Abstract
The late Don R. Swanson was well appreciated during his lifetime as Dean of the Graduate Library School at University of Chicago, as winner of the American Society for Information Science Award of Merit for 2000, and as author of many seminal articles. In this informal essay, I will give my personal perspective on Don’s contributions to science, and outline some current and future directions in literature-based discovery that are rooted in concepts that he developed.
1. Introduction
Don Swanson (1924 – 2012) was well appreciated during his lifetime as Dean of the Graduate Library School at University of Chicago, as winner of the American Society for Information Science Award of Merit for 2000, and as author of many seminal articles (Figure 1). Don became Emeritus in 1996, but did not truly retire until around 2007, when he suffered a series of strokes. Around ten years ago, Tanja Bekhuis (2006) wrote a review article that discussed Don’s contributions and their subsequent influence on bioinformatics and text mining. Recently, Sebastian et al (2017a) have published a comprehensive review from a technical standpoint, and the reader is urged to consult this article for an overview of existing and emerging methods that are being applied to the field of literature-based discovery. Here I give a more personal perspective. In particular, I will include a discussion of problems and issues which were inherent in Don’s thoughts during his life, but which have not yet been fully taken up and studied systematically.
Figure 1.

Don R. Swanson.
The first thing to realize about Don is that Don is not short for Donald. Don was his legal first name. Don’t make that mistake, please – it irritated him no end!
The second thing to realize is that my relationship with Don was idyllically intellectual in nature. I call my collaboration with Don my “Garage Band” period – the term referring to buddies who spend their free time playing rock music in their garages, playing out of sheer enjoyment, and oblivious of the outer world at large. We were unconcerned whether our research would be seen as Important by others, whether it would be published in high impact journals, whether we would secure grant funding, or other non-scientific concerns that too often drive research efforts.
2. Undiscovered Public Knowledge
Perhaps the most influential and enduring contribution that Don has had on information science is the concept of “undiscovered public knowledge” (UPK), which he approached from a very broad, philosophical standpoint (Swanson, 1986a). The philosopher of science Karl Popper had envisioned that man exists in three worlds – World I is the objective, real world which scientists seek to learn about; World II includes the thoughts and mental activities of scientists; and World III consists of the products of scientists, in particular, the published articles that express findings, models, assertions, and so forth (Popper, 1978). Just as man cannot hope to have perfect knowledge of reality (World I), so Don realized that man cannot have perfect knowledge of World III either. Knowledge can be public (e.g., it is published) and at the same time, inaccessible or imperfectly known for one reason or another.
Undiscovered public knowledge encompasses several distinct scenarios:
For example, one may ask: How many articles are published that no one reads – no one at all besides the author and (we hope) the reviewers? Information contained in such articles is, indeed, public yet undiscovered.
How much information is contained in articles that few can find, because the article is poorly indexed by Web of Science or by online search engines? Such articles may have been published without a digital presence, or placed in a journal that has limited circulation or low visibility.
A related type of information loss occurs when someone publishes an important article in an obscure or topically inappropriate journal, so that no one will take the finding seriously even if they see it. Few people have the self-confidence to recognize a breakthrough when it comes without the imprimatur of acceptance by a prestigious journal. An example of this happened quite recently: “This German Retiree Solved One Of World’s Most Complex Maths Problems - And No One Noticed” (Wolchover, 2017). Thomas Royen wrote a paper proving the Gaussian correlation inequality (GCI) and posted a preprint in the arXiv repository; when his work failed to get recognized, he chose to get his proof out in an obscure journal called the Far East Journal of Theoretical Statistics. He might as well have put it in a bottle and thrown it in the ocean!
Some of my own informatics discoveries have been closely related to undiscovered public knowledge. For example, my group discovered that many mammalian microRNAs are derived from genomic repeat elements in the genome (Smalheiser and Torvik, 2005). Although we came to this realization through computational studies (Smalheiser and Torvik, 2004), in fact, in retrospect, the discovery could have been made simply by inspection of the public data available at the UCSC Genome Browser (https://genome.ucsc.edu/). This website brings together dozens of different types of genomic data that are calculated or measured, for example, predicted transcription factor binding regions, cross-species conservation levels, and so on. Each type of data is superimposed on the reference genome, and users can open up and visualize any number of the datasets to observe them in juxtaposition with each other. Two of the data tracks show a) positions of known microRNA genes in red and b) Repeatmasker output, which identifies genomic repeat elements in two shades of grey (Figure 2). If anyone had opened up these two tracks and looked carefully, they would have seen that many of the microRNAs were within the sequences encompassed entirely by specific genomic repeat elements. The fact that no one DID do this indicates that this knowledge was public, yet undiscovered.
Figure 2. Screenshot of UCSC Genome Browser showing the sequence for human mir-95 juxtaposed to tracks for genomic repeats.
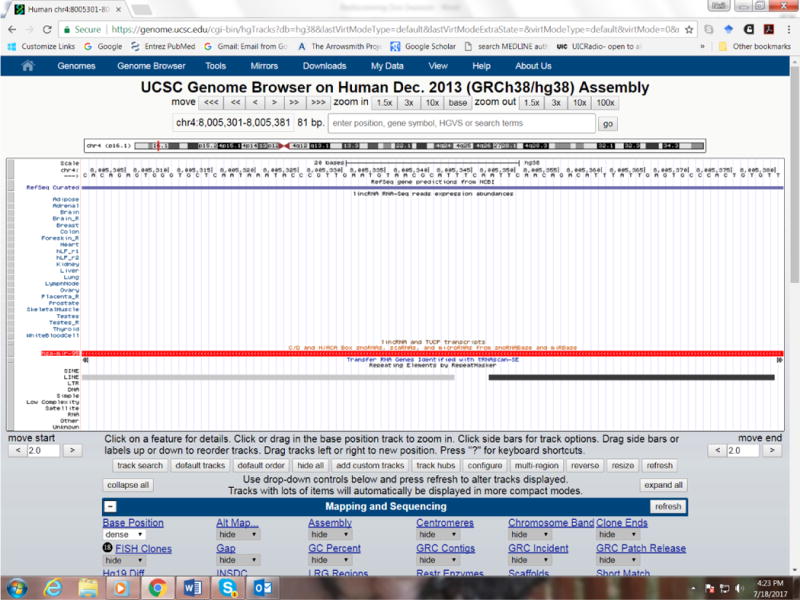
The genomic region of the mir-95 sequence corresponds to two LINE2 elements in opposite orientations. This provides evidence that, when transcribed into RNA, these LINE2 elements bind each other, creating the hairpin secondary structure that permits the processing of this sequence by enzymes (Drosha and Dicer) to form a microRNA (Smalheiser and Torvik, 2005).
