Abstract
Histone H2B monoubiquitination plays a critical role in the regulation of gene transcription. Deregulation of H2B monoubiquitination contributes to human pathologies, such as cancer. Here we report that human USP36 is a novel H2Bub1 deubiquitinase. We show that USP36 interacts with H2B and deubiquitinates H2Bub1 in cells and in vitro. Overexpression of USP36 markedly reduced the levels of H2Bub1 in cells. Using the p21 gene as a model, we demonstrate that depletion of USP36 increases H2Bub1 at the p21 locus, primarily within its gene body. Consistently, knockdown of USP36 induced the expression of p21 and inhibits cell proliferation. Together, our results reveal USP36 as a novel H2B deubiquitinase and shed light on its additional functions in regulating gene expression.
Keywords: Deubiquitinating enzyme, Histone, H2B, H2Bub1, USP36
1. Introduction
Posttranslational modifications of core histones play a key role in regulating chromatin dynamics, gene transcription, and DNA repair [1, 2]. Monoubiquitination of Histone 2b is an evolutionally conserved modification. A single ubiquitin E3, Rad6/Bre1, mediates H2B monoubiquitination at Lys 123 (K123) in yeast whereas the orthologs RNF20 and RNF40 form a complex to mediate H2B monoubiquitination at K120 in human [3, 4]. Monoubiquitinated H2B (H2Bub1) is generally enriched at actively transcribed genes and positively correlated with their expression levels [5, 6]. It has been implicated in nucleosome organization [7], transcription elongation [8, 9], as well as histone cross-talk [1, 10, 11]. H2Bub1 stimulates the function of the histone chaperone FACT and promotes efficient RNA polymerase (Pol) II passage during transcription elongation [8, 9]. Thus, H2B monoubiquitination plays an important role in regulating gene expression. Not surprisingly, deregulation of this modification contributes to human pathologies, including cancer [12–14]. In support of this notion, several studies have found a negative correlation between H2Bub1 levels and tumor progression. Consistently, the RNF20 gene promoter is frequently hypermethylated in breast cancers, indicating that RNF20 may be an important tumor suppressor [15].
On the other hand, H2Bub1 can be reversed by deubiquitinating enzymes (DUBs). In yeast, two DUBs, ubp8 and ubp10, deubiquitinate H2Bub1 [16, 17]. Ubp8 (USP22 in humans) is a subunit of DUB module within the SAGA (Spt-Ada-Gcn5-acetyltransferase) co-activator complex [17] that may be broadly required for transcription [18]. In contrast, ubp10 has been shown to act as a transcriptional repressor and associates with the silent regions of the yeast genome [16]. In addition, ubp8 and ubp10 appear to regulate distinct regions of genes, with ubp8 deubiquitinating H2B close to the transcriptional start sites and ubp10 deubiquitinating H2B within the gene body [19]. Similar functions have been reported for Drosophila DUBs, Nonstop and Scrawny, the ubp8 and ubp10 orthologs, respectively [20, 21]. Like ubp10, Scrawny has been described as a transcriptional repressor [20].
In humans, a handful of DUBs can deubiquitinate H2Bub1 in addition to USP22 [22, 23], including USP44, USP42, USP49, USP3, and USP15 [24–28]. This increase in enzymatic complexity is most likely indicative of a wider breadth of regulatory functions for H2Bub1. Here, we show that human USP36 conserves the function of its yeast and Drosophila orthologs ubp10 and Scrawny to deubiquitinate H2B. Using the model gene CDKN1A (coding for p21), we demonstrate that USP36 deubiquitinates H2Bub1 at the p21 gene body. Consistently, we show that knockdown of USP36 increased p21 levels and drastically suppressed cell proliferation. Together, these results suggest that USP36 is an important regulator of H2Bub1.
2. Material and methods
2.1. Cell Culture, Plasmids, and Antibodies
Human H1299, 293 and HeLa cells were cultured as described [29, 30]. USP36 plasmids were described [29, 31]. Flag-H2B was constructed by inserting H2B cDNA into the pcDNA3-2Flag vector. Anti-USP36 antibody was a gift from Dr. Komada (Tokyo Institute of Technology, Japan) [31]. Anti-H2B, anti-H2Bub1 (Millipore), anti-Flag (Sigma), anti-H3K4me3, anti-H3K36me3 (Abcam) and anti-p21 (NeoMarkers) were purchased.
