Abstract
Activation of complement C5 generates the potent anaphylatoxin C5a and leads to pathogen lysis, inflammation and cell damage. The therapeutic significance of C5 inhibition is demonstrated by Eculizumab, one of the world’s most expensive drugs. However, the mechanism of C5 activation by C5-convertases remains elusive and limits development of therapeutics. Here, we identify and characterize a novel protein family of tick-derived C5 inhibitors. Structures of C5 in complex with the novel inhibitors, the Phase I & II inhibitor OmCI, and an Eculizumab Fab reveal three distinct binding sites on C5 that all prevent activation of C5. The inhibitor binding sites, and ability of all three C5-inhibitor complexes to competitively inhibit the C5-convertase, conflict with earlier steric inhibition models, suggesting a priming event is needed for activation.
Complement eliminates microbes, modulates the adaptive immune system and plays an important role in the clearance of immune complexes and apoptotic cells, inflammation and tissue regeneration 1, 2.
Antibody complexes (classical pathway, CP), lectin complexes (lectin pathway, LP) or spontaneous hydrolysis of the soluble complement protein C3 (alternative pathway, AP) initiate a proteolytic cascade that results in the assembly of enzyme complexes (termed C3 convertases) on surfaces local to the activation stimulus. The C3 convertases consist of either one molecule of complement C3b and the serine protease Bb (the so-called C3bBb convertase, generated by AP) or one molecule of C4b and the serine protease C2a (the so-called C4bC2a convertase, generated by CP and LP).
C3 convertases cleave soluble, circulating C3, leading to the formation of membrane-targeted C3b 3 and release of the anaphylatoxin C3a fragment 4. Membrane bound C3b further assembles into C3bBb that amplifies the activation signal. C3b and its proteolytic fragments trigger phagocytosis and modulates adaptive immune responses via B cell stimulation 2. Additional molecules of C3b (termed C3b’ hereafter) are proposed to associate with the C3 convertases to form C5 convertases (either C3bBbC3b’ or C4bC2aC3b’) when levels of complement activation are high 5, 6, 7, 8, 9. This modulates the activity of the enzyme, which now preferentially cleaves complement component C5, rather than C3, to generate C5b and the potent anaphylatoxin C5a that potently attracts and activates neutrophils, monocytes and mast cells 10. C5b assembles with downstream complement components to generate the membrane attack complex (MAC) that represents the pore forming part of the system responsible for lysing Gram-negative bacteria or inducing tissue inflammation 11. To date the proposed C5 convertase complexes have proved refractory to purification.
Where regulation of complement fails it can turn its destructive capabilities against self and is involved in inflammation, autoimmunity and tissue damage associated with infectious diseases 12, 13. The FDA’s approval of administration of an activation-blocking antibody against C5 (Soliris™; Eculizumab) for patients suffering from paroxysmal nocturnal hemoglobinuria (PNH) or atypical haemolytic uremic syndrome (aHUS) demonstrates the therapeutic utility of blocking the pathway at this point. Inhibition at the level of C5 prevents formation of proinflammatory C5a and MAC but leaves intact generation of the analphylotoxin C3a and cellular opsonisation by C4b and C3b 10, 14. Eculizumab is one of the most expensive drugs in the world, costing ~500k USD per patient per year (https://www.nice.org.uk/news/press-and-media/first-nice-highly-specialised-technologies-guidance-recommends-eculizumab-soliris-for-treating-very-rare-life-threatening-blood-disorder). There is therefore much interest in development of novel C5-targeted therapeutics including a molecule (OmCI; coversin), derived from tick-saliva, whose recombinant form is in clinical trials 15.
A paucity of information about where Eculizumab, OmCI and related drugs bind C5, as well as a lack of structural insights into the mechanisms underlying inhibition, limits further clinical development. Production of novel C5 inhibitors and structures of C5-inhibtor complexes may therefore lead to development of improved therapies for diseases driven by complement. We therefore set out to discover novel C5 inhibitors and determine structures of inhibited C5.
In this study, we identify and characterize a novel protein family of ~8kDa tick-derived C5 inhibitors, termed the RaCI (Rhipicephalus appendiculatus C5 Inhibitor) family. We present structures that reveal a previously unknown, small RaCI family protein fold. We determine a series of three structures for ternary complexes of C5 with two bound inhibitors (OmCI plus three sequence diverse members of the RaCI family). Additionally, we show that Eculizumab binds at a further, unique, site that allows us to assemble and characterize the structure of a quaternary complex C5-OmCI-RaCI-Eculizumab Fab fragment. Our structures reveal that all three classes of inhibitors have distinct binding sites spanning multiple C5 domains. In addition, we design a competition assay that gives results which conflict with previously proposed steric models for OmCI and Eculizumab inhibition. Revisiting the early C5 convertase literature in context of our new data lead us to propose that the C5 convertase components assemble around C5 rather than existing as a pre-assembled entity anchored to the cell surface.
Results
A novel family of C5 inhibitors
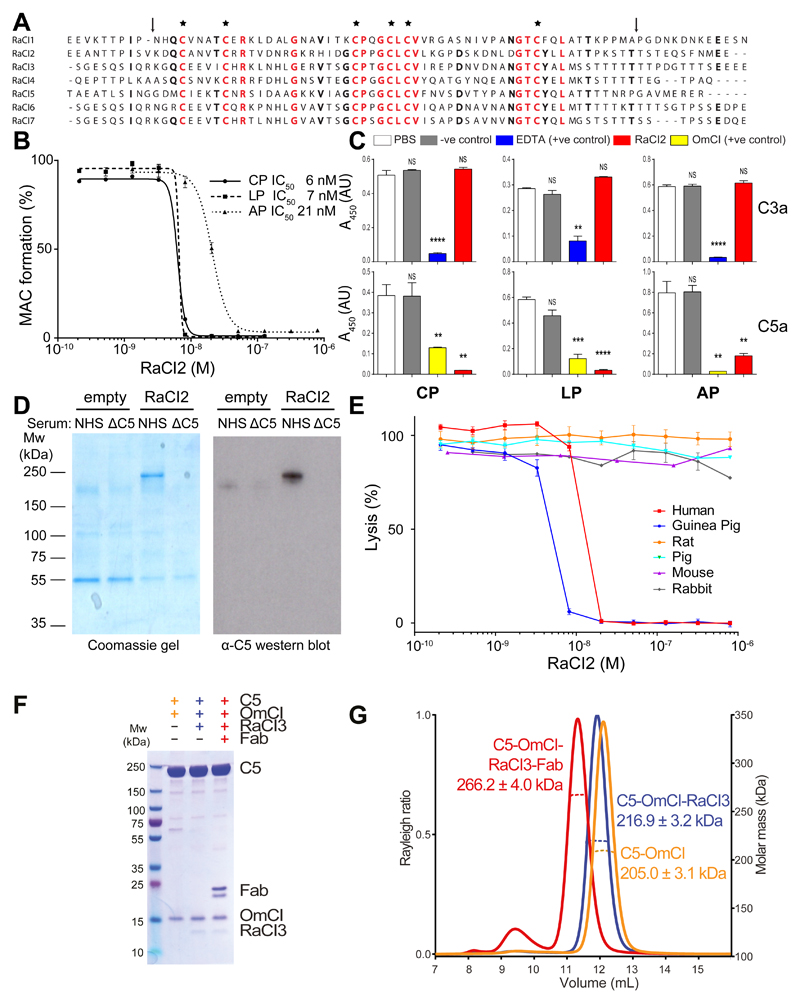
We studied saliva from the hard tick Rhipicephalus appendiculatus to identify new complement inhibitors that could serve to dissect complement activation and its regulatory pathways, as well as acting as potential pharmaceuticals. A complete salivary gland extract from this species was able to prevent complement-mediated haemolysis (Supplementary Figure 1A). The total protein composition of the complete extract is complex. In order to aid identification of the component responsible for the complement inhibitory activity, the salivary gland extracts (SGE) were sequentially fractionated using anion exchange, reversed-phase and size exclusion chromatography. At each stage, the fractions obtained from the chromatographic columns were tested for complement inhibitory activity and fractions with the desired activity were then further fractionated. This eventually generated a fraction highly enriched for inhibition of complement compared to the total extract. This enriched fraction was subjected to LC-MS-MS and expressed sequence tag databases were used to identify peptides 16. No likely candidate genes were identified. We therefore generated a transcriptome from SGE mRNA using RNAseq. Reanalysis of the LC-MS-MS data against our new transcriptome identified a single candidate gene in the enriched fraction that, when over-expressed in insect cells, showed complement inhibiting activity. We named this protein RaCI1 (R. appendiculatus Complement Inhibitor 1). This ~8kDa protein bears no sequence similarity to the previously characterised tick complement inhibitor OmCI (~17kDa). BLAST searches revealed a second RaCI homologue in our R. appendiculatus transcriptome and five additional homologs in other tick species (hereafter termed RaCI2-7) (Fig. 1A). All RaCI1-7 sequences contain six conserved cysteine residues but display low overall sequence conservation (19% identity at the protein level; Supplementary Table 1). Supernatants from insect cell lines expressing each of these homologues tested positive for complement inhibition (Supplementary Fig. 1B), demonstrating that RaCI1-7 share a common function.
