Abstract
Digestive tract cancers (DTCs) is the most common malignant tumors in the world. Despite surgery and medical technology have witnessed the increasing development and sharp advancement in the past decade, DTCs remain a critical concern with high morbidity and mortality. Since a class of small noncoding RNAs termed miRNAs were identified several years ago, increasing studies have attempted to illustrate the relationship between the specific miRNAs dysregulated expression levels and the diseases phenotypic changes. For example, microRNA-483 (miR-483) aberrant expression plays a pivotal part in tumor biology in a variety of human cancer, including DTCs. In this review, we focus on the present key findings from recent profiling studies, discuss the use of miR-483 as a novel biomarker for DTCs. At the same time, we emphasize the significant diversities and technical difficulties must be overcome before clinically relevant signatures arose. It is believed that this might provide researchers an insight into the molecular targeting cancer treatment.
Keywords: MicroRNA, miR-483, digestive tract cancers, regulatory mechanism, clinical value
Introduction
Digestive tract cancers (DTCs) is the most common malignant tumors in the world, and it is a multifactorial and heterogenous disease whose beginning and development can be ascribed to the accumulative effect of genetic predispositions, environment factors, and their complicated interplay 1. Despite the considerable advances in surgery and medical technology in recent times, DTC remains a critical concern with high morbidity and mortality, as it is diagnosed mainly at late stages 2. Since a class of small noncoding RNAs termed microRNAs were recognized several years ago, numerous investigators have probed into the relationship between the specific miRNAs dysregulated expression levels and the diseases phenotypic changes 3. Particularly, emerging evidence suggests that abnormalities in the expression of miRNA are associated with a variety of cancers, including DTCs 4. In this review, we focus on the present key findings from recent profiling studies, discuss the use of miR-483 as a novel biomarker for DTCs, and at the same time, we emphasize the significant diversities and technical difficulties that must be overcome before clinically relevant signatures will arise 5.
The function of miRNA in gene regulation
MicroRNAs (miRNAs) are a family of noncoding RNAs with 20-25 nucleotide-longths, which post-transcriptionally modulate gene expression through canonical base pairing 6. The pioneer discovery of miRNAs was initiated by Lee and his colleagues in 1993, after that, a growing number of miRNAs have been identified 7. Currently, more than 2,500 mature miRNA sequences have been identified in humans according to the miRBase database, and they modulate about one-third of the expression of protein-coding genes at the post transcriptional level 4,8. Recent studies have found that miRNAs play significant roles in diverse biological activities, including physiology in normal cell and pathological conditions, such as abnormal cell cycle progression which contains proliferation, differentiation and apoptosis, especially tumorigenesis 8.
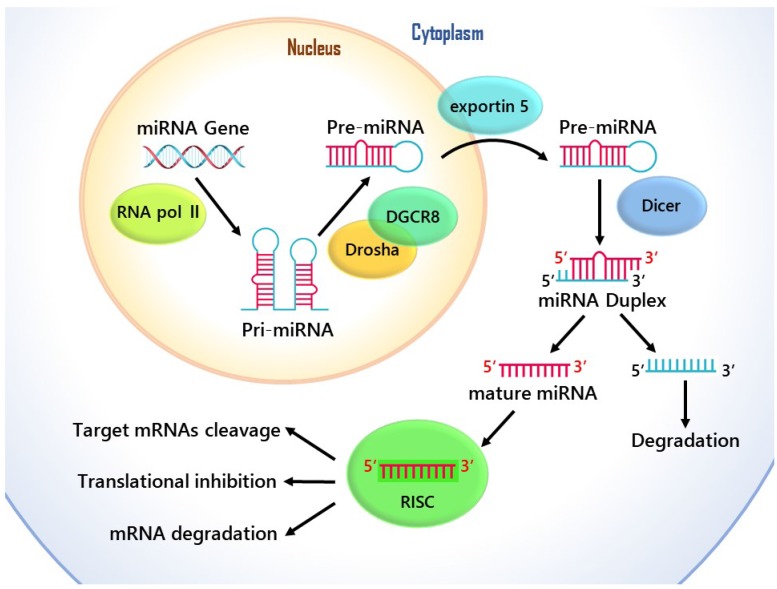
MicroRNAs are single-stranded noncoding genome-encoded small RNA molecules, and they are transcribed by RNA polymerase II into primary transcripts (pri-miRNAs) of variable length (1 kb- 3 kb), and then are processed in the nucleus through Drosha-DGCR8 Complex. This generates hairpin like intermediates named precursor miRNAs (pre-miRNAs), which are nearly 80-to 100- nucleotides long and have two-nucleotide 3' overhangs 8. Pre-miRNAs are transported out of the cell's nucleus by exportin-5 to the cytoplasm for further processing, and they are converted into a 20-25 nucleotide long miRNA duplexes by Dicer 9. Then the guide string of mature miRNA combines with the RNA induced silencing complex (RISC), which is able to mediate target mRNAs cleavage and/or translational inhibition 10 Fig. 1. Although previous research into miRNA function has primarily concerned about the target places in the 3' untranslated regions (UTRs), enough evidence has showed that through complementary interplays with the coding sequence (CDS) and 5'-UTR of mRNAs, miRNAs can also cause the degradation of mRNA and help regulate translational repression 11.
Figure 1.
The cellular and molecular mechanisms of the function of miRNA in gene regulation.