2.2. Transfection, Immunoblot (IB) and co-immunoprecipitation (Co-IP) analyses
Cell transfection, lysate preparation, IB and Co-IP analyses were conducted as previously described [29, 30].
2.3. Histone isolation
Cells were resuspended in extraction buffer (0.5% Triton-X, 1 mM PMSF, 1 mM pepstatin A in PBS) and lysed on ice for 10 min, followed by centrifugation. Pellets were washed once in extraction buffer and resuspended in 0.2 N HCl and incubated at 4 °C overnight. After centrifugation, supernatant containing extracted histones was neutralized by adding 1 M NaOH.
2.4. In vitro deubiquitination assay
Recombinant His-USP361-800 and its C131A mutant proteins were expressed in E. coli and purified using Ni2+-NTA purification method. Histones were incubated with His-USP361-800 or its C131A mutant proteins for 16 hours at 25 °C in deubiquitination buffer containing 50 mM Tris-HCl (pH 8.0) and 10 mM DTT and assayed by IB.
2.4. Glutathione S-transferase (GST) fusion protein-protein interaction assays
Histones (~170 ng) extracted from H1299 cells were incubated with the glutathione-Sepharose 4B beads (GE Healthcare) containing 100 ng of GST-USP361-800, GST-USP361-800/C131A, or GST alone. After washing, bound proteins were analyzed by IB.
2.6. Chromatin Immunoprecipitation (ChIP)-qPCR and RT-qPCR assays
ChIP assays were conducted as previously described [32]. Immunoprecipitated DNA fragments were analyzed by qPCR. The primers used for amplifying the p21 gene locus were: 5′-AGCAGGCTGTGGCTCTGATT-3′ and 5′-CAAAATAGCCACCAGCCTCTTCT-3′ (Up); 5′-AGCCGGAGTGGAAGCAGA-3′ and 5′-AGTGATGAGTCAGTTTCCTGCAAG-3′ (Start); 5′-CCAGGGCTGCGATTAGGAA-3′ and 5′-GTGTCCCTCATGGGTGTGAAT-3′ (Mid); 5′-CCTCCCACAATGCTGAATATACAG-3′ and 5′-AGTCACTAAGAATCATTTATTGAGCACC-3′ (End). RT-qPCR was conducted as described using SYBR Green Mix (Bio-Rad) [30]. All qPCR reactions were carried out in triplicate. Relative gene expression was calculated using the ΔCτ method.
2.7. Micrococcal nuclease (MNase) digestion for chromatin fractionation
Cells were resuspended in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, 1 mM PMSF, 1 mM pepstatin A, 0.05% Triton-X) and incubated on ice for 5 min. After centrifugation and wash, the nuclear pellets were split into 2 tubes. Half of the nuclear pellets was resuspended in buffer A in the presence of 1X MNase buffer, 1% BSA and 1 μl MNase. The other half was untreated. Reactions were incubated for 5 min at 37 °C followed by 20 min at room temperature and stopped by adding 1 mM EGTA. Nuclei digested with or without MNase were then lysed in solution B (3 mM EDTA, 0.2 mM EGTA, 1 mM dithiothreitol and protease inhibitors) for 30 min on ice, followed by centrifugation. Equal volume of soluble and insoluble pellet was assayed by IB.
2.8. EdU Incoporation assay
Cells were incubated with 10 μM EdU for 6 h at 37 °C. The cells were then fixed in 4% paraformaldehyde and permeabilized in PBS containing 0.5% Triton X-100 at room temperature. After washing, 0.5mL of EdU staining buffer (100mM Tris-HCL, 4 mM CuSO4, 5 μM azide dye (Click Chemisty Tools), 100 mM ascorbic acid in PBS) was added to each plate and incubated for 30 min. After wash, the cells were stained with DAPI for 10 min. The number of EdU labeled cells out of the total number of DAPI stained cells was counted using ImageJ software.
2.9. MTS assay to measure cell viability
Cell viability was measured using MTS assay (Promega) following the manufacturer’s instructions. 20 μl of MTS reagent was added to each well and the plate was incubated at 37 °C for 3 hours, followed by measuring absorbance at 490 mm.
2.10. IncuCyte proliferation assay
Cell confluence was monitored in IncuCyte Zoom System (Essen Bioscience) as described [29].