Figure 1. Complement inhibition by RaCI.
(A) Sequence alignment of RaCI family members. RaCI1 is from Rhipicephalus appendiculatus and was purified from salivary gland extract. RaCI5 is a homologue from the R. appendiculatus transcriptome described in this study. RaCI2, fusion of the partial sequences in GO496246 and GO496255, Rhipicephalus microplus; RaCI3, EG364159, Dermacentor andersoni; RaCI4, GR908966, Hyalomma marginatum; RaCI6, EG363232, D. andersoni; RaCI7, EG363717, D. andersoni. Residues conserved in five or more RaCIs are in bold and residues conserved in all RaCIs are colored red. Asterisks indicate conserved cysteines and arrows indicate truncations used in Figure 4D (B) RaCI2 inhibits classical pathway (CP), lectin pathway (LP) and alternative pathway (AP) in a Wieslab assay. Error bars, s.e.m. (n = 3 technical replicates). (C) C3a and C5a levels in supernatants of the Wieslab assay as performed in (B). In the Wieslab samples EDTA treatment (C3a generation) or OmCI (C5a generation) are used as positive controls. The functionally unrelated histamine-binding protein 2, which is structurally related to OmCI, is used as a negative control (C3a & C5a generation). Error bars, s.e.m. (n = 3 Wieslab samples). Significance was calculated using an unpaired two-tailed t-test using PBS as a reference. NS p > 0.05; **p < 0.01; ***p < 0.005; ****p < 0.001. (D) Pull-down assay from serum using beads coupled with RaCI2 or empty beads. NHS, normal human serum; ΔC5, C5-depleted serum. (E) Cross-species activity of RaCI2 in a complement haemolysis assay. Error bars, s.e.m. (n = 3 technical replicates). (F) C5 complexes were assembled as described in the methods, purified by size exclusion chromatography and visualized on a Coomassie-stained SDS-PAGE gel and (G) analyzed using SEC-MALS .
We next measured the activity of one of the homologues (RaCI2) on each of the three major complement activation pathways, using an ELISA-based assay (Wieslab). RaCI homologues were tested for their ability to inhibit all three complement activation pathways (Fig. 1B and Supplementary Fig. 1C) and had IC50 values similar to those previously characterized for the unrelated tick complement inhibitor OmCI. To test whether the RaCI family act upstream or downstream of cleavage of C3, we assessed whether the anaphylatoxins C3a and C5a were formed for each of the three pathways. Our experiments revealed that RaCIs leave C3a levels unaltered but completely abolish formation of C5a in all three pathways (Fig. 1C). We performed a pull-down assay from blood serum to identify the target of RaCI. The pull-down revealed a single protein specifically bound to RaCI. Western blotting using anti-C5 antibodies confirmed that this protein was C5 (Fig. 1D). RaCI thus targets complement after convergence of the three pathways. The higher IC50 in the AP assay is explained by the higher concentration of C5 present (5.6% serum in the AP assay versus 1% serum in CP and LP pathway assays) rather than by any difference in the ability of RaCIs to inhibit the different pathways. The IC50s calculated correlate with the concentration of C5 in the assay and reflect an approximately 1:1 molar ratio of inhibitor to C5. OmCI has been shown to inhibit complement from many different mammalian species 17, 18, therefore we also tested cross-species reactivity for RaCI. RaCI potently inhibits complement from guinea pig but not from pig, rat, mouse or rabbit. In contrast, as expected, OmCI inhibits complement from all these species (Fig. 1E and Supplementary Fig. 1D) 17, 18. This suggests that the OmCI binding site on C5 is more highly conserved across species than that targeted by the RaCI family. Taken together, these data demonstrate that our newly identified RaCI family bind human C5 and block the generation of C5a and MAC, leaving the upstream part of the complement cascade intact.
Crystal structures of C5-inhibitor complexes
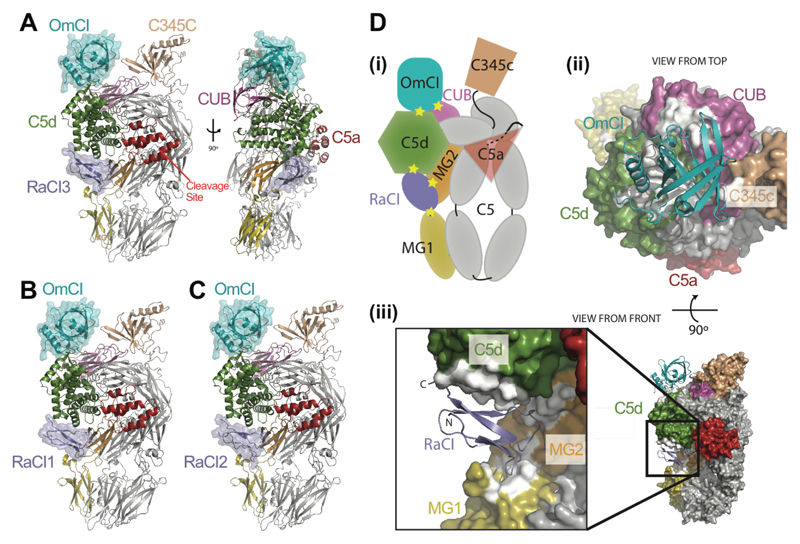
The difference in the ability of RaCl and OmCl to inhibit complement across different species, and the lack of sequence similarity between the two inhibitors led us to hypothesize that RaCl and OmCl bind different sites on C5. To purify C5-inhibitor complexes we first used His-tagged OmCl to isolate C5 from blood serum (Fig. 1F; Materials and Methods). SEC-MALS data demonstrated that C5 and OmCI bind with a 1:1 molar ratio (Fig. 1G), in agreement with previous data 19. Adding an excess of free RaCI to the binary complex (followed by further purification) increased the mass of the complex by ~10kDa, corresponding to one copy of RaCI being incorporated. The formation of a ternary C5-OmCl-RaCl complex demonstrates that RaCI and OmCI bind different epitopes on C5 (Fig. 1 F,G). To gain further insight into the mechanism of inhibition of C5 cleavage and activation, we wanted to test whether the antibody drug Eculizumab binds another distinct epitope on C5 to those bound by the tick inhibitors. We generated a custom-made active Fab fragment from the patented sequence for Eculizumab (hereafter termed EcuFab; Supplementary Fig. 2) and added it to the ternary C5-OmCl-RaCl complex (Fig. 2). This resulted in the formation of an ~50kDa heavier quaternary complex containing C5-OmCl-RaCl-EcuFab in a 1:1:1:1 ratio demonstrating that all three inhibitors target different binding sites on C5.