By causing the proliferation and inhibiting the apoptosis of tumor cells through the negative regulation of their target gene expression, MiRNA was highly likely to promote tumor. 12. Because multiple distinct mRNA molecules are meditated by a single miRNA and a variety of miRNAs are able to target on a single mRNAs, the regulatory networks of MiRNA gene are especially complicated. 13. On account of this, miRNA-mediated inhibition makes the gene expression regulation highly complex 14. This phenomenon indicates that the roles miRNAs and miRNA targets play in disease, cellular differentiation, and homeostasis are of great significance. In addition, more studies show that the in cancer types like lung cancer, esophageal cancer, breast cancer and several other cancers, miRNA dysregulation expression has been detected 15.
The expression of miR-483 in digestive tract cancers
Relying on their downstream targets, MiRNA genes can be either oncogenic or tumor suppressive, which are usually stayed in weak parts and deletion genomic areas 7. Copying from five promoters, IGF2 (insulin-like growth factor II) is a maternally imprinted gene whose activity is both tissue-specific and developmentally regulated 16. Situating within IGF2 gene second intron on chromosome 11p15, the Human miR-483 is considered as co-expression with its IGF2 host gene 17. The stem-loop precursor of miR-483 is processed into two mature forms: miR-483-3p and miR-483-5p, both of them have been shown to exert an impact on cell proliferation and promote tumor development 3,18.
miR-483-3p and miR-483-5p molecules express in different ways and play different roles in a variety of cancer types despite of their similarity. For example, researchers have identified that miR-483-3p was over-expressed in pancreatic cancer 19, Wilms' tumors 20, adrenocortical cancer 21, prostate cancer 22, colorectal cancer 23 and breast cancer 24. Similarly, miR-483-5p has been documented to be up-regulated in oral cancer 25, esophageal cancer 26, gastric cancer 27, Cholangiocarcinoma 28, as well as in colorectal cancer 29 and in adrenocortical cancer 30. miR-483 family molecules oncogenic effects include promoting cell proliferation, transfer, invasion and drug resistance. Nevertheless, few evidence have been proved that the miR-483 family may help eliminate tumor. For instance, miR-483 has been found to be under-expressed in intestinal cancer 31 and hepatocellular cancer 32. Among them, miR-483-5p suppresses metastasis in plasma cell leukemia 33 or angiosarcoma 34, and miR-483-3p is down-regulated in breast cancer 24. In malignant diseases, miR-483-3p and miR-483-5p are frequent aberrations, and this implies that they are closely related to the morbid state.
The targets of miR-483 in different types of cancer
In recent years, it is obvious that the non-protein coding genome plays a pivotal role in the development of functional regulation and disease. Through combining to the 3'untranslated region (UTR) of the target mRNA, MiRNAs modulate their target gene expression, which are usually short. 35. The regulatory axis has been proved in a considerable amount of research. Table 1 unveils the expression patterns, target genes, and functions of miR-483 family molecules in different types of cancer. Relying on their downstream targets, these molecules have been shown to act as both oncogenic and tumor suppressive roles. For instance, in Wilms' tumors, miR-483-3p can target and modulate the proapoptotic protein BBC3/PUMA, the induction of which is mediated by p53, and overexpressed miR-483-3p can protect tumor cells from apoptosis 20. In adrenocortical cancer, the antiapoptotic oncogene PUMA (Protein expression of p53 upregulated modulator of apoptosis) is significantly decreased, and as a target of miR-483-3p, it is regulated by the expression of miR-483-3p which is found over-expressed in adrenocortical cancer 21. In lung cancer, the WNT/β-catenin signaling pathway leads to the over-expression of and the premetastatic function of miR-483-5p has played a part by negatively modulating the direct target genes: RhoGDI1 (Rho GDP dissociation inhibitor alpha) and ALCAM (activated leukocyte cell adhesion molecule), two putative metastasis suppressors 36. miR-483-5p is high expressed in nasopharyngeal cancer, and genes involved in ubiquitin-mediated proteolysis (VHL, UBE3A, and UBE3B) are targeted by miR-483-5p, these pathways may constitute the mechanism underlying miRNA-related tumorigenesis 37. Nevertheless, miR-483 family can also help to eliminate tumor cells by targeting the oncogenic pathway. For example, in gliomas, miR-483-5p is a down-regulated miRNA which is able to restrain glioma cell proliferation through targeting gene ERK1 (extracellular signal-regulated kinase 1) directly, and this implies that miR-483-5p can help to eliminate tumor cells 3.
Table 1.
Summary of the target genes and functions of miR-483 family molecules in diverse cancers.
