2F-1 could be recruited to the H19 promoter to activate H19 expression in breast cancer 33. Our previous study showed that E2F-1 protein levels in frozen PDAC sections trended toward a positive correlation with H19 expression (r=0.339, P=0.098) 28. Taken these data together, we propose that there might exist a complex regulatory loop among H19, miR-675 and E2F-1 in the cell cycle regulation of PDAC (Fig. 5). During the tumourigenesis of PDAC, the aberrant H19 expression would be accompanied by a secondary reduction of miR-675, which makes it unable to down-regulate E2F-1 expression at the post-transcriptional level, resulting in an increased E2F-1 that could be recruited to the H19 promoter to further activate H19 expression. This cascade reaction among H19, miR-675 and E2F-1 creates an unbreakable vicious cycle to promote cancer progression. Loss control of any points of the H19/miR-675/E2F-1 loop would lead to irreversible neoplastic hyperplasia.
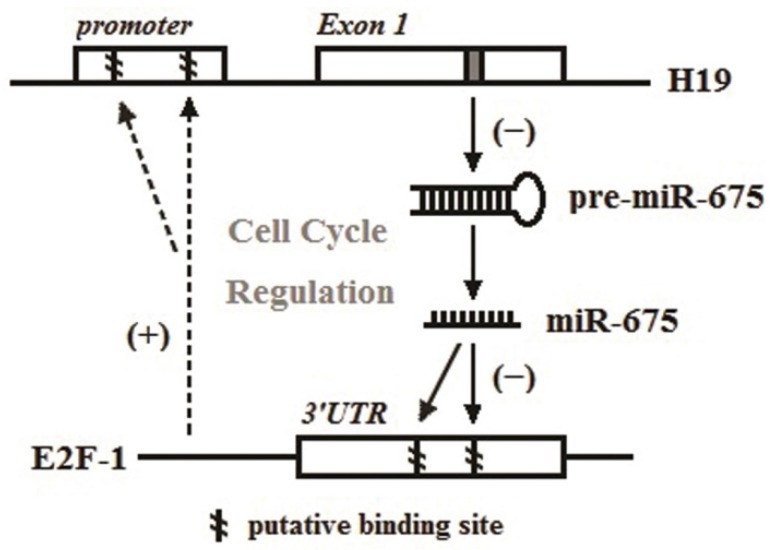
Figure 5.
Pattern graph of the relationship among H19, miR-675 and E2F-1 in the regulation of the cell cycle in PDAC. The dotted arrows represented the findings of Berteaux et al. 33, and the filled arrows represent data established in this work and our previous study 28.
In conclusion, our study revealed that miR-675 could function as a tumour suppressor against PDAC cell proliferation via E2F-1 mediated cell cycle modulation. Additionally, there might exist a H19/miR-675/E2F-1 regulatory loop in cell cycle modulation of human PDAC. Finally, serum miR-675 might have great potential to become the biomarker for not only early diagnosis but also outcome predictions of patients with PDAC.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81372605, 81572339, and 81672353), the National High Technology Research and Development Program of China (SS2015AA020405) and the Scientific Research and Development Program of Beijing Railway Corporation of China (No. J2017Z605). We thank Prof. Zebin Mao at the Department of Biochemistry and Molecular Biology in Health Science Center, Peking University for the assistance and technical support.