Figure 2. Crystal structures of C5-OmCI-RaCI complexes and binding mode of RaCI family and OmCI inhibitors.
(A) Front and side view of C5 in complex with OmCI and RaCI3. Cleavage site, denotes location of cleavage site for release of C5a. Proteins are colored as depicted in panel D(i). All proteins shown as cartoon representations with the two tick inhibitors also shown as semi-transparent surfaces. (B) and (C) crystal structures of C5-OmCI-RaCI1 and C5-OmCI-RaCI2 complexes respectively. (D) Inhibitor binding sites on C5 (i) Cartoon showing domain organization of C5 and interaction sites for both tick-inhibitors (yellow stars). (ii) view onto top of C5 complex with C5 shown as a surface representation and OmCI as a teal cartoon. (iii) zoom view into the RaCI binding site. In both (ii) and (iii) the surface of C5 is colored grey with the exception of the following domains (as in the cartoon shown in (i)); C5a – red, C5d – green, CUB-purple. C5 residues that lie within 4Å of an inhibitor – white.
Crystallization studies were undertaken to gain further insight into the molecular architecture of the inhibited C5 complexes. Crystals for the ternary complex C5-OmCI-RaCI3 were produced that diffracted to 2.6 Å (Table 1; Fig. 2A) and solved by molecular replacement using the existing coordinates for C5 and OmCI. Phases calculated from this partial model clearly revealed density for the RaCI3 component and this was built de novo. RaCI3 is wedged between the MG1, MG2 and C5d domains, while OmCI binds a distinct surface of the C5d and CUB domains (see sections below).
Table 1. X-ray Data collection and refinement statistics.
| C5-OmCI-RaCI3 | C5-OmCI-RaCI1 | C5-OmCI-RaCI2 | |
|---|---|---|---|
| Data collection | |||
| Space group | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| Cell dimensions | |||
| a, b, c (Å) | 104.9, 140.3, 211.3 | 105.3, 140.7, 210.4 | 106.0, 140.2, 209.8 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 66.6-2.6 (2.7-2.6) | 94.1-3.1 (3.3-3.1) | 116.6-3.0 (3.2-3.0) |
| Rmerge | 0.08 (0.79) | 0.12 (0.95) | 0.11 (0.89) |
| I / σI | 9.3 (1.4) | 8.2 (1.4) | 8.1 (1.6) |
| Completeness (%) | 99 (100) | 100 (100) | 97 (96) |
| Redundancy | 3.8 (3.7) | 4.4 (4.3) | 4.6 (4.5) |
| Refinement | |||
| Resolution (Å) | 66.6-2.6 (2.7-2.6) | 94.1-3.1 (3.3-3.1) | 116.6-3.0 (3.2-3.0) |
| No. reflections | 96742 (9419) | 55783 (5464) | 64037 (6268) |
| Rwork / Rfree | 0.231 (0.399) / 0.267 (0.433) | 0.267 (0.426) / 0.281 (0.413) | 0.257 (0.405) / 0.282 (0.419) |
| No. atoms | |||
| Protein | 14432 | 14386 | 14412 |
| Ligand/ion | 68 | 34 | 34 |
| Water | 175 | 0 | 0 |
| B factors | |||
| Protein | 74 | 81 | 74 |
| Ligand/ion | 80 | 90 | 79 |
| Water | 51 | - | - |
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.003 | 0.006 | 0.011 |
| Bond angles (°) | 0.61 | 0.75 | 0.89 |
Number of xtals was 1 for each structure.
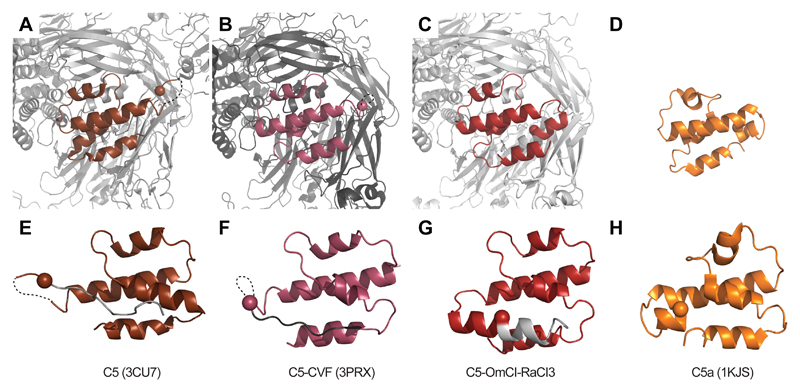
Given the low sequence conservation within the RaCI family, we asked whether they all bind C5 in a similar fashion. The answer was provided by crystallographic structures of C5-OmCI complexes in the presence of RaCI1 and RaCI2 at resolutions of 3.1 Å and 3.0 Å, respectively (Fig. 2B,C). All three RaCI sequences bound the same site on C5. The overall domain arrangement of C5 is the same as that of the previously reported C5 structures 19, with the C345c domain adopting a C3 like orientation, as in the CVF-bound C5 structure 20 (Supplementary Fig. 3,4). The C345c domain has been observed to adopt multiple orientations relative to the main body of C5 in previous crystallographic structures and SAXS data have suggested that the C345c domain is mobile 19, 20, 21. Accordingly, atomic B-factors point to different levels of structural order of C345c within our structures. Of note, the other regions that show the greatest variability compared to previous structures are in the C5a and neighboring MG3 domains (Fig 3). In particular, the C-terminus of the anaphylatoxin, which was previously seen 19, 20 in an extended conformation following an unstructured loop, is now completely modelled and adopts a helical conformation as seen in the NMR structure of human C5a (PDB ID:1KJS; 22). This places the Bb cleavage site (Arg751) in the middle of a helix 36Å distant from the location of the Bb active site proposed on the basis of earlier models of the C5 – convertase interaction 20. Furthermore, the cleavage site is completely buried between helices 1 and 4 of the C5a domain and the MG3 domain, showing that significant rearrangement would be required for proteolysis to occur.
Figure 3. Remodeling of the C5a domain in the inhibited complex.
(A-C) Close-up of the C5a domain (colored) in the C5 structures (grey) as labelled below the panels in the same orientation as the overview shown in Supplementary Figure 4. The peptide that will form the anaphylatoxin after cleavage by Bb is colored in each picture with the location of the alpha-carbon of Arg751 that will form the C-terminus after cleavage highlighted as a sphere. (D-F) These views are rotated 180 degrees from the top panels, essentially looking from within the body of C5, and show only the residues that will form the anaphylatoxin and an additional ten amino acids C-terminal to the cleavage site (grey). The cleavage site, which adopts an extended conformation following an unstructured loop in the C5 and C5-CVF structures 19, 20, is now seen in the middle of a helix and completely buried between helices 1 and 4 of the C5a domain and MG3.