3. Two medical literatures that are logically but not bibliographically connected
The most novel and fruitful type of undiscovered public knowledge discussed by Don occurs when information is not explicitly discussed in any single article at all. Rather, different assertions and findings need to be assembled across documents to create a new coherent assertion, much as different pieces of a puzzle are assembled to create a single picture.
But how to find these pieces residing in scattered places across the literature, and how to assemble them?
Don focused his analyses on first identifying two sets of articles, or literatures, which appear to be complementary (see below) yet are not directly connected to each other. Such literatures are unconnected if they do not have any articles in common, do not have authors in common, and articles in one literature do not cite any articles in the other literature (Swanson, 1987).
In a series of articles in the 1980s, Don analyzed two classic examples of medical literatures that were not (or only slightly) connected, yet contained multiple links of the form “A affects B” in one literature and “B affects C” in the other, such that when they were brought together and assembled, created a persuasive, novel hypothesis. These have become widely analyzed benchmarks for nearly all subsequent studies of literature based discovery.
The first case was the set of articles on Raynaud disease vs. the set of articles on fish oil (Swanson, 1986b). Don noticed that several of the pathological alterations that occur in Raynaud disease corresponded to physiological alternations that are produced by ingesting fish oil, only in opposite directions. That suggests that ingesting fish oil should counteract some of the signs and symptoms of Raynaud disease. Subsequent clinical studies supported this hypothesis (Swanson, 1993).
The second case was the set of articles on dietary Magnesium vs. on migraine headaches (Swanson, 1988). Again, Don noticed that magnesium deprivation has multiple effects in the body that are similar to alterations that are known to worsen migraine headaches, and magnesium itself has effects which should be expected to prevent or treat migraines. For example, magnesium is a calcium channel blocker, and reduces neuronal excitability via opening of NMDA glutamate receptors. Thus, he proposed that supplementation with dietary magnesium may prevent or alleviate migraines. Again, subsequent clinical studies supported this hypothesis (Swanson, 1993).
Don made further analyses of complementary un-connected literatures, both by himself (Swanson, 1990) and in collaboration with me (e.g., Swanson et al, 2001; Smalheiser & Swanson, 1994, 1996a, b, 1998). It is noteworthy that late in his career, Don proposed a link between atrial fibrillation and running (Swanson, 2006). Exercise is known to be a risk factor for atrial fibrillation, and he proposed that this may be mediated by gastroesophageal reflux, which in turn may be alleviated by taking proton pump inhibitors. Besides being another masterful, insightful example of putting together separate pieces of evidence to form a new whole, it is worth mentioning that these analyses were all based on conditions he experienced, himself. He had Raynaud syndrome, and he had migraine headaches. And, his chronic atrial fibrillation eventually caused his strokes and led to his withdrawal from active life.
4. Use of implicit information to bridge disparate literatures
It is important to acknowledge a tension between two different meanings of the term “knowledge discovery”. One meaning, the one I started with, is to assemble pieces of information into new wholes, that represent new/promising/surprising/research directions or provide potentially transformative or breakthrough insights. The other meaning is to analyze and synthesize existing data to impute new but otherwise predictable, everyday information. An example of this is using first names to predict the gender of individuals. Most of the “Jane” and “Linda” individuals will be female, and most of the “Boris” and “John” individuals will be males. But regardless of which type of discovery we are talking about, to my knowledge, all systematic algorithmic methods for knowledge discovery involve linking different literatures or entities via implicit features that they share. In the case of gender prediction, US Census data can be used to associate first names of individuals in the United States with their reported genders; by aggregating the results over all individuals, each first name is associated with a gender balance score (% females/% males). This becomes reference information that is used to impute gender for a given name instance in some other database. The reference information is implicit because it derives from information that is not explicitly present within the database.
Commonly, implicit information is used as a bridge to measure the similarity of two entities. For example, two diseases A and B may be related in terms of how many Medical Subject Headings they share (in articles that describe disease A and disease B, respectively). Or, they may be related in terms of how many single-nucleotide polymorphisms (SNPs) have been shown to affect disease risk in both disease A and B. Or, they may be related in terms of how many clinical signs and symptoms they share. Or, how many single-gene mutations which affect disease A or B affect genes that lie in the same biochemical pathway. There are many possible types of implicit information that connects disease A with disease B, and it is even possible to combine multiple types of information to create a heterogeneous graph in which diseases are nodes and implicit shared items form links between the nodes (Shi et al, 2017).
The use of implicit information is a powerful general technique of knowledge discovery, which has spawned several entire fields in bioinformatics and genomics (Bekhuis, 2006; Zweigenbaum et al, 2007). Don is the father of the field of drug repurposing, which proposes new uses for existing approved drugs (e.g., Weeber et al, 2003; Yang et al, 2017). Prediction of adverse drug effects follows a similar type of logic (e.g., Shang et al, 2014; Hristovski et al, 2016), as does detection of co-morbidities and other relations among drugs, diseases and genes (Frijters et al, 2010; Ding et al, 2013; Vos et al, 2014). Almost all approaches to genomic discovery involve implicit information as well. Furthermore, implicit information is a central concept generally in text mining and natural language processing.
5. The one node search
In Don’s original A-B-C model, implicit information was used in what is known as the “one node search” approach (Figure 3):
Begin with a set of articles that discusses or presents information regarding a problem, e.g., prostate cancer or poverty = literature C.
Look for another literature, unknown at the outset, which has information that can contribute to solving the problem = literature A.
Use words and phrases in the titles of articles in the two literatures = B-terms [use filtering to keep only “important” words in some sense]. The B-terms are the implicit information.
Carry out many searches to create B1, B2, B3…. Literatures.
Tabulate the title words and phrases in each Bi-literature = candidate A-terms and rank them according to how many B-literatures they are in.
Carry out a search using each Ai-term to define the Ai-literature.
An Ai-literature which shares many B-terms with the original C-literature is hypothesized to contain information that may help solve the problem.
Figure 3. Schematic diagram illustrating the One Node Search.