2.11. Lentiviral knockdown
Lentivirus-mediated knockdown was conducted as previously described [29]. The shRNA sequences are 5′-CGTCCGTATATGTCCCAGAAT-3′ (shRNA-1) and 5′-GCGGTCAGTCAGGATGCTATT-3′ (shRNA-2, used for all experiments, except where indicated).
3. Results
3.1. USP36 deubiquitinates H2Bub1
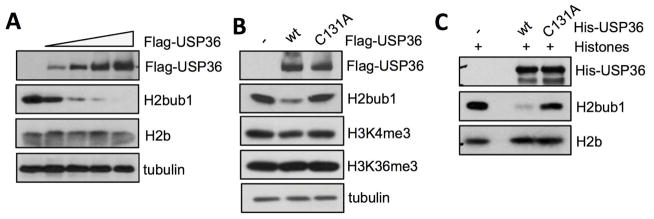
To determine whether USP36 has a conserved function in deubiquitinating H2Bub1, we examined its effect on global H2Bub1 using a monoclonal antibody that specifically recognizes the branch site where ubiquitin is conjugated to H2B [5]. As shown in Fig. 1A, H2Bub1 levels markedly decreased upon overexpression of USP36 in a dose-dependent manner in H1299 cells, while the total H2B levels remained unchanged. To determine whether the decreased H2Bub1 was dependent on the catalytic activity of USP36, we utilized a catalytic-inactive C131A mutant of USP36 (USP36C131A) [33]. Unlike wild-type (wt) USP36, the catalytic mutant did not decrease H2Bub1 levels (Fig. 1B). Similar results were also observed in other tested cell lines (data not shown). To examine whether USP36 could directly deubiquitinate H2Bub1, we performed an in vitro deubiquitination assay. Purified histones were incubated with either a His-tagged wt USP36 N-terminal fragment (His-USP361-800) or its catalytically inactive mutant (His-USP361-800/C131A). As shown in Fig. 1C, only wt USP361-800 was capable of deubiquitinating H2Bub1 in vitro. Together, these data demonstrate that USP36 is a bona fide DUB for H2Bub1.
Figure 1. USP36 Deubiquitinates H2Bub1.
(A) USP36 deubiquitinates H2Bub1 in cells. H1299 cells transfected with increasing doses of Flag-USP36 were assayed by IB. (B) USP36-mediated reduction of H2Bub1 requires its catalytic activity. H1299 cells transfected with Flag-tagged wt USP36 or its catalytically inactive C131A mutant were assayed by IB. (C) USP36 deubiquitinates H2Bub1 in vitro. Histones purified from H1299 cells were incubated with wt His-USP361-800 or the His-USP361-800/C131A mutant purified from bacteria and assayed by IB.
3.2. USP36 interacts with H2B
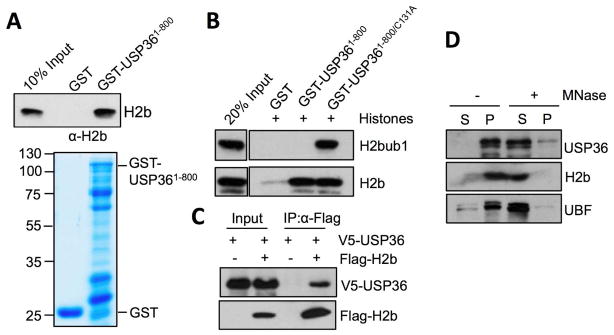
To examine how USP36 deubiquitinates H2Bub1, we tested whether it interacts with H2B using in vitro GST pull-down assays. Purified histones were incubated with bacterially purified GST-USP361-800 or GST only as a control, followed by IB analysis. As shown in Fig. 2A, H2B bound GST-USP361-800, but not GST alone. Interestingly, we only detected the pulldown of H2Bub1 by the GST-USP361-800/C131A mutant, but not the wt GST-USP361-800 (Fig. 2B). This result further supports that USP36 deubiquitinates H2Bub1 (Fig. 1C), as in this setting wt USP36 is capable of removing the modification. We further showed that V5-USP36 was co-immunoprecipitated with Flag-H2B only when both proteins were co-expressed in cells using co-IP assays (Fig. 2C). To test whether endogenous USP36 associated with chromatin, nuclei isolated from H1299 cells were treated with or without MNase, which cleaves DNA surrounding nucleosomes, resulting in solubilization of chromatin bound factors. In the absence of MNase, the majority of endogenous USP36 was found in the insoluble nuclear pellet, whereas in nuclei treated with MNase, the majority of USP36 was solubilized, suggesting that a significant fraction of USP36 is associated with chromatin (Fig. 2D). Together, we conclude that USP36 interacts with H2B in cells and in vitro and that this likely occurs within the context of chromatin.