| Cancers | Family member | Expression | Target genes | Function | References |
|---|---|---|---|---|---|
| Oral cancer | miR-483-5p | over-expressed | - | tumor promotor | 25 |
| Esophageal cancer | miR-483, miR-483-5p | over-expressed | BSG | tumor promotor, drug resistance | 2, 6, 38, 42 |
| Gastric cancer | miR-483, miR-483-5p | over-expressed | - | tumor promotor, histologic transformation, proliferation and invasion | 27, 31, 43 |
| Intestinal cancer | miR-483 | downregulated | - | tumor suppressor | 31 |
| Colorectal cancer | miR-483-3p, miR-483-5p | over-expressed | IGF2, DLC-1, CKB | tumor promotor | 7, 23 ,29, 44 |
| Pancreatic cancer | miR-483-3p | over-expressed | DPC4/Smad4 | tumor promotor | 8, 19, 39, 45 |
| Hepatocellular cancer | miR-483-3p, miR-483-5p | over-expressed | IGF2 | tumor promotor | 46 |
| miR-483, miR-483-5p | downregulated | RAI16 | Bidirectional/ tumor suppressor | 9, 32, 47, 48 | |
| Cholangiocarcinoma | miR-483-5p | over-expressed | - | tumor promotor | 28 |
| Wilms' tumors | miR-483-3p, miR-483-5p | over-expressed | IGF2, BBC3/PUMA | tumor promotor | 16, 17, 18, 20 |
| Adrenocortical cancer | miR-483-3p, miR-483-5p | over-expressed | PUMA | tumor promotor | 21, 30, 49, 50 |
| Nasopharyngeal carcinoma | miR-483-5p | over-expressed | VHL, UBE3A, UBE3B | tumor promotor | 37, 51 |
| Lung Cancer | miR-483-5p | over-expressed | RhoGDI1, ALCAM | tumor promotor | 36, 52 |
| miR-483 | downregulated | MMP3 | tumor suppressor | 53 | |
| Ovarian Cancer | miR-483-5p | over-expressed | - | tumor promotor | 54, 55 |
| Multiple myeloma | miR-483-5p | over-expressed | - | tumor promotor | 15, 55 |
| Plasma cell leukemia | miR-483-5p | downregulated | - | tumor suppressor | 33 |
| Malignant Mesothelioma | miR-483-3p | over-expressed | RBI | tumor promotor | 56 |
| Prostate cancer | miR-483-3p, miR-483-5p | over-expressed | Ago2 | tumor promotor | 22, 35 |
| Breast cancer | miR-483-3p, miR-483-5p | downregulated | - | tumor suppressor | 24 |
| Glioma | miR-483-5p | downregulated | ERK1 | tumor suppressor | 3 |
| Pheochromocytoma | miR-483-5p | over-expressed | - | tumor promotor | 57, 58 |
| Angiosarcoma | miR-483-5p | downregulated | - | tumor suppressor | 34 |
miRNA biosynthetic machinery deregulation leads to differential expression of miRNAs and downstream target genes. In connection with these reports, some results also showed that miR-483 upstream regulatory mechanism is of significance 8. In colorectal cancer, miR-483 was co-transcribed with the IGF2 gene driven by the IGF2 promoter, and both of miR-483 and IGF2 were increased in colorectal cancer tissues than healthy individuals 23. Furthermore, epigenetic features, including DNA methylation, histone modifications, and miRNAs, have been described as early events in carcinogenesis recently 31. In Wilms' tumor, it has been found that miR-483-3p was regulated by IGF2 via an epigenetic pathway, and in many biological situations like proliferation and tissue regeneration, this is a quite common mechanism. Broad dysregulation of miR-483 has been observed in various forms of cancer, which shows that it has an impact on tumorigenesis. Clearly, there is a dearth of the investigation into the role of miR-483 in cancer.
The pathways and interactions of miR-483 involved in digestive tract cancers
Since a wealth of studies exhibited the expression of miR-483 family in different types of cancer, it is important for us to make a deeper understanding of oncogenesis in digestive tract cancers. Recent studies have explored the molecular modification result in abnormally expression of miR-483 family and found several possible mechanisms. As a multifunctional highly glycosylated integral transmembrane protein, Basigin (BSG/CD147) has been shown that acts as a key modulator in the pathogenesis of cancer invasion and metastasis. Li et al. found that the miR-483-5p binding site polymorphism in the 3'-UTR of the BSG gene is related to increased susceptibility of esophageal cancer, and the miR-483-5p/BSG axis may play an important role in esophageal cancer carcinogenesis 2. In pancreatic cancer, DPC4/Smad4 has been identified as a tumor-suppressor gene and is regarded as a target of miR-483-3p, and the miR-483-3p over-expression not only inhibits DPC4/Smad4 protein level, but also represses the expression of DPC4/Smad4 downstream targets like PAI, p21 and Msx2 19. It has been reported that miR-483-3p is over-expressed in hepatocellular carcinoma that carry mutations in Wnt/β-catenin signaling pathway genes and in hepatocellular carcinoma with wild-type TP53 gene. Transcribing via the β-catenin (CTNNB1)/USF1 complex, miR-483-3p targets of TP53 and PUMA, which are the important downstream apoptotic factor. Through the pathway, miR-483-3p could induce apoptosis resistance of hepatocellular carcinoma 67. In another research by this author, researchers found that O-GlcNAc transferase(OGT) is an important factor which meditates numeral cellular processes and repression of its activity brings about low-expression of miR-483-3p. Moreover, they identified that cellular glucose availability is participated in the regulation of miR-483-3p and demonstrated OGT activity as a molecular mechanisms link between miR-483-3p expression and glucose. Combined with their earlier research results, the glucose/OGT/miR-483-3p signaling pathway plays a vital part in the tumorgenic mechanisms of liver cancer and may involve in the drug resistance 68. The DLC-1 gene has been revealed as a candidate tumor suppressor, and in colorectal cancer, miR-483-3p contributes to cancer cells proliferation via suppressing its target DLC-1 gene 23. On the contrary, miR-483 has also been reported as tumor inhibitor in other types of cancer through oncogenes repression losses or intervention in pathways that administrate cellular processes. For instance, miR-483-5p is downregulated in hepatocellular cancer, through MAPK/ERK and TGF-β pathways dependent mechanism, RAI16, as a target gene of miR-483-5p, is regulated by miR-483-5p which enhances HCC tumorigenesis in vitro and in vivo 32.