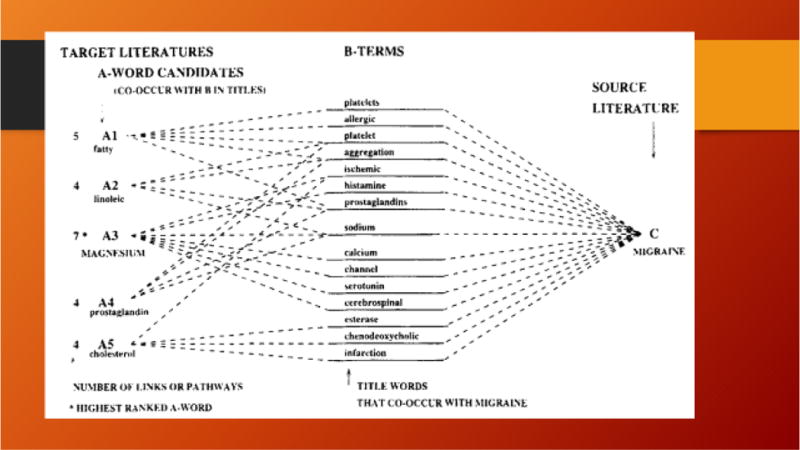
Reprinted from Swanson & Smalheiser (1997) with permission.
Despite its conceptual appeal, the one node search has several nuances and limitations in practice, and many variations of the ABC model have been explored (see reviews in Bruza & Weeber, 2008; Smalheiser, 2012b; Sebastian et al, 2017a):
For example, different words that have essentially the same meaning (lexical variants, synonyms, abbreviations, and alternative spellings) should ideally be counted and treated as a single B-term. Conversely, Preiss and Stevenson (2016) have demonstrated that word sense disambiguation, i.e., to separate different senses of the same word as used in different instances, can improve performance of discovery systems.
Titles don’t capture all information in an article. Words contained in the abstract and full-text will also contribute information, albeit these terms will also contribute significant noise (Cohen et al, 2010).
Words and phrases are not the only, or necessarily the best, type of information to employ for linking literatures. Many other investigators have used concepts, MeSH terms, entities and relations extracted from text (reviewed in Bruza & Weeber, 2008; Sebastian et al, 2017a).
Similarly, ranking Ai-literatures according to the number of Bi-terms in their titles is a relatively crude and nonrobust measure. The hope is the B-terms will point to the existence of causal mechanisms that link the literatures, but this is not necessarily the case. Other investigators have proposed ranking measures based on e.g., mutual information, relations, and/or network properties, including citations ((e.g., Wren, 2004; van der Eijk et al, 2004; Smalheiser, 2012b; Ding et al, 2013; Hristovski et al, 2015; Cameron et al, 2015).
The one node search involves multiple searches and calculations of title words and phrases, which introduces computational complexity. In practice, investigators generally restrict the number or type of B-terms to be used for linking, with either semantic or statistical criteria. Furthermore, rather than searching for all possible A-literatures that might exist, generally they are restricted to being in some predefined semantic category (such as drugs).
Presenting many Ai-literatures for the investigator, even when ranked, causes great cognitive complexity, since each candidate A-literature requires detailed manual examination to assess.
6. The Two Node Search
Perhaps the most important limitation of the one node search is not technical, but sociological: The one node search is intended to help investigators who are looking for a new hypothesis – yet most investigators are already drowning in a sea of existing potential hypotheses and findings, and their goal is not to find still more hypotheses, but rather to decide which of the existing ones is most promising to pursue. Thus, in my own work, I have emphasized the importance of the two node search strategy, which can be summarized as follows:
An investigator already has a hypothesis (or an experimental finding) that links A and C, but which has not been explicitly investigated directly in any single published article.
He or she carries out a two node search between the set of articles that discuss A and the set of articles that discuss C, and examines the shared title words and phrases Bi.
The goals are to rank the list of Bi-terms to hone in on the most relevant and promising links, and to examine possible mechanisms that link A to C.
To create a quantitative model that would allow us to rank Bi-terms, I assembled a team of neuroscientists, who used the two node search tool freely in the course of their scientific work. Vetle Torvik and I chose 8 of their searches as a gold standard, in which Bi-terms were manually marked as being relevant for linking A to C. Each Bi-term (marked as relevant or not relevant) was scored according to eight features (Table 1). These features are domain-independent insofar as they do not rely on any reported knowledge about entities, facts or relations; rather, they are based on statistical properties such as the frequency of the term within MEDLINE (Table 1; Torvik & Smalheiser, 2007). As a negative control training set, we chose random pairs of query literatures (having similar size and topics as the gold standard set), and scored all Bi-terms in the negative set. We created a logistic regression model, based on a weighted sum of these features, to predict the probability that a given Bi-term would be marked as relevant, i.e., that it would be deemed relevant by users for linking A and C in a meaningful manner (Torvik & Smalheiser, 2007).
Table 1.
Eight features used to characterize each B-term.
| 1. Does the B-term occur in more than one paper within literatures A and C? |
| 2. Do the AB and BC sub-literatures share any MeSH terms? |
| 3. Does the B-term map to at least one UMLS semantic category? |
| 4. Does the B-term have a high literature cohesion score? |
| 5. Is the B-term moderately frequent within MEDLINE as a whole? |
| 6. Did the B-term first appear recently within MEDLINE as a whole? |
| 7. Is the B-term highly characteristic within literature A or C? |
| 8. Do the words within the B-term all occur on the customized 1400 word stoplist? |
Reprinted from Torvik & Smalheiser (2007) with permission. See this reference for definitions and details regarding how the features were numerically scored.
The two node search interface at http://arrowsmith.psych.uic.edu makes it easy for investigators to carry out two node searches among PubMed articles.
The two node search also provides an aggregate measure of the implicit semantic similarity of any two literatures, based upon the body of Bi-terms, taken as a whole. Suppose we perform a two node search and find that there are 1263 terms on the B-list, of which 402 are predicted to be relevant (i.e., the estimated probability of relevance is >0.5). The ratio 402/1263 = 0.32 is called the pR score, and it provides an overall measure of the shared implicit information between the peanut butter and health literacy literatures. Randomly chosen pairs of literatures tend to have pR scores around 0.07, whereas literatures that are very closely related in terms of topics tend to have pR scores of 0.4-0.5. We have used the pR score as an important feature for literature-based discovery (Peng et al, 2017).
