Extended Data Figure 9. Glia-specific knockdown of srl has little to no effect on EGFR-PI3K-induced tumor growth but glia-specific knockdown ERR inhibits F3-T3-induced tumor growth.
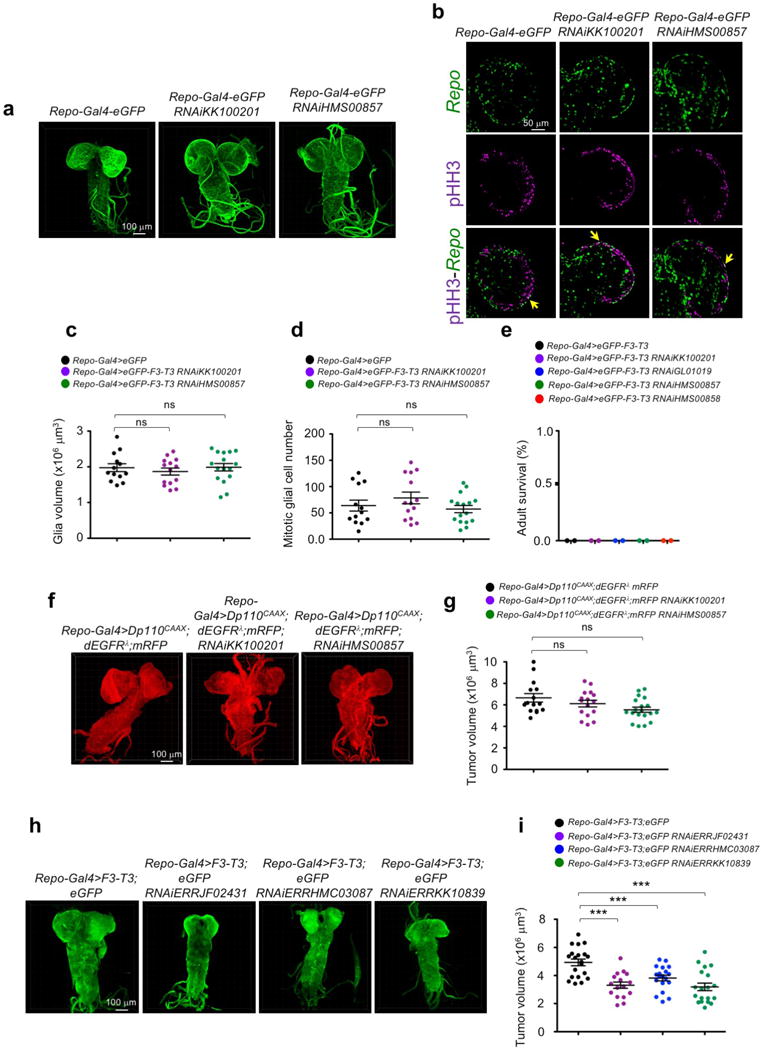
a, Optical projections of whole brain-ventral nerve cord complexes from larvae with control and srl-deficient glia. b, Glia-specific srl knockdown in larval brains did not significantly impact the overall glial population (Repo+ cells) nor the mitotic index of glial cells (Repo+; pHH3+ cells, yellow arrows). c, Quantification of glia volume in larval brains with control and srl-deficient glia; n=13 for repo-Gal4>eGFP; n=14 for repo-Gal4>eGFP;RNAiKK100201; and n=16, for repo-Gal4>eGFP;RNAiHMS00857. Data are Mean±s.d. P: ns, two-tailed t-test, unequal variance. d, Quantification of proliferating glia number (Repo+; pHH3+ cells) in larval brains with control and srl-deficient glia; n=13 for repo-Gal4>eGFP; n=14 for repo-Gal4>eGFP;RNAiKK100201; and n=16 for repo-Gal4>eGFP;RNAiHMS00857. Data are Mean±s.d. P: ns, two-tailed t-test, unequal variance. e, Adult lethality in repo-Gal4>F3-T3 and repo-Gal4>F3-T3;RNAi-srl larvae (n=100). f, Optical projections of control and srl-deficient brain tumors from repo-Gal4>Dp110CAAX;dEGFRλ;mRFP larvae. g, Quantification of tumor volume in control and srl-deficient tumors; n=15 for repo-Gal4>Dp110CAAX;dEGFRλ;mRFP; n=16 for repo-Gal4>Dp110CAAX;dEGFRλ;mRFP;RNAiKK100201; n=19 for repo-Gal4>Dp110CAAX;dEGFRλ;mRFP;RNAiHMS00857. Data are Mean±s.d. P: ns, two-tailed t-test, unequal variance. h, Optical projections of brain tumors from Drosophila larvae repo-Gal4>F3-T3 and repo-Gal4>F3-T3;RNAi-ERR. RNAi-mediated knockdown of ERR reduces the volume of F3-T3-induced glial tumors. i, Quantification of tumor volume in the control and ERR-deficient tumors; n=20 for repo-Gal4>F3-T3; n=16 for repo-Gal4>F3-T3;RNAiJF02431; n=19, for repo-Gal4>F3-T3;RNAiHMC03087; n=19, for repo-Gal4>F3-T3;RNAiKK10839). P: ***<0.001, two-tailed t-test, unequal variance. In all experiments n are biologically independent animals. Experiments in a, b, f, h were repeated twice with similar results.