References
- 1.Siegel R L, Miller K D, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Pachnis V, Belayew A, Tilghman S M. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci U S A. 1984;81(17):5523–5527. doi: 10.1073/pnas.81.17.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dey B K, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28(5):491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsang W P, Ng E K, Ng S S. et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31(3):350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 5.Liu C, Chen Z, Fang J. et al. H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumour Biol. 2016;37(1):263–270. doi: 10.1007/s13277-015-3779-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Z, Song L, He J. et al. Ectopic expressed long non-coding RNA H19 contributes to malignant cell behavior of ovarian cancer. Int J Clin Exp Pathol. 2015;8(9):10082–10091. [PMC free article] [PubMed] [Google Scholar]
- 7.Yu L L, Chang K, Lu L S. et al. Lentivirus-mediated RNA interference targeting the H19 gene inhibits cell proliferation and apoptosis in human choriocarcinoma cell line JAR. BMC Cell Biol. 2013;14:26. doi: 10.1186/1471-2121-14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma C, Nong K, Zhu H. et al. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35(9):9163–9169. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhu M, Chen Q, Liu X. et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281(16):3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Yu B, Li J. et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauderlique-Le R H, Vennin C, Brocqueville G. et al. Enrichment of Human Stem-Like Prostate Cells with s-SHIP Promoter Activity Uncovers a Role in Stemness for the Long Noncoding RNA H19. Stem Cells Dev. 2015;24(10):1252–1262. doi: 10.1089/scd.2014.0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H T, Choi B H, Niikawa N. et al. Frequent loss of imprinting of the H19 and IGF-II genes in ovarian tumors. Am J Med Genet. 1998;80(4):391–395. doi: 10.1002/(sici)1096-8628(19981204)80:4<391::aid-ajmg16>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 13.Lustig-Yariv O, Schulze E, Komitowski D. et al. The expression of the imprinted genes H19 and IGF-2 in choriocarcinoma cell lines. Is H19 a tumor suppressor gene? Oncogene. 1997;15(2):169–177. doi: 10.1038/sj.onc.1201175. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y, Wang Y, Luan W. et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9(1):e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu G, Xiang T, Wu Q F. et al. Long Noncoding RNA H19-Derived miR-675 Enhances Proliferation and Invasion via RUNX1 in Gastric Cancer Cells. Oncol Res. 2016;23(3):99–107. doi: 10.3727/096504015X14496932933575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He D, Wang J, Zhang C. et al. Down-regulation of miR-675-5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol Cancer. 2015;14:73. doi: 10.1186/s12943-015-0342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y Q, Weng J, Li S Q. et al. MiR-675 Promotes the Growth of Hepatocellular Carcinoma Cells Through the Cdc25A Pathway. Asian Pac J Cancer Prev. 2016;17(8):3881–3885. [PubMed] [Google Scholar]
- 18.Vennin C, Spruyt N, Dahmani F. et al. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6(30):29209–29223. doi: 10.18632/oncotarget.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhai L L, Wang P, Zhou L Y. et al. Over-expression of miR-675 in formalin-fixed paraffin-embedded (FFPE) tissues of breast cancer patients. Int J Clin Exp Med. 2015;8(7):11195–11201. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y W, Zhang H, Duan C J. et al. miR-675-5p enhances tumorigenesis and metastasis of esophageal squamous cell carcinoma by targeting REPS2. Oncotarget. 2016;7(21):30730–30747. doi: 10.18632/oncotarget.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M, Wen T F, He L H. et al. A six-microRNA set as prognostic indicators for bile duct cancer. Int J Clin Exp Med. 2015;8(10):17261–17270. [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L, An X, Li Z. et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc Res. 2016;111(1):56–65. doi: 10.1093/cvr/cvw078. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Zhang Y, Wei H. et al. The mir-675-5p regulates the progression and development of pancreatic cancer via the UBQLN1-ZEB1-mir200 axis. Oncotarget. 2017;8(15):24978–24987. doi: 10.18632/oncotarget.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz N A, Andersen K K, Roslind A. et al. Prognostic microRNAs in cancer tissue from patients operated for pancreatic cancer-five microRNAs in a prognostic index. World J Surg. 2012;36(11):2699–2707. doi: 10.1007/s00268-012-1705-y. [DOI] [PubMed] [Google Scholar]
- 25.Zhuang M, Gao W, Xu J. et al. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448(3):315–322. doi: 10.1016/j.bbrc.2013.12.126. [DOI] [PubMed] [Google Scholar]
- 26.Tsang W P, Ng E K, Ng S S. et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31(3):350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 27.Matouk I J, Halle D, Gilon M. et al. The non-coding RNAs of the H19-IGF2 imprinted loci: a focus on biological roles and therapeutic potential in Lung Cancer. J Transl Med. 2015;13:113. doi: 10.1186/s12967-015-0467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma L, Tian X, Wang F. et al. The long noncoding RNA H19 promotes cell proliferation via E2F-1 in pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2016;17(10):1051–1061. doi: 10.1080/15384047.2016.1219814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Bu D, Ma Y. et al. H19 Overexpression Induces Resistance to 1,25(OH)2D3 by Targeting VDR Through miR-675-5p in Colon Cancer Cells. Neoplasia. 2017;19(3):226–236. doi: 10.1016/j.neo.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steck E, Boeuf S, Gabler J. et al. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J Mol Med (Berl) 2012;90(10):1185–1195. doi: 10.1007/s00109-012-0895-y. [DOI] [PubMed] [Google Scholar]
- 31.Ohshima K, Inoue K, Fujiwara A. et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5(10):e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka M, Oikawa K, Takanashi M. et al. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4(5):e5532. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berteaux N, Lottin S, Monte D. et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. 2005;280(33):29625–29636. doi: 10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]