6.1. The one node search reconceptualized as a series of two node searches
Don’s original web-based one node search tool is no longer available. I have implemented a simpler version (at http://arrowsmith.psych.uic.edu/cgi-bin/arrowsmith_uic/one-node.cgi) in which the investigator starts with a literature that represents a problem to be solved (e.g., Huntington disease). Next, the user will be prompted to choose a category of Medical Subject Headings (MeSH) to search within, which encompass a set of literatures describing entities (or classes of entities) that represent possible approaches or solutions to the problem. (Alternatively, the user can choose the Free Format option, to enter any list of PubMed search queries manually, one on each line.) For example, to search among different classes of drugs according to their molecular mechanism using the MeSH Tree option, the user would drill down from Chemicals and Drugs to Chemical Actions and Uses to Pharmacologic Actions to finally, Molecular Mechanisms of Pharmacological Action [D27.505.519]. This category includes about twenty classes of drugs, including Alkylating Agents [D27.505.519.124], Angiotensin Receptor Antagonists [D27.505.519.162], Antacids [D27.505.519.170], Antifoaming Agents [D27.505.519.178], and so on. Once the user chooses this MeSH term category, the software will carry out a series of two-node searches, each consisting of A = Huntington disease vs. C = one of the drug classes. These two-node searches are characterized according to the total number of articles in A and C (and nAC, the intersection of A and C), as well as the total number of B-terms. Finally, the searches are ranked according to pR, the percentage of B-terms that are predicted to be relevant for meaningful linkage. The two-node search results are all individually stored temporarily by job ID so users can go back without needing to re-run the search each time. Thus, carrying out a one node search is simply a matter of carrying out a series of two node searches, one for each MeSH term within the category of interest (Smalheiser, 2012b). This greatly simplifies the computational issues involved.
7. Examples from the front lines of scientific investigation
A variety of investigators have used literature-based discovery methods to propose specific hypotheses which were then tested experimentally. Some of these studies introduced new LBD methodology (e.g., Wren et al, 2004), whereas others used the public Arrowsmith two node search interface. Dong et al (2016) investigated links between anandamide and gastric cancer. Maver et al (2013) identified novel treatments for neovascularization in diabetic retinopathy. Miller et al (2012) found mechanisms to link hypogonadism and diminished sleep quality in aging men. Cairelli et al (2013) proposed a possible explanation for the “obesity paradox” whereby obese patients have better outcomes in intensive care. Manev & Manev (2010) studied a 5-lipoxygenase-leptin-Alzheimer connection. Kell (2009) used LBD to assess abnormal iron chelation as a common pathogenetic factor in a variety of diseases.
In my own laboratory studies, separately from Don, I have also put together assertions and knowledge from disparate literatures to formulate hypotheses that I have tested and verified experimentally. Unlike the examples stated above, in which we or others deliberately searched for complementary literatures, the latter examples arose haphazardly during the course of laboratory investigations.
For example, we had discovered that an enzyme, dicer, which is known to cleave double-stranded RNA to form small RNAs, is expressed and even highly enriched at postsynaptic densities present at synaptic contacts in the central nervous system (Lugli et al, 2005). However, paradoxically, although the dicer protein was present, it appeared to lack enzymatic activity. On the other hand, we knew that treating purified dicer protein with certain proteases in a test tube will cause dicer to form fragments that show greatly enhanced catalytic acitivity. And, there was an extensive body of studies that had shown that a naturally-occurring protease called calpain is activated during synaptic stimulation and cleaves a variety of other proteins in a controlled manner. Putting the two lines of studies together, we predicted that during synaptic stimulation, calpain might cleave dicer such that the activated, cleaved form of dicer would exhibit enzymatic activity. This was confirmed in experiments carried out in mouse brain tissue (Lugli et al, 2005).
Another example of connecting two disparate literatures to create a novel testable hypothesis occurred when we proposed that a phenomenon called RNA interference, which had been studied in worms and other lower organisms, might be involved in mediating learning and memory in the mammalian brain (Smalheiser et al, 2001). It took us a decade to find provisional experimental evidence that this may, indeed, be the case (Smalheiser, 2012a, 2014).
Finally, a third example occurred when we noticed detailed similarities between a class of small vesicles (called secretory exosomes) – secreted by many cell types and reported to contain microRNAs and other types of RNAs – and the structures called synaptic spinules that form at synapses during periods of intense synaptic stimulation (Smalheiser, 2007). This led to the hypothesis that neurons may transfer RNAs and proteins across synapses in an activity-dependent manner (Smalheiser, 2007).
It should be acknowledged that none of these three examples involved computer-generated or automatic LBD algorithms, or even employed an explicit A-B-C model. Instead, both Don’s and my discoveries have largely been made by manual examination of complementary literatures and assembling of quite complex information into coherent wholes (Smalheiser, 2012b). Thus, it should be kept in mind that although most LBD research has focused on situations that are readily recognized by text mining and that follow standard templates (e.g., A affects B and B affects C), these situations represent only the “low hanging fruit”, and more sophisticated models of discourse and assertion will be needed to deal with the rest.
8. New Directions in Literature-based Discovery
8.1. Storytelling
One and two node A-B-C search strategies all consider a single intermediate link between two literatures. Perhaps the most straightforward extension of this idea is to construct and assess multi-step paths that exist between two sets of articles (e.g., Hossain et al, 2012; Sebastian et al, 2015; Baek et al, 2017). Multiple paths can also be constructed to connect entities, authors, and so on. This can be conceptualized variously as an exercise in storytelling, as navigating paths within graphs or networks, or as detecting functional mechanisms.
8.2. “Gaps” – linking two sub-fields that reside inside of a larger field of investigation
My own group has focused recently on linking sub-fields that reside within a larger field of investigation. For example, consider the field of prostate cancer research. Some articles study experimental tumors in mice; some follow people for effects of diet and smoking on risk; some study molecular changes inside tumor cells; some are medicinal chemistry studies, modifying drugs for better solubility or potency or fewer side effects. Not all people in the field of prostate cancer research read all these articles! More to the point, not all topics are explored in all combinations within the prostate cancer field.
If two topics appear at moderately high frequencies within the prostate cancer field and are totally independent of each other, one would expect that they should co-occur in some articles simply by chance. When two MeSH terms co-occur, they often indicate that there is some direct or implicit relationship between them. Specifically, if two topics (defined as MeSH terms) are expected to co-occur in at least 10 articles within a given field, but do not co-occur in any articles at all, we call the pair of topics a “gap”. As reported recently (Peng et al, 2017), gaps can arise for several different reasons. A few gaps reflect idiosyncracies in the rules given to MEDLINE indexers, such that certain closely related MeSH terms are rarely applied to the same article. Some gaps represent “low hanging fruit”, i.e., research directions that have not yet been investigated but are known to be promising and are likely to be followed up on in the near future. Other gaps may indicate the presence of undiscovered public knowledge – that is, investigators may be unaware of connections that exist among different sub-areas of a single field. We are continuing to investigate the phenomenon of gaps and attempting to use them as a means of discovering new, promising research directions.