The RaCI – C5 interface
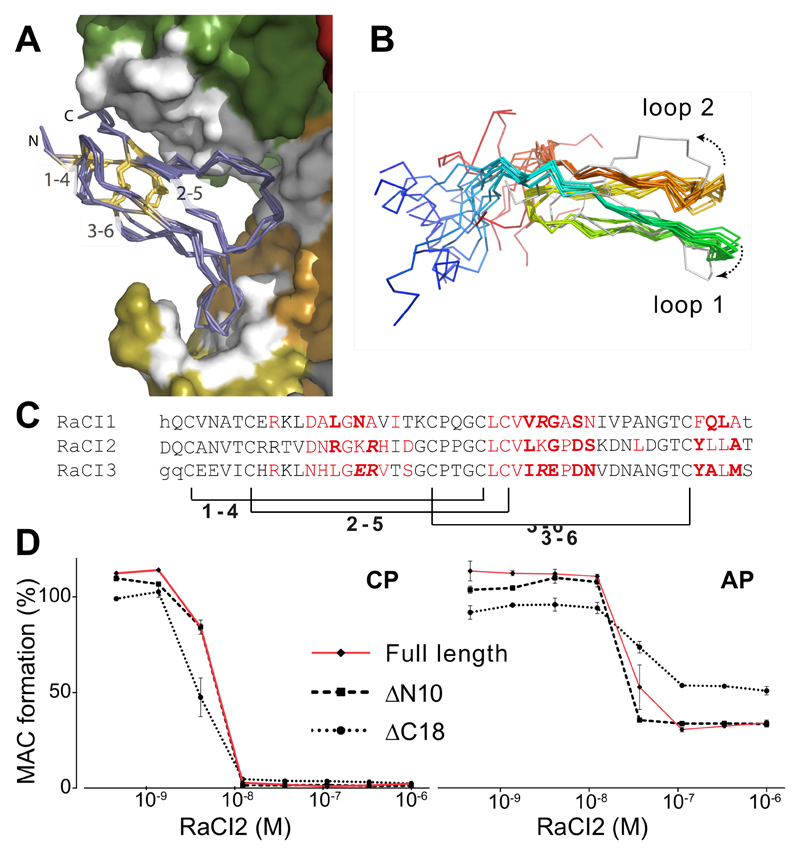
RaCI binds within the cleft created between the MG1, MG2, and C5d domains (Fig. 2, Supplementary Figs. 5-8). Only the central ~50 amino acids of the RaCI molecules adopt an ordered fold and are observed in the crystallographic density. The three RaCI molecules are identically folded, consisting of two main loops that are tied together at the base by three conserved disulphide bridges (Fig. 4A). The fold is unique to the RaCI family since no close structural homologues could be identified in the Protein Data Bank, although the fold is somewhat reminiscent of the disulphide-rich structure of small toxins from snake venom 23. Three stretches of poorly conserved amino acids make extensive contacts with C5 (Fig. 4C), with a number of RaCI-C5 interactions being driven by main chain atoms from RaCI. Extensive use of backbone interactions and a conserved overall fold allows for a high sequence divergence between the RaCI homologues (Fig. 4C).
Figure 4. The structurally conserved core of the RaCI family members is sufficient for activity.
(A) Overlay of RaCI1, 2 & 3 in the C5 binding site (colored as in Fig. 3D). The RaCI proteins are shown as alpha carbon traces with the N- and C-termini and disulphide bonds highlighted. (B) An ensemble of the 10 lowest energy NMR structures of RaCI2 (colored blue to red from N- to C-termini) shows two conserved loops and highly mobile N- and C-terminal regions. The ensemble is overlaid on the structure of RaCI2 (grey) taken from the C5-OmCI-RaCI2 complex. (C) Alignment of RaCI1-3 ordered core residues. Residues that were not seen in the electron density are in lower case. Residues that make van der Waals interactions in red, hydrogen bonds in bold and salt bridges are in italics. Brackets indicate conserved disulphides. (D) Elisa-based activation assay (CP – classical pathway, AP – alternative pathway) to test inhibitory effect of RaCI2 mutants that lack N- and C-termini (lacking 10 and 18 amino acids respectively, see Figure 1A for truncation sites) Error bars, s.e.m. (n = 3 technical replicates).
The relative lack of a canonical hydrophobic core led us to speculate that the RaCI fold might be inherently flexible. To understand any conformational changes that RaCls undergo upon C5 binding, we solved the solution structure of apo-RaCI2 using NMR spectroscopy (Fig. 4B, Table 2). The overlay of the top ten ensemble structures for the apo-RaCI2 revealed the same two ordered loops observed in the structure seen in the context of the C5-RaCI2 complex. However, in the C5-bound structure the tip of loop 1 is bent and loop 2 is twisted compared to the apo structure (Fig. 4B), suggesting that these loops change conformation upon binding of RaCl2 to C5. These regions are also characterized by higher intrinsic mobility (as assessed by residual dipolar coupling measurements) for residues that change orientation upon binding (data not shown). As seen in the C5 complex, the N- and C-terminus of RaCI2 are also disordered in the solution structure (Fig. 4B). We made truncated mutants of RaCI2 to assess whether these poorly conserved and flexible residues are required for inhibition. The ability of the mutants to inhibit both the CP and AP were tested (Fig. 4D). Both mutants retained activity with similar potency to the full-length protein, demonstrating that it is the core of RaCI that is required for inhibition of C5 cleavage and activation.
Table 2.
NMR and refinement statistics for RaCI
| Residues 19-88 | Residues 34-78 | |
|---|---|---|
| NMR distance and dihedral constraints | ||
| Distance constraints | ||
| Total NOE | 609 | 436 |
| Intra-residue | 385 | 265 |
| Inter-reside | ||
| Sequential (|i - j| = 1) | 140 | 94 |
| Medium-range (|i - j| < 4) | 26 | 22 |
| Long-range (|i - j| > 5) | 61 | 58 |
| Intermolecular | 0 | 0 |
| Hydrogen bonds | 0 | 0 |
| Total dihedral angle restraints | 62 | 42 |
| Structure statistics | ||
| Violations (mean ± s.d.) | ||
| Distance constraints (Å) | 0.21±0.25 | 0.22±0.26 |
| Dihedral angle constraints (°) | 8.2±3.8 | 8.2±3.8 |
| Max. dihedral angle violation (°) | 17.3 | 17.3 |
| Max. distance constraint violation (Å) | 1.58 | 1.58 |
| Deviations from idealized geometry | ||
| Bond lengths (°) | 0.001 | 0.001 |
| Bond angles (°) | 0.2 | 0.2 |
| Impropers (°) | 0 | 0 |
| Average pairwise r.m.s. deviation over 10 structures (Å) | ||
| Heavy | 4.0 | 0.8 |
| Backbone | 4.3 | 1.4 |
The OmCI –C5 interface
In our crystal structures, OmCI makes contacts with the C5d, CUB and C345c domains of C5 (Fig. 2D, Supplementary Fig. 7-8). Seven amino acids near the βC-βD loop and at the C-terminus of OmCI make three hydrogen bonds and van der Waals interactions with residues Thr952-Glu958 in the CUB domain. The interactions with the C5d domain are more extensive and include two salt bridges and up to eleven hydrogen bonds. Four discontinuous patches in the OmCI sequence interact with residues between Lys1213 and Asn1241 of the C5d domain. Most contacts occur in the βH-α3 region, previously demonstrated to be critical for binding to C5 24. As in the earlier structures of C5 19, 20 portions of the C345c domain are still relatively disordered but a single amino acid from this domain (Phe1631) forms a point of contact with OmCI residues 62 and 63.
OmCI is a bifunctional protein that simultaneously binds C5 and the LTB4 leukotriene 25. Our structures demonstrate the feasibility of simultaneous C5 and LTB4 binding by OmCI (Supplementary Fig. 9). The distinct binding sites are located in opposite faces of OmCl and the LTB4 binding pocket is accessible in the C5-OmCl complex. As seen in earlier structures of OmCI 25, 26, density within the barrel suggests that the C5-bound OmCI pocket is occupied, although the weak density suggests a mixed population or partial occupancy and an atomic model therefore cannot be built.