Figure 2. USP36 interacts with H2B.
(A). USP36 binds to H2B in vitro. Purified histones were incubated with GST-USP361-800 or GST only, followed by IB with anti-H2B antibody. (B) USP36 binds to H2Bub1 in vitro. Purified histones were incubated with GST-USP361-800, GST-USP361-800/C131A or GST only, followed by IB. (C) USP36 interacts with H2B in cells. H1299 cells transfected with V5-USP36 alone or together with Flag-H2B were subjected to co-IP with anti-Flag followed by IB. (D). USP36 associates with chromatin. Purified histones were treated without or with Mnase and assyed by IB. The soluble (S) and insoluble (P) fractions are indicated.
3.3. USP36 deubiquitinates H2Bub1 within the p21 gene body
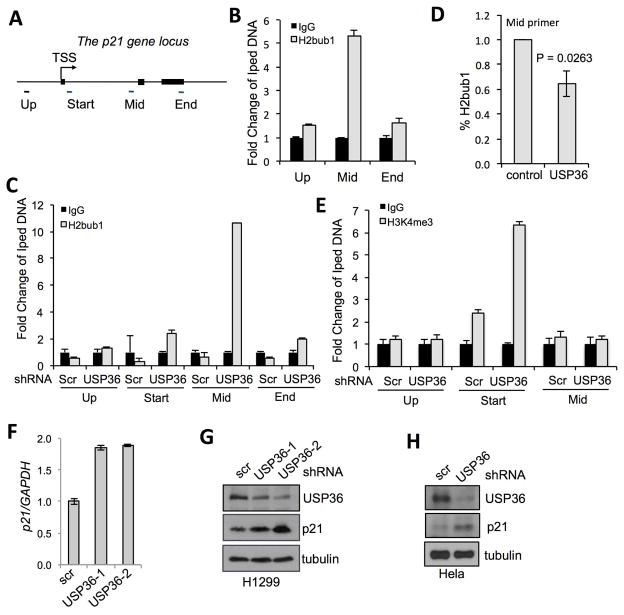
In order to test the function of USP36 deubiquitination of H2Bub1 in regulating gene expression, we focused on the CDKN1A (gene coding for the cell cycle inhibitor p21) locus as it has been widely used as a model to study the function of H2Bub1 [5, 34, 35]. H2Bub1 is often enriched within the gene bodies of actively transcribed genes, such as the p21 gene [5, 34, 35]. Thus, we hypothesized that USP36 may deubiquitinate H2Bub1 at the p21 gene locus and regulate its expression. To this end, we carried out chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR) assays using primer sets amplifying the upstream, middle or end region of the p21 gene (Fig. 3A). Consistent with published results [5], we observed that H2Bub1 is enriched in the p21 gene body (Fig. 3B). We then examined whether knockdown of USP36 regulates H2Bub1 levels at this locus. H1299 cells were infected with lentiviruses encoding shRNA against USP36, followed by ChIP using anti-H2Bub1 antibody. As shown in Fig. 3C, knocking down USP36 substantially increased H2Bub1 levels within the middle region of p21 with only modest effects on region just downstream of the transcriptional start site and the far 3′ end of the p21 gene. Consistently, overexpression of USP36 modestly, but significantly (P<0.05), reduced the levels of H2Bub1 in the middle region of the p21 gene (Fig. 3D). As it has been shown that H2Bub1 is associated with the active histone mark H3K4me3, particularly enriched at promoters and the 5′ end of genes [10, 34], we next assayed the H3K4me3 modification in p21 gene locus. Our results showed an increase in H3K4me3 at the region just adjacent to the start site after USP36 knockdown (Fig. 3E). We also detected slightly decreased global levels of H3K4me3 upon USP36 overexpression (Fig. 1B). In contrast, H3K36 methylation, which is not thought to be involved in histone cross-talk, remained unchanged (Fig. 1B). These results suggest that increased H2Bub1 at the p21 gene body is associated with the increased levels of H3K4me3 near the transcription start site upon knockdown of USP36.