8.3. Discovery via analogy
A popular and important approach in literature-based discovery (and text mining in general) is the semantic representation of words, concepts, relations or predications by vectors (Gordon & Dumais, 1998; Cole & Bruza, 2005; Widdows & Cohen, 2015), either high-dimensional vectors (Cohen & Widdows, 2009) or low-dimensional vectors (Mikolov et al, 2013; Pennington et al, 2014). One of the endearing features of semantic vector representations is that vectors that lie near each other exhibit similar meanings or similar relations. For example, the relation “King :: Queen” is implemented by subtracting the vector for King from the vector for Queen, resulting in a difference vector (King – Queen) that embodies the relation. Other vectors that encode similar relations, e.g. “Man :: Woman” also lie near this difference vector. In particular, one can pose the question “King :: Queen as Man :: X?” and solve for X by identifying the difference vector which includes Man and lies closest to (King – Queen). Trevor Cohen has extensively explored the use of an analogy model for literature based discovery based on vector proximity (e.g., Mower et al, 2016; Cohen & Widdows, 2009, 2017; Cohen et al, 2010).
8.4. Link prediction
Many discoveries involve combining new concepts or bridging disparate fields. One may hope to identify such publications by looking for newly published articles that contain novel combinations of text terms (Packalen & Bhattacharya, 2015), novel combinations of Medical Subject Headings (Mishra & Torvik, 2016; Peng et al, 2017), or whose reference lists cite novel combinations of journals (Uzzi et al, 2013). This leads to a model of literature-based discovery that is based on link prediction on networks. For example, Kastrin et al (2016) model LBD as considering all pairs of MeSH terms that have never co-occurred within a single article before, and seek to learn the factors that best predict the likelihood of an article appearing in the near future that is indexed by both of the MeSH terms. Sebastian et al (2017b) combined text and citation networks for link prediction.
8.5. Scientific arbitrage
Don often referred to literature-based discovery as an exercise in “scientific arbitrage”, in which certain ideas or findings are under-valued in one scientific arena, and gain in value by applying them to another field. (In fact, I believe he performed arbitrage in financial markets too!). In his final published article (Swanson, 2011), Don discussed the problem of identifying neglected, dead, or discarded findings and hypotheses as sources of new knowledge. Neglected findings, which are explicitly stated in one or more articles yet not well cited or followed up upon, may reflect a variety of issues: The articles in which they appeared may not be easy to find (particularly in full-text form), the findings themselves may have been refuted by later studies, or they may simply have been ahead of their times. The use of text mining to identify these neglected findings, and predict which (if any) ought to be resurrected and rehabilitated, remains an open question for further investigation.
A particular type of neglected finding is what I have called “negative consensus” (Smalheiser & Gomes, 2014), in which the investigators in a given field mention that a particular event or happenstance does NOT occur in nature. Sometimes this is documented by definitive experimental studies, in which case one would expect that negative assertions would cite the negative evidence. Often, however, the negative assertions simply reflect prevailing dogma or investigators’ expectations or “common sense”, and such cases do not cite any supporting evidence at all. My (somewhat contrarian) view is that negative consensus statements that lack experimental testing are in fact good subjects for further research A small input of experimental testing may challenge the prevailing paradigm or dogma that made the finding seem so unlikely. For example, we noted that the protein Argonaute binds DNA in the test tube, yet investigators have simply assumed that it binds RNA within living cells – in part, this is because Argonaute is thought to reside in the cytoplasm whereas cytoplasmic DNA is thought not to exist. However, Argonaute does have functions in the nucleus, and there are indeed reports that extrachromosomal DNA exists in both nucleus and cytoplasm. Hence, the idea that Argonaute may bind DNA is not absurd but is well worth investigating (Smalheiser and Gomes, 2014). I believe that it is worthwhile to develop text mining tools that can identify negative consensus statements and help investigators decide which are likely to be promising to study. Agrawal et al (2011) have compiled a database of biomedical negated sentences, which might be mined to identify those assertions that are reliably negative across multiple documents.
8.6. The penumbra of a field as a source of new knowledge
A scientist working in a field (say, Alzheimer disease) is acutely aware that some lines of investigation are “mainstream” and reside in the core of the field, whereas other lines of work are marginal, either because they are new, or not considered interesting or credible, or because they are pursued by people who are not themselves recognized full-time Alzheimer researchers. For example, studies of amyloid or tau protein aggregates are intensively studied and are published in high-impact journals as well as in journals devoted to aging and Alzheimer disease. In contrast, studies of gut microbes (the so-called microbiota) are not a mainstream topic in Alzheimer disease, at least not yet. Standard techniques such as text mining, summarization, and clustering, together with citation analysis, can help to identify which articles, topics, keywords, and concepts reside in the core of a given field and which reside in the periphery, or penumbra.
Initially, literature-based discovery techniques sought to make linkages across literatures, without asking whether the links predominantly involve the cores or the peripheries of the literatures. Don’s first inclination was to filter out B-terms that did not have adequate frequency of mentions in each literature, implying that he was focusing on the cores (Swanson & Smalheiser, 1997). In contrast, Kostoff et al (2009), Petrič et al (2010), and Workman et al (2016) have argued that low-frequency terms which reside in the penumbra of one or both fields may sometimes be more promising for finding links that are interesting and unexpected.
8.7 Evidence synthesis and reproducibility in science
In the early days of literature-based discovery, when assembling ideas, assertions and published findings, we did not worry much about the reliability of each reported item, or how many articles obtained similar results. If a paper reported that protein A binds protein B in adult female rat lung, the extracted assertion would be “protein A binds protein B” without worrying much about its scope or generalizability to other situations. The goal has been to identify interesting and promising hypotheses, which after all need to be experimentally confirmed on their own terms.
Over the past ten years, however, it has become clear that a significant minority (if not the majority) of published findings are hard to replicate and have low reliability, due to a combination of flaws in experimental design, small sample sizes, naïve data analysis practices, and over-interpretation of statistical testing (e.g., Rzhetsky et al, 2006; Ioannadis, 2005; Smalheiser, 2017). Thus, going forward, it will be important not merely to identify terms and concepts for linking, but to assess the reliability of the articles that contain them and to filter or rank them accordingly. Kilicoglu (2017) has recently proposed that text mining may aid in at least four ways, namely, plagiarism/fraud detection, ensuring adherence to reporting guidelines, managing information overload and accurate citation/enhanced bibliometrics.
Even more broadly, literature-based discovery is moving closer to the field of evidence synthesis, which collects reported findings across multiple studies (e.g., the set of all clinical trials that have employed nonsteroidal anti-inflammatory agents for chronic arthritic knee pain) and attempts to reach a consensus, if possible. This field employs techniques such as systematic review, meta-analysis, and summarization. Although most of this work is currently done manually, there is a recent push for the use of automated text mining tools to accelerate the process (Jonnalagadda et al, 2015; O’Mara-Eves et al, 2015). In fact, text mining-based detection of reliable trends in the literature, i.e. detecting when “signal” is truly above “noise”, is itself a type of literature-based discovery, albeit explicit (rather than implicit) assertions are being mined.