Structure of the C5-OmCl-RaCl-EcuFab complex
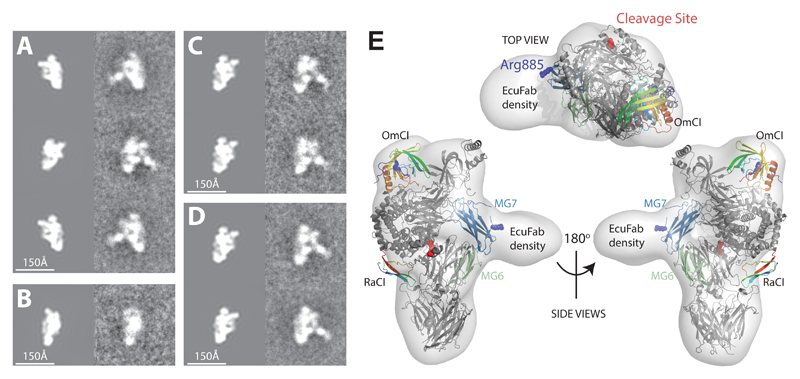
It is not known where Eculizumab binds C5. Patient data have suggested that Arg885 is positioned in or near the epitope, as Arg885His or Arg885Cys polymorphisms induce drug resistance 27, 28. Antibody – peptide studies have also implicated three short peptides as potentially involved, although these data are hard to interpret due to the fact that two of the peptides identified are buried in the folded structure and the three regions are structurally discontinuous, separated by distances much larger than those that can be incorporated in a single antibody epitope [https://www.google.com/patents/US6355245]. We therefore sought to solve the structure of the C5-OmCl-RaCl-EcuFab complex. Attempts to obtain crystals failed. We therefore turned to Transmission Electron Microscopy (TEM) and single-particle analysis 29. 5,587 projection images of different views of the C5-OmCl-RaCl-EcuFab complex preserved in negative stain were acquired and an ab initio 3D reconstruction was calculated, without using the X-ray model as a search reference at any point in the analysis. Fitting of the C5-OmCI-RaCI complex structures into the low resolution EM envelope produced a good fit and revealed additional density due to the presence of bound EcuFab. In Figure 5, we present the fit of the C5 complex to the 3D reconstruction and a comparison of reference-free class averages with projections of a 16-Å-resolution EM density map, simulated from the X-ray structure. The agreement between the projections of the X-ray model and the class averages is excellent and shows additional density due to EcuFab binding the interface between the two lobes of the bipartite C5 structure in two orthogonal side-views (Fig. 5A,B). The reference-free class averages for similar projection directions point to a moderate flexibility in the positioning of EcuFab relative to the C5 core (Fig. 5C,D), which may explain why the density due to EcuFab is of slightly lower resolution than the remaining EM density. The EcuFab epitope is centered on the C5 MG7 domain with both Arg885 and the only surface exposed peptide previously implicated as forming part of the epitope (Lys879-Cys-883) lying within the binding site (Fig. 5E).
Figure 5. Eculizumab inhibits C5 activation by binding to a site centered on the MG7 domain.
Left panel (A-D): Comparison of projections of a 16-Å-resolution EM density map (left column), simulated from the C5 complex X-ray structure with reference-free class averages (right column). (A) Side-views related by ~180° rotation shows the specificity of binding of EcuFab to only one side of the C5 complex. (B) View related by ~90° rotation with respect to the top and bottom image pairs in (A) reveals, as expected, no additional density due to EcuFab which is not visible in this orientation, but shows good agreement with the X-ray model. (C & D) Reference-free class averages for similar projection directions point to a moderate flexibility in the positioning of EcuFab relative to the C5 core. (E) Fit of the C5 complex X-ray structure to the low-resolution EM envelope. Top view (top panel) and side views (bottom panel) of the EM-density map (transparent) with the C5 complex X-ray structure docked. Polymorphisms at Arg885 render patients resistant to Eculizumab. The cleavage site is more than 50 Å distant from the EcuFab binding site.
Inhibition of the C5 convertases
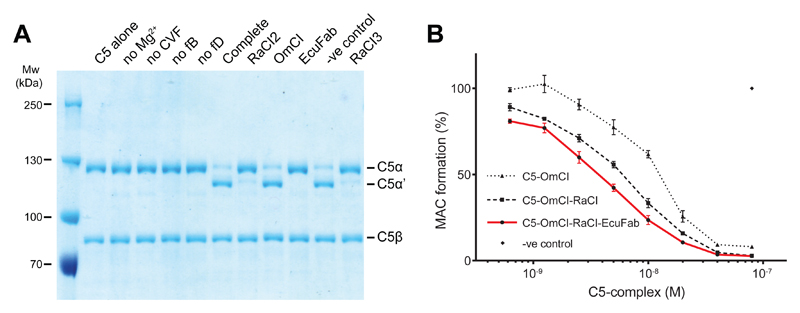
The mechanism of conversion of C5 to C5a and C5b by the C5 convertases C3bBbC3b’ or C4bC2aC3b’ is poorly understood, due in part to their very short half-life. Current structural models are based on the highly stable and presumed to be analogous Cobra Venom Factor (CVF)-Bb complex (CVFBb; CVF is homologous to C3b), which acts in solution without requiring a second C3b’ molecule. Current models for the C5 convertase are based on structures of C3b, C4b, C5, C5-CVF, CVFB and C3bBb 19, 20, 30, 31, 32, 33, and suggest that the MG4, MG5 and MG7 domains of C5 bind the Bb-bound C3b molecule, positioning the active site of the catalytic Bb subunit in the vicinity of the scissile Arg751-Leu752 bond. Neither OmCI nor RaCI bind near the C5a domain, or near the proposed C5 convertase-binding site. Thus inhibition by direct steric hindrance of convertase binding can be ruled out, yet both potently inhibit endogenous convertase-mediated activation of C5 (Fig. 1B; Supplementary Fig. 1C). We tested whether OmCI, RaCI and EcuFab could inhibit CVFBb convertase activity (Fig. 6A). Both RaCI and EcuFab inhibited cleavage by the CVFBb convertase whilst, as previously reported 15, OmCI did not block CVFBb activity (Fig. 6A). The difference in the ability of the three inhibitors to interfere with a CVF-based convertase contrasts strongly with their equal abilities to inhibit the native C3bBbC3b’ and C4bC2aC3b’ convertases (Supplementary Figs. 1,2; Fig. 1B).
Figure 6. All the inhibited C5 complexes can themselves inhibit cleavage of C5 by the native convertase but not all the inhibitors can inhibit cleavage by a CVF convertase.
(A) Coomassie stained gel of C5 conversion by the CVFBb convertase in the presence of RaCI, OmCI, EcuFab or negative control (histamine-binding protein 2). Lanes 3-6 lack the component indicated; Complete shows the pattern of cleavage when CVFBb is assembled (by incubating CVF, fB and fD) and given C5 substrate to act upon in the presence of Mg2+. Final five lanes show the effect of addition of the specified inhibitor or control protein to the C5 substrate prior to incubation with CVFBb. C5: CVF: fB: fD were present at a final molar ratio of 1: 0.1: 0.1: 0.01. (B) Purified C5-OmCI, C5-OmCI-RaCI and C5-OmCI-RaCI-EcuFab complexes are added to an Elisa-based classical pathway activation assay to test for competition with native C5 Error bars, s.e.m. (n = 3 technical replicates).
Next we designed a competition experiment in which highly purified inhibited complexes were added to an ELISA-based CP assay. All binary and ternary complexes of C5 and the tick inhibitors competed with native C5 demonstrating that they still bind the C5 convertase (Fig. 6B, Supplementary Figure 10). Unlike RaCI and OmCI the monoclonal antibody Eculizumab has been suggested to act sterically by preventing C5 – convertase interactions through binding to the MG7 domain 28, a binding mode now confirmed by our EM data (Supplementary Fig. 11). Surprisingly, complexes of C5 with EcuFab bound (C5-OmCI-RaCI-EcuFab, Fig. 6B; C5-EcuFab, Supplementary Fig.5) also inhibited MAC formation in the competition assay suggesting that, unexpectedly, EcuFab does not prevent binding of the inhibited C5 complex to the convertase. Taken together our results demonstrate that C5-CVF interactions do not provide a suitable model for interactions between C5 and the endogenous C5 convertases.