The clinical value of miR-483 in digestive tract cancers
mir-483 family molecules have always been found in digestive tract cancer tissues, including Oral cancer, Esophageal cancer, Gastric cancer, Intestinal cancer, Colorectal cancer, Pancreatic cancer, Hepatocellular cancer and Cholangiocarcinoma, albeit in various levels. Table 2 presents the clinicopathological association of miR-483 family molecules in digestive tract cancers. For instance, colorectal cancer exhibits large differences in miRNA expression levels compared with normal tissue. Compared to the normal adjacent tissues, both miR-483-3p and miR-483-5p are synchronously increased in colorectal cancer tissues, which shows that the miR-483 family may serve as a new target for therapy and a potential biomarker for the recognition of colorectal cancer 23. In the field of esophageal cancer research, miR-483 expression was found to be over-active in cancer tissues, which shows that the miR-483 level measurement is helpful in distinguishing esophageal cancer from high-risk group 38. Compared with side normal tissue, miR-483-3p is aberrantly over-active in pancreatic cancer tissues as a member of miR-483 family, indicating that aberrant expression of miR-483-3p is an important indicator in pancreatic cancer in the early stage and is related to tumor distinction and prognosis 19. As another part of miR-483 family, miR-483-5p is over-active specifically in Cholangiocarcinoma 28. Apart from the diagnostic potential, it has been reported that miR-483 family molecules are in association with digestive tract cancers clinicopathological situation, making the using of these molecules in prognostic application possible. For example, in terms of clinical outcome, high miR-483-3p expression is related to high mortality in pancreatic cancer, and it has a considerably higher mortality than that with low expression. Furthermore, miR-483-3p is suggested to be a predictor of poor disease outcome 39. miR-483-5p expression was built up considerably in oral cancer patients, which was associated with tumor nodal metastasis (TNM) stage and lymph nodal metastases significantly. Compared with those with low expression, patients whose serum have high miR-483-5p may have a relatively low survival rate and therefore, high serum miR-483-5p expression was adequate enough to indicate the overall survival rate 25. In esophageal cancer, high expression of miR-483 had a poorer overall survival and a less median survival, and it may predict less sensitivity to chemotherapy. Otherwise, downregulation of miR-483 may reverse drug resistance and it may increase drug accumulation 38. Hence, miR-483 family molecules in digestive tract cancers may consider as biomarkers which can apply clinically in diagnosis, prognosis, or prediction of treatment efficacy.
Table 2.
Summary of the clinicopathological significance of miR-483 family molecules in digestive tract cancers.
| Cancers | Family member | Diagnostic potential | Clinicopathological or prognostic association | References |
|---|---|---|---|---|
| Oral cancer | miR-483-5p | Differentiate normal and cancer | nodal metastases and late cancer stages | 25 |
| Esophageal cancer | miR-483, miR-483-5p | Differentiate normal, pre-cancer and cancer | Associated with shorter survival and multidrug-resistance | 2, 6, 38, 42 |
| Gastric cancer | miR-483, miR-483-5p | Differentiate normal and cancer | - | 27, 31, 43 |
| Intestinal cancer | miR-483 | Differentiate normal and cancer | - | 31 |
| Colorectal cancer | miR-483-3p, miR-483-5p | Differentiate normal, pre-cancer and cancer | - | 7, 23 ,29, 44 |
| Pancreatic cancer | miR-483-3p | Differentiate normal, pre-cancer and cancer | Associated with shorter survival | 8, 19, 39, 45 |
| Hepatocellular cancer | miR-483-5p | Differentiate normal and cancer | - | 9, 32, 46, 47, 48 |
| Cholangiocarcinoma | miR-483-5p | Differentiate normal and cancer | - | 28 |
Prospects of miR-483 in DTCs
The potential applications of miR-483 as a useful biomarker
MiRNAs are known to be excreted from many types of cells in both exosome-dependent and exosome-independent ways, and they have been confirmed to be extremely stable and are detectable in biopsy tissues, blood, wastes and other humoral such as tears, saliva, milk and urine 40. Therefore, it has paved the way for employment of miRNAs as novel biomarkers for many diseases 15. When looking at the literature concerning miR-483 family, some researches have detected that digestive tract cancer patients whose peripheral blood had miR-483 molecules and connected with clinical importance. One research showed that miR-483-5p is significantly boosted in the colorectal cancer tissues, and the serum miR-483-5p level is also increased in colorectal cancer patients compared to the normal samples 23. Concordantly, miR-483-5p expression is also considerably upregulated in oral cancer patient sera than in controls, and thus it is associated with diagnosis, clinicopathological features, and prognosis of oral cancer patients 41. miR-483-5p is over-expressed in Cholangiocarcinoma (CC) when compared with primary sclerosing cholangitis (PSC), and the study provides a basis for the use of miR-483-5p as a new biomarker for the diagnosis of PSC and CC 28. In terms of pancreatic cancer, the miR-483-3p plasma expression level is considerably higher in patients than healthy controls, and this indicates that the measurement of miR-483-3p from plasma helps to distinguish pancreatic cancer from healthy subjects 25. The above results suggest that the miR-483 family can discriminate cancer patients from high-risk individuals, and they may serve as circulating biomarkers for multiple cancers.
Obstacles
Whether intrinsic or acquired, chemotherapeutic drug resistance of cancer cells is thought to be a major obstacle obstructing successful treatment of cancer, which may lead to ultimate cancer death. Although inadequate delivery of the drug to the tumor can contribute to chemo resistance, cellular reprogramming also plays a major role in establishing this resistance. By turning off the chemo sensitivity related genes and turning on the chemo resistance related genes, tumor cells can effectively evade the drug and resist to apoptosis 59.