9. Discussion and Conclusions
The recent advent of Big Data has provided massive, openly available datasets that provide rich fodder for literature based discovery, as well as serving as training sets for machine learning approaches to discovery. Furthermore, major Big Data techniques include linking datasets together and combining heterogeneous datasets (including electronic medical records and data warehouses), both of which are increasingly tractable with current computational resources, and both of which are fundamental to obtaining implicit information used for discovery. The new directions discussed in this review (e.g., outliers, analogies, negative consensus, and others) go beyond the A-B-C model and open up the field to an exciting variety of models of discovery.
Historically, the big stumbling-block of literature-based discovery has been the fact that its models seek to predict novel, untested, even surprising findings, which inherently are difficult to score as “right” or “wrong” without costly experimentation. This has bedeviled methodological studies that seek to improve predictive performance. Existing benchmarks are relatively few (Sebastian et al, 2017a). Time-slicing is an alternative technique in which articles up to a certain date are used to construct a hypothesis, and then the literature is examined a few years later to determine whether that hypothesis is tested or at least mentioned in the literature (Yetisgen-Yildiz et al, 2009). Some of the new research directions that I have discussed in this article are easier to evaluate than the classic one or two node searches. For example, link prediction seeks to predict which pairs (of, say, MeSH terms) are most likely to appear together in the same article in the future, which can be assessed quantitatively without considering the “truth” of the article. It is gratifying that the techniques of literature-based discovery have been absorbed into the mainstream of bioinformatics, medical informatics, and computer science, whose practitioners find abundant value even in predicting findings that are relatively non-surprising and incremental. For example, if protein A is known to have a certain function, and protein X is similar to protein A in several respects, then protein X may be hypothesized to share functions with A. Different discovery models of protein functions can be assessed on how well they predict functions across a database of known proteins, without relying on having experimental data for the unknown or novel proteins.
The general scientific public is still not aware of the availability of tools for literature-based discovery. Our Arrowsmith project site maintains a suite of tools that are free and open to the public (http://arrowsmith.psych.uic.edu), as does BITOLA which is maintained by Dmitar Hristovski (http://http://ibmi.mf.uni-lj.si/bitola), and Epiphanet which is maintained by Trevor Cohen (http://epiphanet.uth.tmc.edu/). Bringing user-friendly tools to the public should be a high priority, since even more than advancing basic research in informatics, it is vital that we ensure that scientists actually use discovery tools and that these are actually able to help them make experimental discoveries in the lab and in the clinic.
Acknowledgments
My informatics research is supported by NIH grants R01LM010817 and P01AG039347.
References
- Agarwal S, Yu H, Kohane I. BioNØT: a searchable database of biomedical negated sentences. BMC Bioinformatics. 2011 Oct 27;12:420. doi: 10.1186/1471-2105-12-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH, Lee D, Kim M, Lee JH, Song M. Enriching plausible new hypothesis generation in PubMed. PLoS One. 2017 Jul 5;12(7):e0180539. doi: 10.1371/journal.pone.0180539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekhuis T. Conceptual biology, hypothesis discovery, and text mining: Swanson’s legacy. Biomedical Digital Libraries. 2006;3:2. doi: 10.1186/1742-5581-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruza P, Weeber M, editors. Literature-based Discovery. Springer-Verlag; Berlin Heidelberg: 2008. [Google Scholar]
- Cairelli MJ, Miller CM, Fiszman M, Workman TE, Rindflesch TC. Semantic MEDLINE for discovery browsing: using semantic predications and the literature-based discovery paradigm to elucidate a mechanism for the obesity paradox. AMIA Annu Symp Proc. 2013 Nov 16;2013:164–73. [PMC free article] [PubMed] [Google Scholar]
- Cameron D, Kavuluru R, Rindflesch TC, Sheth AP, Thirunarayan K, Bodenreider O. Context-driven automatic subgraph creation for literature-based discovery. J Biomed Inform. 2015 Apr;54:141–57. doi: 10.1016/j.jbi.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen KB, Johnson HL, Verspoor K, Roeder C, Hunter LE. The structural and content aspects of abstracts versus bodies of full text journal articles are different. BMC Bioinformatics. 2010 Sep 29;11:492. doi: 10.1186/1471-2105-11-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T, Whitfield GK, Schvaneveldt RW, Mukund K, Rindflesch T. EpiphaNet: An Interactive Tool to Support Biomedical Discoveries. J Biomed Discov Collab. 2010 Sep 21;5:21–49. [PMC free article] [PubMed] [Google Scholar]
- Cohen T, Widdows D. Empirical distributional semantics: methods and biomedical applications. J Biomed Inform. 2009 Apr;42(2):390–405. doi: 10.1016/j.jbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T, Widdows D. Embedding of semantic predications. J Biomed Inform. 2017 Apr;68:150–166. doi: 10.1016/j.jbi.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole R, Bruza P. Discovery Science. Springer; Berlin/Heidelberg: 2005. A bare bones approach to literature-based discovery: an analysis of the Raynaud’s/Fish-oil and migraine-magnesium discoveries in semantic space; pp. 84–98. [Google Scholar]
- Ding Y, Song M, Han J, Yu Q, Yan E, Lin L, Chambers T. Entitymetrics: measuring the impact of entities. PLoS One. 2013 Aug 29;8(8):e71416. doi: 10.1371/journal.pone.0071416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Liu Y, Zhu W, Mou Q, Wang J, Hu Y. Simulation of Swanson’s literature-based discovery: anandamide treatment inhibits growth of gastric cancer cells in vitro and in silico. PLoS One. 2014 Jun 20;9(6):e100436. doi: 10.1371/journal.pone.0100436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frijters R, van Vugt M, Smeets R, van Schaik R, de Vlieg J, Alkema W. Literature Mining for the Discovery of Hidden Connections between Drugs, Genes and Diseases. PLoS Computational Biology. 2010;6(9):e1000943. doi: 10.1371/journal.pcbi.1000943. http://doi.org/10.1371/journal.pcbi.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MD, Dumais S. Using latent semantic indexing for literature based discovery. Journal of the American Society for Information Science. 1998;49(8):674–685. [Google Scholar]