Discussion
We here demonstrate how three clinically important proteins 34 target C5 revealing why patients carrying an Arg885His or Cys polymorphism cannot be treated with Eculizumab 27 and how OmCI binds LTB4 and C5 simultaneously. Additionally, we identify a new family of C5 inhibitors that contains highly divergent sequences that nevertheless adopt a conserved fold to target the same site on C5. RaCI exhibits cross-species reactivity, potentially enabling study of therapeutic efficacy in animal models of disease, unlike the human-specific Eculizumab. All three inhibitor classes are similarly potent complement inhibitors in vitro, but RaCI (~8 kDa) is half the size of OmCI (~17 kDa) and much smaller than Eculizumab (~150kDa), and we show that its size may be further reduced (~5 kDa) to aid drug administration by removal of flexible N- and C-termini The non-related tick proteins OmCI and RaCI target different sites on C5 (C5d-CUB and C5d-MG1-MG2, respectively) and both sites are distinct from that used by Eculizumab. Preliminary data have already demonstrated that OmCI may provide an efficient alternative therapeutic for patients carrying polymorphisms in the Eculizumab binding site (ash.confex.com/ash/2015/webprogram/Paper79479.html) and it is likely that the RaCI family members will prove equally potent.
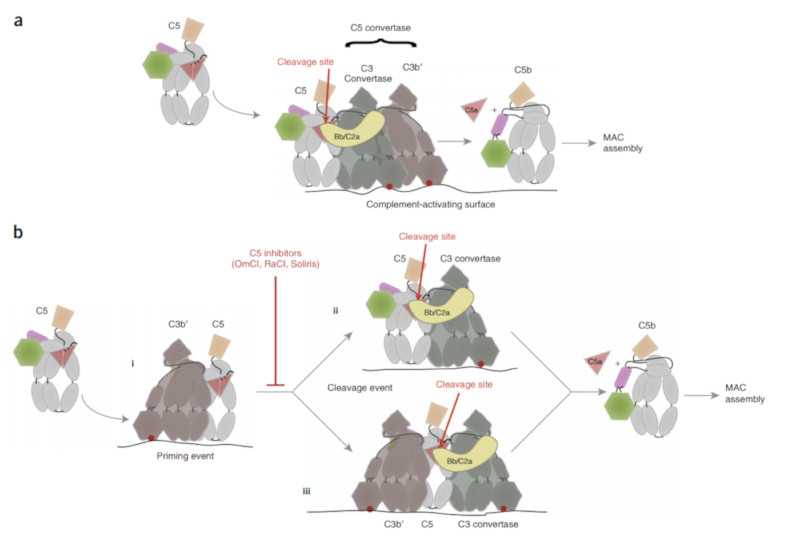
It is reasonably well understood how C3 convertases cleave C3, but the analogous C5 convertases remain elusive. C5 cleavage requires both the C3 convertase (C4bC2a or C3bBb) and an additional surface-bound molecule of C3b’ generated by the C3 convertase 6, 7, 8, 9, 35. C5 is presumed to bind the endogenous convertase as it binds CVF 20 even though the CVF convertase is not dependent on additional C3b’ or surfaces. The exact role of the C3b’ molecule in the C5 convertase activity is disputed. Some studies have shown that C3b’ can covalently attach through its C3d domain to C4b or C3b in the C3 convertase, suggesting that the resulting ternary complexes bind and cleave C5 36, 37 although this has recently been shown not to be required for cleavage 38. Based on crystal structures of C3bBb, C5-CVF and C4b 20, 31, 32 it has been proposed that the reaction of C3b’ with the C3 convertase induces a conformational change in the convertase moiety that leads to a substrate preference for C5; C5 binding is presumed to occur on the opposite edge of the C3 convertase, analogous to the binding of C5 to CVF, which positions the Bb or C2a protease in proximity to the cleavage site (Fig. 7A; 31).
Figure 7. The structures of the inhibited C5 complexes and competition assay yield an altered view of the C5 activation pathway.
Cartoons are used to represent key aspects of current and proposed models of C5 activation with the point of covalent attachment to the surface of the C3b or C4b molecules indicated by the red dot. C3b’ – brown; C3 convertase – grey with enzyme component (Bb or C2a) - yellow. C5 and C5b colored as in Fig 3. (A) Depiction of current model for activation where a preformed C5 convertase consisting of C3BbC3b’ or C4bC2aC3b’ is pre-assembled on the surface and interactions with C5 are made via the C3 convertase component. (B) Our data suggest that a first interaction (i) is with the surface bound C3b’ molecule and that this either results in conformational changes in C5 that render it susceptible to cleavage by the C3 convertases (ii) or that simultaneous interactions by C5 with the C3b’ and the C3 convertases are required to promote cleavage (iii).
However, our competition assays show that EcuFab-bound complexes still inhibit conversion of C5 despite our structural evidence that the EcuFab-bound C5 would be unable to interact with the convertase in the mode implied by the C5-CVF based convertase models. This implies there must be a distinct interaction event, utilising a different surface to the EcuFab binding site, prior to any C5-CVF-like mode of convertase interaction. This is consistent with an earlier model for the endogenous C5 convertase, in which C5 was proposed to directly bind C3b’ (coupled to the surface through its thioester), possibly concomitant with a conformational change that enables cleavage by soluble or surface bound C3 convertase 35. It would also explain why a high density of C3b’ coupled to the surface is required for cleavage by the endogenous convertase 7, 38 as you would need two distinct C5 interaction events on the surface (Fig. 7B). Whether C5 remains bound to C3b’ while simultaneously binding the C3 convertase or whether a molecule of C5 with a short-lived altered conformation transfers to a C3 convertase remains to be investigated. Both scenarios explain why high densities of surface-coupled C3b’ are required.
In addition to lacking a requirement for C3b’ and surfaces, the CVF-convertase clearly differs from the endogenous convertase as OmCI, the Staphylococcus aureus inhibitor SSL7 and point mutations in the C345c domain of C5 all inhibit endogenous C5 convertases but not CVF-based convertases (this study, 15, 38). Our new model proposes that cleavage of C5 by endogenous convertases requires two, distinct, interactions at the surface (Fig. 7B). Presumably CVF-convertases do not require the priming interaction to promote productive interactions with the enzyme, perhaps due to higher affinity C5 binding by CVF 39. Our structures suggest that the EcuFab will sterically prevent interactions with CVF but that RaCI will not act in the same way (Supplementary Fig. 11). Instead, binding of RaCI may lock C5 into a conformation that cannot interact with either the CVF convertases or the C3 convertase component of the endogenous pathway. Alternatively, since the C5a scissile bond in our doubly inhibited C5 (Fig. 3C, F) is inaccessible for cleavage by Bb RaCI may act by preventing a structural ‘priming’ event that rearranges this region to a conformation more similar to that seen in the C5 – CVF complex (Fig. 3B, E) 20. It is harder to propose mechanisms for those inhibitors and mutations which differentially effect endogenous and CVF convertases. However, as for RaCI, they may lock C5 in a conformation which cannot interact with the endogenous C3 convertase, but that this lock is not sufficiently strong to prevent interactions with the more tightly binding CVF convertase 39. Although we do not know the nature of the priming event in the endogenous convertase activity we note that the single point of contact of OmCI with the C345c domain of C5 involves a residue (Phe1631), that when mutated, reduces the ability of the endogenous convertase but not the CVF convertase to cleave C5 40. This suggests that the priming event may involve rearrangements of the highly mobile C345c domain.