Since the discovery of the first miRNA was associated with drug resistance by targeting the target gene 60, the increasing number of other miRNAs have been confirmed to be associated with drug resistance spanning almost all classes of drugs 61. Recently, the research about mechanisms of drug resistance has emphasized miRNAs as significant modulators of the death of cells. miRNAs may regulate one or several mRNAs to participate in core apoptotic signaling pathway, and the loss of function mutation in miRNA could change the levels of drug targets in live cells. Thus, downregulated pro-apoptotic miRNAs expression and/or upregulated anti-apoptotic miRNAs expression in tumor cells could be connected with high apoptosis limitation and drug resistance 62. Studies have shown that the miR-483 family plays a role in reducing chemo resistance by targeting these genes involved in resistance development. Zhou et al. suggested that the miR-483 expression is significantly upregulated in esophageal squamous cell cancer tissues, and there is a reverse relationship between the expression level of miR-483 and overall survival. In depth analysis revealed that miR-483 could inhibit Puma, which might promote apoptosis by interacting and inhibiting the antiapoptotic factors BCL2 and BCLXL 63. Downregulate miR-483 expression in esophageal cancer cells can confer sensitivity to both P-glycoprotein related and non-P-glycoprotein related drugs, and it may increase the Adriamycin (ADR) accumulation and decrease the amount of released ADR. These findings suggest that downregulation of miR-483 may contribute to the development of anticancer drug resistance of esophageal cancer 6. Although the area of miRNAs pharmacogenomics is still new, and studies are lacking on large randomized clinical trials, further study of miRNA signatures in biopsies of tumor has potential to predict drug resistance 62. The knowledge of all the miRNAs associated with drug resistance will be a key to designing drugs with greater efficacy and safety, and has significant health and economic implications 61.
Actually, despite enormous efforts and the comprehensive knowledge of attractive miRNA targets in many disease conditions, apart from drug resistance, several other obstacles remain to be overcame for the transition of miRNAs from basic to clinical application as utilization of miRNA-based therapeutic strategies for treatment 64. Firstly, the successful therapeutic agent delivery to target tissue is restricted by anatomical, physical, pharmacokinetic, and the barriers of pharmacodynamics. This may be solved in a range of delivery strategies and targeting approaches including cholesterol-conjugated miRNA, nanoparticle-conjugated miRNA, liposome encapsulation, antibody-based targeted therapy and aptamer-based targeted drug delivery. As ubiquitously present nucleases can downgrade the miRNA-mimics and anti-miRNAs inside human body, and synthetic modification is necessary for these agents to become stable. Secondly, miRNA-based therapies, which include antisense strategies, have the potential for suppressing the miRNA and exerting off-target effects. Thus, target miRNAs should be taken into account so as to make sure that high degree of specificity intended endogenous target could be modulated by miRNA-based therapeutics in order to minimize off-target effects. Finally, miRNA-therapeutics administration and carrier medium is affected by independent off-target. For example, targeted nanoparticles may give rise to deleterious results via miRNA sequence independent off-target effects. Both inherent and adaptive weapons of the human immune systems may detect miRNA agents, and for therapeutic components, chemical modification is essential so as to avoid the off-target effects of immunostimulatory. Furthermore, these agents may alter physiological processes or may break the organ system function due to the accumulation, clearance, and excretion of these molecules. Hence, to it is necessary to take these therapeutic barriers into consideration so as to utilize miRNA-based therapeutics successfully 65. Although making miRNA replacement therapy routine is not an easy task, it should be noted that miRNAs were found in mammals just a decade ago. As time goes by, it will be known to all whether fast rate of discovery has deepened the understanding of the genetics, and the miRNA function will be interpreted into applications like miRNA replacement therapy for diseases, especially for cancer 66.
Conclusion
In summary, microRNAs represent crucial regulators of tumor cell cycle progression which contains proliferation, differentiation and apoptosis, especially tumorigenesis. miR-483 is a new star miRNA in human cancer which is located within the IGF2 gene second intron, and it has two ripe forms: miR-483-3p, miR-483-5p. Recent research indicates that the dysregulation of miR-483 expression has been detected in a diversity cancer types specially in digestive tract cancers (DTCs), and miR-483 may interact with cell signaling pathways and control cellular homeostasis. Emerging evidence suggests that miR-483 molecules are extremely stable and are detectable in biopsy tissues or different kinds of body fluid, and the miR-483 family could become a novel diagnostic and prognostic tool for treatment strategy in cancer therapy. With some obstacles remain to be overcame, it is to be hoped that more research can be conducted on miRNAs stability optimization, delivery systems improvement as well as miRNA therapeutics control.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No: 81171923).