- Hossain MS, Gresock J, Edmonds Y, Helm R, Potts M, Ramakrishnan N. Connecting the dots between PubMed abstracts. PLoS One. 2012;7(1):e29509. doi: 10.1371/journal.pone.0029509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristovski D, Kastrin A, Dinevski D, Rindflesch TC. Constructing a graph database for semantic literature-based discovery. Stud Health Technol Inform. 2015;216:1094. [PubMed] [Google Scholar]
- Hristovski D, Kastrin A, Dinevski D, Burgun A, Žiberna L, Rindflesch TC. Using literature-based discovery to explain adverse drug effects. J Med Syst. 2016 Aug;40(8):185. doi: 10.1007/s10916-016-0544-z. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. Why most published research findings are false. PLoS Med. 2005 Aug;2(8):e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonnalagadda SR, Goyal P, Huffman MD. Automating data extraction in systematic reviews: a systematic review. Syst Rev. 2015 Jun 15;4:78. doi: 10.1186/s13643-015-0066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastrin A, Rindflesch TC, Hristovski D. Link prediction on a network of co-occurring MeSH Terms: Towards literature-based discovery. Methods Inf Med. 2016 Aug 5;55(4):340–6. doi: 10.3414/ME15-01-0108. [DOI] [PubMed] [Google Scholar]
- Kell DB. Iron behaving badly: inappropriate iron chelation as a major contributor to the aetiology of vascular and other progressive inflammatory and degenerative diseases. BMC Medical Genomics. 2009;2:2. doi: 10.1186/1755-8794-2-2. http://doi.org/10.1186/1755-8794-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilicoglu H. Biomedical text mining for research rigor and integrity: tasks, challenges, directions. Brief Bioinform. 2017:bbx057. doi: 10.1093/bib/bbx057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostoff RN, Block JA, Solka JL, Briggs MB, Rushenberg RL, Stump JA, Johnson D, Lyons TJ, Wyatt JR. Literature-related discovery. Annual Review of Information Science and Technology. 2009;43(1):1–71. [Google Scholar]
- Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005 Aug;94(4):896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- Manev H, Manev R. Benefits of neuropsychiatric phenomics: example of the 5-lipoxygenase-leptin-Alzheimer connection. Cardiovasc Psychiatry Neurol. 2010;2010:838164. doi: 10.1155/2010/838164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maver A, Hristovski D, Rindflesch TC, Peterlin B. Integration of Data from Omic Studies with the Literature-Based Discovery towards Identification of Novel Treatments for Neovascularization in Diabetic Retinopathy. BioMed Research International. 2013;2013:848952. doi: 10.1155/2013/848952. http://doi.org/10.1155/2013/848952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Advances in neural information processing systems. 2013:3111–3119. [Google Scholar]
- Miller CM, Rindflesch TC, Fiszman M, Hristovski D, Shin D, Rosemblat G, Zhang H, Strohl KP. A closed literature-based discovery technique finds a mechanistic link between hypogonadism and diminished sleep quality in aging men. Sleep. 2012 Feb 1;35(2):279–85. doi: 10.5665/sleep.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Torvik VI. Quantifying conceptual novelty in the biomedical literature. Dlib Mag. 2016;22:9–10. doi: 10.1045/september2016-mishra. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower J, Subramanian D, Shang N, Cohen T. Classification-by-Analogy: Using Vector Representations of Implicit Relationships to Identify Plausibly Causal Drug/Side-effect Relationships. AMIA Annual Symposium Proceedings. 2016;2016:1940–1949. [PMC free article] [PubMed] [Google Scholar]
- O’Mara-Eves A, Thomas J, McNaught J, Miwa M, Ananiadou S. Using text mining for study identification in systematic reviews: a systematic review of current approaches. Syst Rev. 2015 Jan 14;4:5. doi: 10.1186/2046-4053-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packalen M, Bhattacharya J. Neophilia ranking of scientific journals. NBER Working Papers No. 2015;21579 doi: 10.3386/w21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Bonifield G, Smalheiser NR. Gaps within the Biomedical Literature: Initial Characterization and Assessment of Strategies for Discovery. Frontiers in Research Metrics and Analytics. 2017;2:3. doi: 10.3389/frma.2017.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper KR. Three worlds The Tanner Lecture on Human Values. The University of Michigan; Ann Arbor: 1978. http://tannerlectures.utah.edu/_documents/a-to-z/p/popper80.pdf, accessed on July 17, 2017. [Google Scholar]
- Pennington J, Socher R, Manning CD. Glove: Global vectors for word representation. EMNLP. 2014 Oct;14:1532–1543. [Google Scholar]
- Petrič I, Cestnik B, Lavrač N, Urbančič T. Outlier detection in cross-context link discovery for creative literature mining. The Computer Journal. 2010;55(1):47–61. [Google Scholar]
- Preiss J, Stevenson R. The effect of word sense disambiguation accuracy on literature based discovery. BMC Medical Informatics and Decision Making. 2016;16(Suppl 1):57. doi: 10.1186/s12911-016-0296-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzhetsky A, Iossifov I, Loh JM, White KP. Microparadigms: chains of collective reasoning in publications about molecular interactions. Proc Natl Acad Sci U S A. 2006 Mar 28;103(13):4940–5. doi: 10.1073/pnas.0600591103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian Y, Siew EG, Orimaye SO. Emerging approaches in literature-based discovery: techniques and performance review. The Knowledge Engineering Review. 2017a;32 [Google Scholar]
- Sebastian Y, Siew EG, Orimaye SO. Learning the heterogeneous bibliographic information network for literature-based discovery. Knowledge-Based Systems. 2017b;115:66–79. [Google Scholar]
- Shang N, Xu H, Rindflesch TC, Cohen T. Identifying plausible adverse drug reactions using knowledge extracted from the literature. Journal of Biomedical Informatics. 2014;52:293–310. doi: 10.1016/j.jbi.2014.07.011. http://doi.org/10.1016/j.jbi.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Li Y, Zhang J, Sun Y, Philip SY. A survey of heterogeneous information network analysis. IEEE Transactions on Knowledge and Data Engineering. 2017;29(1):17–37. [Google Scholar]
- Smalheiser NR. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol Direct. 2007 Nov 30;2:35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR. The search for endogenous siRNAs in the mammalian brain. Exp Neurol. 2012a;235:455–463. doi: 10.1016/j.expneurol.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR. Literature‐based discovery: Beyond the ABCs. Journal of the Association for Information Science and Technology. 2012b;63(2):218–224. [Google Scholar]