In conclusion, our study suggests that the C5 convertase is not a pre-assembled complex. Our data provide strong support for a model in which substantial conformational changes in C5 take place after binding to surface bound C3b’, priming C5 for cleavage by the bona fide C3 convertases. The wide variety of inhibitor structures and binding sites on C5 now revealed suggests that inhibition of this stage of complement activation may be achieved via many mechanistically distinct inhibition strategies potentially allowing fine tuning of therapeutic effects.
Online Methods
RaCI purification and identification
Adult Rhipicephalus appendiculatus ticks were fed on rabbits for three days and glands dissected by Mirko Slovak at the Institute of Zoology (Bratislava, Slovakia). Animal studies were approved by the State Veterinary and Food Administration of the Slovak Republic under decision number 1335/15-221. Dissected glands were stored at -80 °C until further use. Approximately 300 glands from males resuspended in PBS were homogenized in a 1 ml dounce homogeniser (Fisher Scientific). Cleared extract was dialyzed against 25 mM NaPi pH 7.0 and loaded onto a MonoQ 5/50 HR column (GE). The pH of the flowthrough was set to ~1 using 1 M HCl and protein fractionated on a Dynamax 300 Å C8 column (Rainin) using 0.1% Trifluoroacetic acid (buffer A) and 80% acetonitrile + 0.1% Trifluoroacetic acid (buffer B). A gradient of 5-80% buffer B over 20 column volumes (CV) was used. Collected fractions were freeze-dried and resuspended in PBS. Fractions that inhibited complement were pooled and separated on a TSKgel G3000 SWxl column (Tosoh Bioscience) with PBS as running buffer. The fractions were monitored for activity and the most active was selected for in-solution trypsin digestion followed by LC-MS-MS analysis.
Transcriptome sequencing and assembly
Fifty salivary glands from males and females were each dissolved in 1 ml Trizol (Ambion). 100 μl 1-Bromo-3-chloropropane was added and the sample was vigorously mixed. After centrifugation at 12,000 x g for 15 minutes at 4 °C, the colorless upper phase containing the RNA was transferred to a fresh tube after which an equal volume of 70% ethanol was added and the sample was vortexed. Total RNA was extracted using the PureLink RNA Mini Kit (Ambion) following the manufacturer’s protocol. RNA was eluted in 30 μl RNase-free water and the quality of the RNA was assessed on a bio-analyzer (Bio-Rad). The mRNA fraction was selected from approximately 6 μg of total RNA using poly-T beads before conversion to cDNA. Second strand cDNA synthesis incorporated dUTP. The cDNA was end-repaired, A-tailed and adapter-ligated. Different adapters were used for RNA from male and female ticks. Prior to amplification, samples underwent uridine digestion. Mixed female and male samples were 100bp paired-end sequenced in two lanes on a HiSeq sequencer (Illumina). Sequences were assembled de novo using the Trinity software package with default settings 41. This resulted in 118,614 female and 81,358 male-specific transcripts. From these, six translation frames were generated which were used as a database for LC-MS-MS analysis of purified salivary proteins.
Heterologous protein expression and purification
His-RaCI and non-tagged homologues were expressed in Drosophila melanogaster S2 suspension cells (Expres2ion Biotechnologies, Denmark). Codon optimized genes (GeneArt, Germany) were cloned into pExpreS2-2 vectors including a BiP signal peptide or the native signal peptide (Supplementary Table 2). Transfections to obtain stable cell lines were carried out following manufacturer’s instructions and eight days after transfection the spent medium was tested for complement inhibiting activity.
RaCI family members expressed in insect cells were glycosylated and therefore we expressed RaCI1-4 in Escherichia coli Shuffle T7 cells (New England Biolabs). Codon optimized genes without the signal peptide were cloned into pETM14 vectors (European Molecular Biology Laboratory, Germany), using NcoI or BspHI and NotI restriction sites to yield 3C-cleavable His-tagged proteins (Supplementary Table 2). Cells were grown till OD600nm = 0.6 - 0.8 and induced with 0.5 mM IPTG followed by expression at 21 °C for 16 hours. Cells were lysed in 150 mM NaCl, 50 mM NaPi pH 8.0 and proteins were purified on HisTrap HP columns (GE). His-tags were removed with HRV 3C protease, followed by a reverse HisTrap column step. RaCIs were subsequently dialysed against 2 mM Cystine, 1 mM Cysteine, 20 mM Ethanolamine pH 9.0, twice for 24-28 hours at 4 °C to aid the formation of disulfides. RaCI1-3 were then purified on a MonoQ GL column (GE) using 20 mM Ethanolamine pH 9.0 and a gradient of NaCl (RaCI1; 0-100 mM, RaCI2; 50-350 mM, RaCI3; 0-250 mM). RaCI4 was purified on a MonoS GL column (GE) using 20mM MES pH 5.5 with a 0-250 mM NaCl gradient. Protein was dialyzed against PBS and frozen till further use. Truncated RaCI2 variants used in Figure 4D (Supplementary Table 2) were produced as wild type but Cysteine/Cystine dialysis was followed by a Superdex 75 10/30 column (GE) step with PBS as running buffer.
His-tagged OmCI carrying two glycosylation mutations (N78Q/N102Q) (Supplementary Table 2) was expressed using the Kluyveromyces lactis protein expression kit (New England Biolabs) following manufacturer’s instructions. The vector was linearized with SacII prior to transformation. Expression was performed in 10 g/L yeast extract, 20 g/L peptone supplemented with 2% galactose. After 3 or 4 days of expression spent media was filtered and loaded onto a cOmplete His-Tag Purification Column (Roche) followed by a wash and elution step with PBS supplemented with 250 mM Imidazole. Eluted OmCI was loaded on a Superdex 75 16/60 column (GE), and peak fractions were pooled, dialyzed and frozen until further use.
Purification of C5-complexes
Out-dated human serum was used for preparation of C5-complexes for crystallization, MALS and some of the competition assays. Serum was thawed and cleared by centrifugation and filtration. 4 mg His-tagged OmCI was incubated with 200 ml serum at room temperature for 30 minutes. C5-OmCI complexes were loaded on 2 x 5 ml Ni-NTA columns (Qiagen). Columns were washed with 30 CV PBS, 20 mM Imidazole followed by elution with 3 CV PBS, 250 mM Imidazole. After dialysis against PBS the complexes were further purified on a MonoQ 10/100 GL column (GE) with 0-35% gradient of PBS (buffer A) and PBS, 1 M NaCl (buffer B) over 30 CV. Fractions containing pure complexes were dialyzed against 100 mM NaCl, 20 mM HEPES pH 7.5. After dialysis 5-fold molar excess of RaCI was added, samples were incubated for 30 minutes at room temperature and resolved on a Superdex 200 10/30 HR column (GE) with 100 mM NaCl, 20 mM HEPES pH 7.5. Ternary complexes were used for crystallization.
For competition experiments, pure C5 (Complement Technologies, USA) was incubated with a 2-fold molar excess of RaCI, OmCI or EcuFab to create the binary complexes. EcuFab is a custom made Fab fragment using the manufacturer’s framework (Ab00296-10.6, Absolute Antibody, UK) and includes the VL and VH sequences of Eculizumab (European Patent Office: EP0758904 A1). Excess inhibitor was removed with a Superdex 200 5/50 column (GE) in PBS. For MALS and competition experiments, C5-OmCI complexes purified from serum were incubated with 3-fold molar excess of RaCI3 and EcuFab. Excess inhibitor was removed by gel filtration on a Superdex 200 10/30 HR column (GE) with 100 mM NaCl, 20 mM HEPES pH 7.5. Peak fractions were dialyzed against PBS.