References
- 1.Liu C, Jiang Z, Deng Q X. et al. Meta-analysis of association studies of CYP1A1 genetic polymorphisms with digestive tract cancer susceptibility in Chinese. [J] Asian Pacific Journal of Cancer Prevention Apjcp. 2014;15(11):4689–95. doi: 10.7314/apjcp.2014.15.11.4689. [DOI] [PubMed] [Google Scholar]
- 2.Li H Y, Liu Y C, Bai Y H, SNP at miR-483-5p-binding site in the 3'-untranslated region of the BSG gene is associated with susceptibility to esophageal cancer in a Chinese population. [J] Genetics & Molecular Research Gmr; 2016. p. 15. (2) [DOI] [PubMed] [Google Scholar]
- 3.Li W, Ming S, Hou S. et al. MiR-483-5p suppresses the proliferation of glioma cells via directly targeting ERK1[J] Febs Letters. 2012;586(9):1312–1317. doi: 10.1016/j.febslet.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 4.Amirkhah R, Schmitz U, Linnebacher M. et al. MicroRNA-mRNA interactions in colorectal cancer and their role in tumor progression.[J] Genes Chromosomes & Cancer. 2015;54(3):129–141. doi: 10.1002/gcc.22231. [DOI] [PubMed] [Google Scholar]
- 5.Andorfer C A, Necela B M, Thompson E A. et al. MicroRNA signatures: clinical biomarkers for the diagnosis and treatment of breast cancer.[J] Trends in Molecular Medicine. 2011;17(6):313. doi: 10.1016/j.molmed.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y, Hong L. Prediction value of miR-483 and miR-214 in prognosis and multidrug resistance of esophageal squamous cell carcinoma. [J] Genetic Testing & Molecular Biomarkers. 2013;17(6):470. doi: 10.1089/gtmb.2012.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yong F L, Law C W, Wang C W. Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer[J] BMC Cancer. 2013;13(1):280. doi: 10.1186/1471-2407-13-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rachagani S, Macha M A, Menning M S. et al. Changes in microRNA (miRNA) expression during pancreatic cancer development and progression in a genetically engineered KrasG12D; Pdx1-Cre mouse (KC) model[J] Oncotarget. 2015;6(37):40295–40309. doi: 10.18632/oncotarget.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Zhao L J, Tan Y X. et al. Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma.[J] World Journal of Gastroenterology Wjg. 2012;18(38):5442–5453. doi: 10.3748/wjg.v18.i38.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Li Z, Chan M T. et al. The roles of microRNAs in Wilms' tumors. [J] Tumor Biology. 2016;37(2):1–6. doi: 10.1007/s13277-015-4514-8. [DOI] [PubMed] [Google Scholar]
- 11.Guo Z W, Xie C, Yang J R. et al. MtiBase: a database for decoding microRNA target sites located within CDS and 5′UTR regions from CLIP-Seq and expression profile datasets[J] Database the Journal of Biological Databases & Curation. 2015;2015:bav102. doi: 10.1093/database/bav102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Liang H, Bai M. et al. miR-135b Promotes Cancer Progression by Targeting Transforming Growth Factor Beta Receptor II (TGFBR2) in Colorectal Cancer. [J] Plos One. 2015;10(6e):0130194. doi: 10.1371/journal.pone.0130194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sluijter J P G. MicroRNAs in Cardiovascular Regenerative Medicine: Directing Tissue Repair and Cellular Differentiation[J]. Isrn Vascular Medicine. 2013; 2013. [Google Scholar]
- 14.Malizia A P, Wang D Z. miRNA in Cardiomyocyte Development[J] Wiley Interdisciplinary Reviews Systems Biology & Medicine. 2011;3(2):183–90. doi: 10.1002/wsbm.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu X, Zhao M, Wu S. et al. Circulating microRNA 483-5p as a novel biomarker for diagnosis survival prediction in multiple myeloma. [J] Medical Oncology. 2014;31(10):1–8. doi: 10.1007/s12032-014-0219-x. [DOI] [PubMed] [Google Scholar]
- 16.Liu M, Roth A, Yu M. et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. [J] Genes & Development. 2013;27(23):2543–2548. doi: 10.1101/gad.224170.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson J A, Bryan K, Williams R. et al. miRNA profiles as a predictor of chemoresponsiveness in Wilms' tumor blastema. [J] Plos One. 2013;8(1):e53417. doi: 10.1371/journal.pone.0053417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, Li Z, Chan M T. et al. The roles of microRNAs in Wilms' tumors. [J] Tumor Biology. 2016;37(2):1–6. doi: 10.1007/s13277-015-4514-8. [DOI] [PubMed] [Google Scholar]
- 19.Hao J, Zhang S, Zhou Y. et al. MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer. [J] Febs Letters. 2011;585(1):207–213. doi: 10.1016/j.febslet.2010.11.039. [DOI] [PubMed] [Google Scholar]
- 20.Veronese A, Lupini L, Consiglio J. et al. Oncogenic role of miR-483-3p at the IGF2/483 locus.[J] Cancer Research. 2010;70(8):3140–3149. doi: 10.1158/0008-5472.CAN-09-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Özata DM, Caramuta S, Velázquezfernández D. et al. The role of microRNA deregulation in the pathogenesis of adrenocortical carcinoma[J] Endocr Relat Cancer. 2011;18(6):643. doi: 10.1530/ERC-11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korzeniewski N, Tosev G, Pahernik S. et al. Identification of cell-free microRNAs in the urine of patients with prostate cancer.[J] Urologic Oncology Seminars & Original Investigations. 2015;33(1):16.e17. doi: 10.1016/j.urolonc.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Cui H, Liu Y, Jiang J. et al. IGF2-derived miR-483 mediated oncofunction by suppressing DLC-1 and associated with colorectal cancer[J] Oncotarget. 2016;7(30):48456–48466. doi: 10.18632/oncotarget.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, Liu D, Li W. et al. Identification of featured biomarkers in breast cancer with microRNA microarray[J] Archives of Gynecology & Obstetrics. 2016;294(5):1–7. doi: 10.1007/s00404-016-4141-7. [DOI] [PubMed] [Google Scholar]