Figure 3. USP36 deubiquitinates H2Bub1 within the p21 gene.
(A). A schematic diagram of the p21 gene locus. Filled boxes indicate exons. Primers for amplifying different regions relative to transcriptional start site were indicated. (B). H2Bub1 levels are enriched within the p21 gene body. 293 cells were subjected to ChIP using control IgG or anti-H2Bub1, followed by qPCR detection using indicated primers. (C). Knockdown of USP36 increases H2Bub1 in the p21 gene body. H1299 cells infected with scrambled (scr) or USP36 shRNA were assayed by ChIP using anti-H2Bub1 or IgG, followed by qPCR. (D). Overexpression of USP36 reduces the levels of H2Bub1 in the p21 gene body. H1299 cells transfected with control or Flag-USP36 followed by ChIP using anti-H2Bub1 and qPCR detection of the p21 gene body. (E). Knockdown of USP36 increases H3K4me3 near the transcription start sites of the p21 gene. H1299 cells infected with scr or USP36 shRNA were assayed by ChIP using anti-H3K4me3 or control IgG, followed by qPCR. (F) (G). Knockdown of USP36 increases p21 expression in cells. H1299 cells were infected with scr or USP36 shRNA-encoding lentiviruses, followed by detection of p21 mRNA by qPCR (F) and p21 protein by IB (G). (H) HeLa cells infected with scr or USP36 shRNA were assayed by IB.
3.4. USP36 regulates the expression of p21
We next examined whether knockdown of USP36 affects p21 levels. RT-PCR analysis indeed showed that knockdown of USP36 by two individual shRNAs increased the levels of p21 mRNA (Fig. 3F) and protein (Fig. 3G) in H1299 cells. These effects were not cell-type specific, as similar results were also observed in other tested cell lines including HeLa cells (Fig. 3H). Together, these data suggest that USP36 negatively regulates p21 expression through deubiquitination of H2Bub1 at the gene body.
3.5. Knockdown of USP36 suppresses cell proliferation
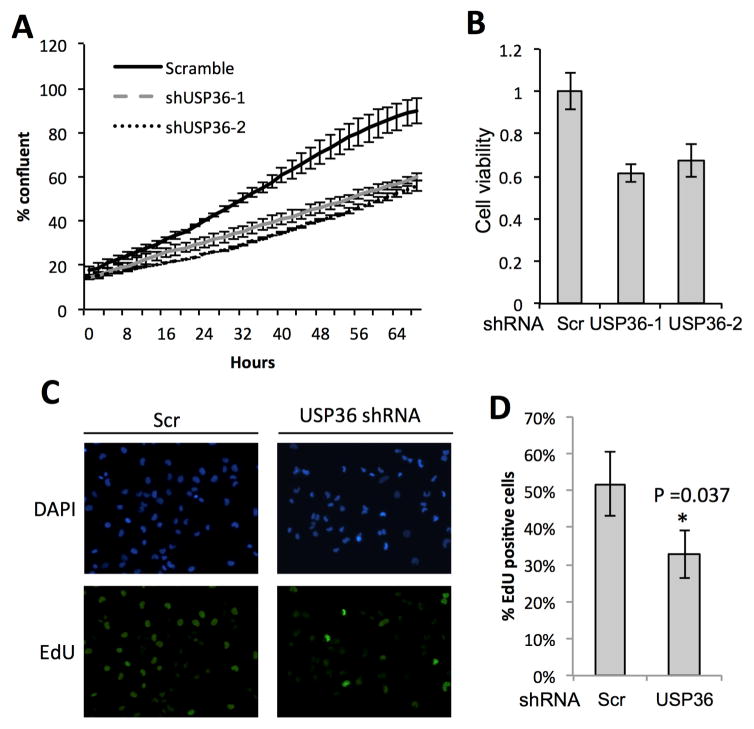
Previous studies have shown that H2Bub1 levels are negatively correlated with cancer progression [12–14]. Consistently, RNF20 has tumor suppressive properties and was silenced through hypermethylation in breast cancers [13]. In contrast, USP22, an H2Bub1 deubiquitinase, supports oncogenesis and was shown to be a negative prognostic factor in a “death by cancer gene signature” [36]. To analyze whether USP36 suppression of p21 expression regulates cell proliferation, we performed USP36 knockdown, followed by cell proliferation assays using the Incucyte system to monitor for cell confluence over time. As shown in Fig. 4A, knockdown of USP36 by two individual shRNAs reduced the confluence of H1299 cells by more than 50% at the end of the assay. Using MTS assays to compare the viability of scrambled to USP36 shRNA transfected cells, we also found that knockdown of USP36 significantly decreases the viability in H1299 cells (Fig. 4B). Cell proliferation assays using EdU labeling further showed that knockdown of USP36 significantly suppressed proliferation of H1299 cells (Fig. 4C and 4D), consistent with the increased expression of p21.