- Smalheiser NR. The RNA-centred view of the synapse: non-coding RNAs and synaptic plasticity. Philos Trans R Soc Lond B Biol Sci. 2014 Sep 26;369(1652) doi: 10.1098/rstb.2013.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR. Data Literacy: How to make your experiments robust and reproducible. Elsevier; 2017. in press. [Google Scholar]
- Smalheiser NR, Gomes OL. Mammalian Argonaute-DNA binding? Biol Direct. 2014 Dec 4;10:27. doi: 10.1186/s13062-014-0027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Manev H, Costa E. RNAi and brain function: was McConnell on the right track? Trends Neurosci. 2001 Apr;24(4):216–8. doi: 10.1016/s0166-2236(00)01739-2. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Swanson DR. Assessing a gap in the biomedical literature: magnesium deficiency and neurologic disease. Neurosci Res Commun. 1994;15:1–9. [Google Scholar]
- Smalheiser NR, Swanson DR. Indomethacin and Alzheimer’s Disease. Neurology. 1996a;46:583. doi: 10.1212/wnl.46.2.583. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Swanson DR. Linking estrogen to Alzheimer’s Disease: an informatics approach. Neurology. 1996b;47:809–810. doi: 10.1212/wnl.47.3.809. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Swanson DR. Calcium-independent phospholipase A2 and schizophrenia. Arch Gen Psychiat. 1998;55:752–753. doi: 10.1001/archpsyc.55.8.752. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI. A population-based statistical approach identifies parameters characteristic of human microRNA-mRNA interactions. BMC Bioinformatics. 2004 Sep 28;5:139. doi: 10.1186/1471-2105-5-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005 Jun;21(6):322–6. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Swanson DR. Undiscovered public knowledge. Library Quarterly. 1986a;56:103–118. [Google Scholar]
- Swanson DR. Fish oil, Raynaud’s Syndrome, and undiscovered public knowledge. Perspect Biol Med. 1986b;30:7–18. doi: 10.1353/pbm.1986.0087. [DOI] [PubMed] [Google Scholar]
- Swanson DR. Two medical literatures that are logically but not bibliographically connected. J Am Soc Inform Sci. 1987;38:228–233. [Google Scholar]
- Swanson DR. Migraine and magnesium: eleven neglected connections. Perspect Biol Med. 1988;31:526–557. doi: 10.1353/pbm.1988.0009. [DOI] [PubMed] [Google Scholar]
- Swanson DR. omatomedin C and arginine; implicit connections between mutually-isolated literatures. Perspect Biol Med. 1990;33:157–186. doi: 10.1353/pbm.1990.0031. [DOI] [PubMed] [Google Scholar]
- Swanson DR. Intervening in the life cycles of scientific knowledge. Library Trends. 1993;41:606–631. [Google Scholar]
- Swanson DR. Atrial fibrillation in athletes: Implicit literature-based connections suggest that overtraining and subsequent inflammation may be a contributory mechanism. Med Hypotheses. 2006;66(6):1085–92. doi: 10.1016/j.mehy.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Swanson DR. Literature-based resurrection of neglected medical discoveries. J Biomed Discov Collab. 2011 Apr 20;6:34–47. doi: 10.5210/disco.v6i0.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson DR, Smalheiser NR. An interactive system for finding complementary literatures: a stimulus to scientific discovery. Artificial Intelligence. 1997;91:183–203. [Google Scholar]
- Swanson DR, Smalheiser NR, Bookstein A. Information discovery from complementary literatures: categorizing viruses as potential weapons. JASIST. 2001;52:797–812. [Google Scholar]
- Torvik VI, Smalheiser NR. A quantitative model for linking two disparate sets of articles in Medline. Bioinformatics. 2007;23(13):1658–1665. doi: 10.1093/bioinformatics/btm161. [DOI] [PubMed] [Google Scholar]
- Uzzi B, Mukherjee S, Stringer M, Jones B. Atypical combinations and scientific impact. Science. 2013;342:468–472. doi: 10.1126/science.1240474. [DOI] [PubMed] [Google Scholar]
- van der Eijk CC, van Mulligen EM, Kors JA, Mons B, van den Berg J. Constructing an associative concept space for literature‐based discovery. Journal of the Association for Information Science and Technology. 2004;55(5):436–444. [Google Scholar]
- Vos R, Aarts S, van Mulligen E, Metsemakers J, van Boxtel MP, Verhey F, van den Akker M. Finding potentially new multimorbidity patterns of psychiatric and somatic diseases: exploring the use of literature-based discovery in primary care research. Journal of the American Medical Informatics Association: JAMIA. 2014;21(1):139–145. doi: 10.1136/amiajnl-2012-001448. http://doi.org/10.1136/amiajnl-2012-001448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber M, Vos R, Klein H, de Jong-van den Berg LTW, Aronson AR, Molema G. Generating Hypotheses by Discovering Implicit Associations in the Literature: A Case Report of a Search for New Potential Therapeutic Uses for Thalidomide. Journal of the American Medical Informatics Association: JAMIA. 2003;10(3):252–259. doi: 10.1197/jamia.M1158. http://doi.org/10.1197/jamia.M1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdows D, Cohen T. Reasoning with vectors: a continuous model for fast robust inference. Logic Jnl IGPL. 2015 Oct;23(2):141–73. doi: 10.1093/jigpal/jzu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren JD. Extending the mutual information measure to rank inferred literature relationships. BMC Bioinformatics. 2004 Oct 7;5:145. doi: 10.1186/1471-2105-5-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren JD, Bekeredjian R, Stewart JA, Shohet RV, Garner HR. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 2004 Feb 12;20(3):389–98. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]
- Wolchover N. A long-sought proof, found and almost lost. Quanta Magazine. 2017 Mar 28; https://www.quantamagazine.org/statistician-proves-gaussian-correlation-inequality-20170328.
- Workman TE, Fiszman M, Cairelli MJ, Nahl D, Rindflesch TC. Spark, an application based on Serendipitous Knowledge Discovery. J Biomed Inform. 2016 Apr;60:23–37. doi: 10.1016/j.jbi.2015.12.014. [DOI] [PubMed] [Google Scholar]
- Yang HT, Ju JH, Wong YT, Shmulevich I, Chiang JH. Literature-based discovery of new candidates for drug repurposing. Brief Bioinform. 2017 May 1;18(3):488–497. doi: 10.1093/bib/bbw030. [DOI] [PubMed] [Google Scholar]
- Yetisgen-Yildiz M, Pratt W. A new evaluation methodology for literature-based discovery systems. J Biomed Inform. 2009 Aug;42(4):633–43. doi: 10.1016/j.jbi.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Zweigenbaum P, Demner-Fushman D, Yu H, Cohen KB. Frontiers of biomedical text mining: current progress. Briefings in Bioinformatics. 2007;8(5):358–375. doi: 10.1093/bib/bbm045. http://doi.org/10.1093/bib/bbm045. [DOI] [PMC free article] [PubMed] [Google Scholar]