- 25.Xu H, Yang Y, Zhao H. et al. Serum miR-483-5p: a novel diagnostic and prognostic biomarker for patients with oral squamous cell carcinoma[J] Tumor Biology. 2016;37(1):447–453. doi: 10.1007/s13277-015-3514-z. [DOI] [PubMed] [Google Scholar]
- 26.Wu C, Wang C, Guan X. et al. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma[J] Plos One. 2014;9(3e):92292. doi: 10.1371/journal.pone.0092292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Jin W, Yan Y. et al. Erratum to: loss of hsa-miR-337-3p expression is associated with lymph node metastasis of human gastric cancer[J] Journal of Experimental & Clinical Cancer Research. 2014;33(1):18. doi: 10.1186/1756-9966-32-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernuzzi F, Marabita F, Lleo A. et al. Serum microRNAs as novel biomarkers for primary sclerosing cholangitis and cholangiocarcinoma[J] Clinical & Experimental Immunology. 2016;185(1):61. doi: 10.1111/cei.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaedcke J, Grade M, Camps J. et al. The Rectal Cancer microRNAome - microRNA Expression in Rectal Cancer and Matched Normal Mucosa[J] Clinical Cancer Research. 2012;18(18):4919. doi: 10.1158/1078-0432.CCR-12-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soon P S H, Tacon L J, Gill A J. et al. miR-195 and miR-483-5p Identified as Predictors of Poor Prognosis in Adrenocortical Cancer[J] Clinical Cancer Research, An Official Journal of the American Association for Cancer Research. 2009;15(24):7684. doi: 10.1158/1078-0432.CCR-09-1587. [DOI] [PubMed] [Google Scholar]
- 31.Assumpção M B, Moreira F C, Hamoy I G. et al. High-Throughput miRNA Sequencing Reveals a Field Effect in Gastric Cancer and Suggests an Epigenetic Network Mechanism[J] Bioinformatics & Biology Insights. 2015;9:111–117. doi: 10.4137/BBI.S24066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, Zhao L J, Yang Y. et al. Retinoic acid induced 16 enhances tumorigenesis and serves as a novel tumor marker for hepatocellular carcinoma.[J] Carcinogenesis. 2012;33(12):2578–85. doi: 10.1093/carcin/bgs289. [DOI] [PubMed] [Google Scholar]
- 33.Lionetti M, Musto P, Di M M. et al. Biological and clinical relevance of miRNA expression signatures in primary plasma cell leukemia[J] Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2013;19(12):3130. doi: 10.1158/1078-0432.CCR-12-2043. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Kuang D, Zhao X. et al. miR-497-5p inhibits cell proliferation and invasion by targeting KCa3.1 in angiosarcoma. [J] Oncotarget. 2016;7(36):58148. doi: 10.18632/oncotarget.11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer D, Wahlfors T, Mattila H. et al. MiRNA Profiles in Lymphoblastoid Cell Lines of Finnish Prostate Cancer Families[J] Plos One. 2015;10(5e):0127427. doi: 10.1371/journal.pone.0127427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Q, Xu Y, Yang C. et al. miR-483-5p promotes invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1 and ALCAM. [J] Cancer Research. 2014;74(11):3031–42. doi: 10.1158/0008-5472.CAN-13-2193. [DOI] [PubMed] [Google Scholar]
- 37.Zheng X H, Cui C, Ruan H L. et al. Plasma microRNA profiling in nasopharyngeal carcinoma patients reveals miR-548q and miR-483-5p as potential biomarkers. [J] Chinese Journal of Cancer. 2014;33(7):330. doi: 10.5732/cjc.013.10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Hong L. Prediction value of miR-483 and miR-214 in prognosis and multidrug resistance of esophageal squamous cell carcinoma. [J] Genetic Testing & Molecular Biomarkers. 2013;17(6):470. doi: 10.1089/gtmb.2012.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Sun Y, Wu H. et al. Elevated miR-483-3p expression is an early event and indicates poor prognosis in pancreatic ductal adenocarcinoma. [J] Tumor Biology. 2015;36(12):9447–9456. doi: 10.1007/s13277-015-3690-x. [DOI] [PubMed] [Google Scholar]
- 40.Yong F L, Law C W, Wang C W. Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer[J] BMC Cancer. 2013;13(1):280. doi: 10.1186/1471-2407-13-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu Y C, Chang J T, Chan E C. et al. miR-196, an Emerging Cancer Biomarker for Digestive Tract Cancers[J] Journal of Cancer. 2016;7(6):650–655. doi: 10.7150/jca.13460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warnecke-Eberz U, Chon S H, Hölscher A H. et al. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor[J] Tumor Biology. 2015;36(6):4643. doi: 10.1007/s13277-015-3112-0. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Nadauld L, Ootani A. et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture[J] Nature Medicine. 2014;20(7):769. doi: 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Della V S G, Falcetta F, Carlomagno C. et al. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. [J] International journal of radiation oncology, biology, physics. 2012;83(4):1113. doi: 10.1016/j.ijrobp.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 45.Abue M, Yokoyama M, Shibuya R. et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer[J] International Journal of Oncology. 2015;46(2):539–47. doi: 10.3892/ijo.2014.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen J, Wang A, Wang Q. et al. Exploration of Genome-wide Circulating MicroRNA in Hepatocellular Carcinoma (HCC): MiR-483-5p as a Potential Biomarker[J] Cancer Epidemiology Biomarkers & Prevention. 2013;22(12):2364. doi: 10.1158/1055-9965.EPI-13-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin J C, Jin X L, Zhang X. et al. Effect of OSW-1 on microRNA expression profiles of hepatoma cells and functions of novel microRNAs[J] Molecular Medicine Reports. 2013;7(6):1831–7. doi: 10.3892/mmr.2013.1428. [DOI] [PubMed] [Google Scholar]