Figure 4. Depletion of USP36 decreases cell proliferation.
(A) H1299 cells infected with scr or USP36 shRNA lentiviruses for 48 hours were replated and assayed for cell confluence using the IncuCyte system. (B) H1299 cells infected with scr or USP36 shRNA were assayed for cell viability using MTS assay. (C) (D). EdU incorporation assays. H1299 cells were infected with scr or USP36 shRNA. At 72 hour post-infection, the cells were pulse-labeled with EdU for 4 hours followed by EdU staining. Representative images (C) and the quantification (D) were shown.
4. Discussion
DUBs regulate virtually all aspects of ubiquitin biology with effects ranging from altered protein stability to changes in protein function and modified gene expression [37]. One key DUB substrate is H2Bub1, whose deubiquitination plays a critical role in modulating chromatin dynamics and gene expression [7, 38]. In this study, we identified USP36 as a novel H2Bub1 deubiquitinase. USP36 interacts with H2B and deubiquitinates H2Bub1 in cells and in vitro. The interaction of USP36 with H2B likely occurs within the context of chromatin as we found that a significant pool of USP36 is enriched on chromatin. We showed that knockdown of USP36 results in a reduction of H2Bub1 in the p21 gene body and increased the levels of p21 mRNA and protein, suggesting a repressive role for USP36 at the p21 locus.
Functionally, we show that knockdown of USP36 significantly inhibited cell proliferation and these effects are correlated with the induction of p21 expression. USP36 is a nucleolar DUB and has recently been shown to play a critical role in ribosomal biogenesis by deubiquitinating and stabilizing several nucleolar proteins critical for ribosome biogenesis, including nucleophosmin, fibrillarin [31] and RNA polymerase I subunit RPA194 [31, 39]. We found that USP36 also deubiquitinates and stabilizes c-Myc, a key oncogene required for cell proliferation and a master regulator of ribosomal biogenesis [29]. It is interesting to test whether USP36 also regulates rRNA synthesis by deubiquitinating H2Bub1 at the rDNA loci in the nucleolus. Together with its role in silencing the p21 gene, USP36 is clearly a key regulator of cell growth and proliferation and may have oncogenic activity. Consistent with this notion, we have shown that USP36 is overexpressed in multiple types of human cancers [29].
As H2Bub1 is regulated by other DUBs, determining the relationship between USP36 and other DUBs regulating H2Bub1, particularly USP22 as it is predicted to have oncogenic function, will be interesting to flesh out. It is possible that different DUBs regulate distinct target genes or regulate H2Bub1 based on its position within a gene, as it has been shown in yeast where ubiquitination at promoters inhibits recruitment of the basal transcription machinery but promotes elongation within the gene body [19]. Regulation of H2Bub1 by multiple DUBs at overlapping targets could be a mechanism for fine-tuning expression at a particular locus. For example, USP22 was shown to promote transcription of p53 target genes, in particular p21 [22]. In this context, the role of USP22 and USP36 are seemingly opposed. Future studies should not only clarify this point but should also aim to integrate the various DUBs regulating H2Bub1 into a larger framework. Global analysis of USP36’s impact on H2Bub1 and gene expression will therefore be critical.
In summary, we have found that USP36 is a novel H2B DUB. Given that USP36’s overexpression has significant impact on global H2Bub1 levels and there is a negative correlation between global levels of H2Bub1 and cancer progression [12–14], our study uncovers yet another oncogenic function for USP36 and justifies its consideration as a therapeutic target.
Supplementary Material
Highlights.
USP36 is a novel H2B deubiquitinase
Knockdown of USP36 increases H2BUb1 at the p21 gene body
Knockdown of USP36 induces p21 expression and inhibits cell proliferation
Acknowledgments
We thank Dr. Masayuki Komada for providing anti-USP36 antibody. We thank other members of the Dai and Sears’ laboratory for active discussion. This work was supported by NIH/NCI grants R01 CA160474 to M-S. D. and R01 CA186241 to M-S. D. and R.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 3.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 4.Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 6.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 7.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh FK, Kulaeva OI, Patel SS, Dyer PN, Luger K, Reinberg D, Studitsky VM. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc Natl Acad Sci U S A. 2013;110:7654–7659. doi: 10.1073/pnas.1222198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Kim JA, McGinty RK, Nguyen UT, Muir TW, Allis CD, Roeder RG. The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol Cell. 2013;49:1121–1133. doi: 10.1016/j.molcel.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnsen SA. The enigmatic role of H2Bub1 in cancer. FEBS Lett. 2012;586:1592–1601. doi: 10.1016/j.febslet.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Prenzel T, Begus-Nahrmann Y, Kramer F, Hennion M, Hsu C, Gorsler T, Hintermair C, Eick D, Kremmer E, Simons M, Beissbarth T, Johnsen SA. Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res. 2011;71:5739–5753. doi: 10.1158/0008-5472.CAN-11-1896. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Zhu L, Guo T, Wang Y, Yang J. Decreased H2B monoubiquitination and overexpression of ubiquitin-specific protease enzyme 22 in malignant colon carcinoma. Hum Pathol. 2015;46:1006–1014. doi: 10.1016/j.humpath.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, Vojtesek B, Berger SL, Oren M. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol. 2005;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, Devys D. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 2014;28:1999–2012. doi: 10.1101/gad.250225.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 2011;25:2242–2247. doi: 10.1101/gad.177220.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buszczak M, Paterno S, Spradling AC. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science. 2009;323:248–251. doi: 10.1126/science.1165678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 2008;27:394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 24.Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, Golomb L, Pribluda A, Zhang F, Haj-Yahya M, Feldmesser E, Brik A, Yu X, Hanna J, Aberdam D, Domany E, Oren M. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell. 2012;46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hock AK, Vigneron AM, Vousden KH. Ubiquitin-specific peptidase 42 (USP42) functions to deubiquitylate histones and regulate transcriptional activity. J Biol Chem. 2014;289:34862–34870. doi: 10.1074/jbc.M114.589267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long L, Thelen JP, Furgason M, Haj-Yahya M, Brik A, Cheng D, Peng J, Yao T. The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination. J Biol Chem. 2014;289:8916–8930. doi: 10.1074/jbc.M114.551754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, Citterio E. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Jones A, Joo HY, Zhou D, Cao Y, Chen S, Erdjument-Bromage H, Renfrow M, He H, Tempst P, Townes TM, Giles KE, Ma L, Wang H. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev. 2013;27:1581–1595. doi: 10.1101/gad.211037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun XX, He X, Yin L, Komada M, Sears RC, Dai MS. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci U S A. 2015;112:3734–3739. doi: 10.1073/pnas.1411713112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun XX, Challagundla KB, Dai MS. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2012;31:576–592. doi: 10.1038/emboj.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endo A, Matsumoto M, Inada T, Yamamoto A, Nakayama KI, Kitamura N, Komada M. Nucleolar structure and function are regulated by the deubiquitylating enzyme USP36. Journal of cell science. 2009;122:678–686. doi: 10.1242/jcs.044461. [DOI] [PubMed] [Google Scholar]
- 32.Dai MS, Arnold H, Sun XX, Sears R, Lu H. Inhibition of c-Myc activity by ribosomal protein L11. EMBO J. 2007;26:3332–3345. doi: 10.1038/sj.emboj.7601776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Endo A, Kitamura N, Komada M. Nucleophosmin/B23 regulates ubiquitin dynamics in nucleoli by recruiting deubiquitylating enzyme USP36. J Biol Chem. 2009;284:27918–27923. doi: 10.1074/jbc.M109.037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang F, Yu X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell. 2011;41:384–397. doi: 10.1016/j.molcel.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glinsky GV. Death-from-cancer signatures and stem cell contribution to metastatic cancer. Cell Cycle. 2005;4:1171–1175. doi: 10.4161/cc.4.9.2001. [DOI] [PubMed] [Google Scholar]
- 37.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 39.Richardson LA, Reed BJ, Charette JM, Freed EF, Fredrickson EK, Locke MN, Baserga SJ, Gardner RG. A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell reports. 2012;2:372–385. doi: 10.1016/j.celrep.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.