- 48.Wang W, Zhao L J, Tan Y X. et al. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma[J] Carcinogenesis. 2012;33(5):1113. doi: 10.1093/carcin/bgs113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chabre O, Libé R, Assie G. et al. Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. [J] Endocrine-related cancer. 2013;20(4):579. doi: 10.1530/ERC-13-0051. [DOI] [PubMed] [Google Scholar]
- 50.Szabó D R, Luconi M, Szabó P M. et al. Analysis of circulating microRNAs in adrenocortical tumors[J] Laboratory Investigation. 2014;94(3):331–9. doi: 10.1038/labinvest.2013.148. [DOI] [PubMed] [Google Scholar]
- 51.Luan J, Wang J, Su Q. et al. Meta-analysis of the differentially expressed microRNA profiles in nasopharyngeal carcinoma[J] Oncotarget. 2016;7(9):10513–10521. doi: 10.18632/oncotarget.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C, Ding M, Xia M. et al. A Five-miRNA Panel Identified from a Multicentric Case-control Study Serves as a Novel Diagnostic Tool for Ethnically Diverse Non-small-cell Lung Cancer Patients[J] Ebiomedicine. 2015;2(10):1377–85. doi: 10.1016/j.ebiom.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Z G, Jin J Y, Zhang A M. et al. MicroRNA profile of tumorigenic cells during carcinogenesis of lung adenocarcinoma[J] Journal of Cellular Biochemistry. 2015;116(3):458–66. doi: 10.1002/jcb.24999. [DOI] [PubMed] [Google Scholar]
- 54.Yu X, Zhang X, Bi T. et al. MiRNA expression signature for potentially predicting the prognosis of ovarian serous carcinoma[J] Tumor Biology. 2013;34(6):3501–3508. doi: 10.1007/s13277-013-0928-3. [DOI] [PubMed] [Google Scholar]
- 55.Bi C, Chung T H, Huang G. et al. Genome-wide pharmacologic unmasking identifies tumor suppressive microRNAs in multiple myeloma[J] Oncotarget. 2011;6(28):26508–18. doi: 10.18632/oncotarget.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guled M, Lahti L, Lindholm P M. et al. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma—A miRNA microarray analysis[J] Genes Chromosomes & Cancer. 2009;48(7):615. doi: 10.1002/gcc.20669. [DOI] [PubMed] [Google Scholar]
- 57.Meyerrochow G Y, Jackson N E, Conaglen J V. et al. MicroRNA profiling of benign and malignant pheochromocytomas identifies novel diagnostic and therapeutic targets. [J] Endocrine-related cancer. 2010;17(3):835. doi: 10.1677/ERC-10-0142. [DOI] [PubMed] [Google Scholar]
- 58.Patterson E, Webb R, Weisbrod A. et al. The microRNA expression changes associated with malignancy and SDHB mutation in pheochromocytoma[J] Endocrine Related Cancer. 2012;19(2):157–166. doi: 10.1530/ERC-11-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Humphries B, Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. [J] Oncotarget. 2015;6(9):6472. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mishra P J, Humeniuk R, Mishra P J. et al. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance[J] Proceedings of the National Academy of Sciences of the United States of America. 2007;104(33):13513–8. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mishra P J. The miRNA-drug resistance connection: a new era of personalized medicine using noncoding RNA begins. [J] Pharmacogenomics. 2012;13(12):1321. doi: 10.2217/pgs.12.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lima R T, Busacca S, Almeida G M. et al. MicroRNA regulation of core apoptosis pathways in cancer[J] European Journal of Cancer. 2011;47(2):163–74. doi: 10.1016/j.ejca.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 63.David S, Meltzer S J. MicroRNA involvement in esophageal carcinogenesis[J] Current Opinion in Pharmacology. 2011;11(6):612–6. doi: 10.1016/j.coph.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kentaro I, Yuichi I. MicroRNA In Lung Cancer: Novel Biomarkers and Potential Tools for Treatment: [J] Journal of Clinical Medicine. 2016;5(3):36. doi: 10.3390/jcm5030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma S B, Ruppert J M. MicroRNA-Based Therapeutic Strategies for Targeting Mutant and Wild Type RAS in Cancer[J] Drug Development Research. 2015;76(6):328. doi: 10.1002/ddr.21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henry J C, Azevedo-Pouly A C P, Schmittgen T D. microRNA Replacement Therapy for Cancer[J] Pharmaceutical Research. 2011;28(12):3030. doi: 10.1007/s11095-011-0548-9. [DOI] [PubMed] [Google Scholar]
- 67.Veronese A, Visone R, Consiglio J. et al. Mutated beta-catenin evades a microRNA-dependent regulatory loop[J] Proceedings of the National Academy of Sciences of the United States of America. 2011;108(12):4840–4845. doi: 10.1073/pnas.1101734108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pepe F, Pagotto S, Soliman S. et al. Regulation of miR-483-3p by the O-linked N-acetylglucosamine transferase links chemosensitivity to glucose metabolism in liver cancer cells[J] Oncogenesis. 2017;6(5e):328. doi: 10.1038/oncsis.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]