Haemolysis assay
Sheep red blood cells (TCS Biosciences) were washed with PBS and sensitized with Anti-Sheep Red Blood Cell Stroma antibody (cat. no. S1389, Sigma-Aldrich) in 142 mM NaCl, 50 mM Sodium 5,5-diethylbarbiturate, 0.1% gelatin, 0.15 mM CaCl2, 0.5 mM MgCl2, pH 7.35 (GVB2+). 50 μl cells (5 x 108 cells/ml) were mixed with an equal volume of diluted serum in GVB2+ and incubated at 37°C for 1 hour while shaking. 100 μl of PBS was added and non-lysed cells were pelleted by centrifugation. 100 μl of the supernatant was transferred to a clean plate and haemolysis was quantified at A405nm. Cells with GVB2+ instead of serum were used as blank and cells with serum only used for normalization (100% activity). Mouse haemolysis assays were carried out with fresh serum and 1 x 108 sensitized cells/ml in the presence of 5% MCAR (Complement Technology, USA). Final serum dilutions used were: 1/80 (human), 1/20 (mouse), 1/40 (rabbit), 1/160 (rat), 1/40 (pig) and 1/640 (guinea pig). Human serum was from healthy volunteers, mouse serum was a kind gift from David R. Greaves (University of Oxford, UK), pig serum was a kind gift from Tom E. Mollnes (Oslo University Hospital, Norway), rat and guinea pig serum were from Complement Technology (USA) and rabbit serum was from Pal Freeze (USA).
Complement ELISA
The effect of RaCI2 on all three pathways was tested using a Wieslab complement system screen (Euro Diagnostica, Sweden) following manufacturer’s instructions. Measurements were subtracted with buffer only and normalised for serum only samples (100% activity). Other classical and alternative pathway assays were carried out as described by A. Roos et al. 42, with the following modifications. 2 μg/ml IgM (Bio-Rad) and 1.8 mg/ml LPS from Salmonella typhosa (Sigma-Aldrich) were used to coat ELISA plates. Human serum was diluted 1/100 (classical pathway) or 1/10 (alternative pathway). MAC deposition was detected with primary antibodies against C5b-9 (aE11, Abcam), and secondary anti-mouse IgG HRP conjugate (Promega). HRP activity was detected with 2,2'-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) liquid substrate (Sigma-Aldrich) and the reaction was stopped by adding an equal volume of 1% SDS. The absorption was measured at 405nm. Measurements were subtracted with buffer only samples and normalized for serum only samples (100% activity). The competition assays were performed using size-exclusion purified inhibited C5 complexes (see purification of C5 complexes above) added to the reaction at the concentrations specified.
C3a and C5a detection
C3a and C5a levels were determined in supernatants from Wieslab assays in the presence of 0.13 μM (classical and lectin pathways) or 0.8 μM (alternative pathway) inhibitor, which resulted in maximum inhibition. C3a and C5a levels were detected using MicroVue kits (Quidel) following manufacturer’s instructions. For C3a detection, supernatants from Wieslab assays were diluted 40-fold (classical and lectin) and 500-fold (alternative) with diluent. For C5a detection, supernatants from Wieslab assays were diluted 4-fold (classical and lectin) and 20-fold (alternative) with diluent. Background subtraction was done with diluent only samples.
Pull-down assay
RaCI2 and OmCI at 1 mg/ml in PBS were coupled to Pierce NHS-Activated magnetic beads (Thermo Scientific) following manufacturer’s instructions. 20 μl beads were incubated with human serum in the presence of 10 mM EDTA, for 30 minutes at room temperature. Beads were washed three times with 1 ml PBS supplemented with 0.005% Tween-20 and once with 100 μl PBS. Bound proteins were recovered by boiling the beads in non-reducing SDS-PAGE loading buffer, prior to separation on SDS-PAGE gel and analysis by western blot. C5-depleted serum (Complement Technology, USA) was included as a control. Western blots were incubated with 1/2,000 diluted polyclonal anti-C5 antibodies (Calbiochem) followed by 1/2,500 diluted anti-rabbit HRP (Promega). C3 and C4 were detected with anti-C3 (1/50,000) and anti-C4 (1/150,000) polyclonal antibodies (both from Complement Technology, USA) and anti-goat HRP antibodies (1/10,000) (Promega). Blots were incubated with ECL substrate (Promega) and imaged with film.
SEC-MALS
100 μg (1 mg/ml) of protein was injected onto a Superdex 200 10/30 column (GE Healthcare) and the elution with PBS was monitored using a Dawn Helios II (Wyatt Technology) and an Optilab TrEX (Wyatt Technology). Data were analyzed using ASTRA software (Wyatt Technology).
Cobra venom factor convertase assays
C5, factor B (fB) and factor D (fD) were purchased from Complement Technology (USA) and Cobra Venom Factor (CVF) from Naja naja kaouthia was purchased from Quidel. CVF, fB and fD were mixed with a 1:1:0.1 molar ratio in PBS and 5mM MgCl2 and incubated for 1 hour at 37°C. C5 was mixed with a 5-fold molar excess of inhibitor in PBS and incubated for 30 minutes at room temperature. Pre-assembled CVF convertases were added to C5-complexes with a 1:10 molar ratio, incubated at 37 °C for 20 hours and separated on a 4-20% gradient gel (Bio-Rad) in the presence of 10 mM dithiothreitol.
NMR Data Collection and Structure Determination
All NMR data was collected on Bruker Avance II 500MHz triple resonance, pulse field gradient system equipped with cryoprobe. All experiments were performed on 13C,15N labelled RaCI2 in PBS (pH7.4). Assignments were performed using both full length (pMJ99) and truncated form (pMJ111) (Table 2). Assignment experiments included CBCANH/CBCA(CO)NH, HNCA/NH(CO)CA, HNCO, HNHA, HBHA(CO)NH, CCCONH and HCCH TOCSY. All data were processed using NMRPipe43 and assigned using Sparky (Goddard & Kneller, Sparky3, UCSF https://www.cgl.ucsf.edu/home/sparky/). NOE cross-peaks were observed through 13C and 15N NOESY-HSQC, processed using NMRPipe and peak picked using Sparky. Distance restraints were obtained through a combination of CYANA44 auto assignment and manual assignment. Initial CYANA 44 produced restraints distance restraints were used in CS ROSETTA 45 with the chemical shift data to produce an initial structure for residues Gln34-Leu78, excluding the flexible N-terminal and C-terminal regions. The resulting CSROSETTA44 structure was then used to initialise the CYANA44 based structure, using auto assigned and manually assigned NOE based distance restraints, as well as talos+ based angle restraints.
For relaxation experiments all NMR data was collected on Bruker Avance II 600MHz triple resonance, pulse field gradient system equipped with cryoprobe. All experiments were performed on 13C,15N labelled protein in PBS (pH7.4). T1 and T2 measurements were collected using delays of (20, 125, 250, 375, 500, 625, 750, 875ms) and (10, 30, 50, 70, 90, 110, 130, 150ms) respectively. Those peaks with exponential decay, baseline resolution and NOE greater than 0.65 were fit to an axially symmetric diffusion tensor and order parameters (S2) for the residues extracted. Coordinates and restraints have been deposited in the PDB (code 5IEC) and BMRB (code 30024) respectively.
Crystallization, X-ray data collection, Structure determination and refinement
4.8 mg/ml C5-OmCI-RaCI2 or C5-OmCI-RaCI3 in buffer containing 100 mM NaCl, 20 mM HEPES pH 7.5 was mixed with an equal volume of mother liquor containing 10% PEG 20K, 2% v/v 1,4 Dioxane, 0.1 M Bicine pH 9.0, and crystallized in 200 nl drops using the vapor diffusion method at 21 °C. Crystals were cryoprotected in mother liquor supplemented with 30% ethylene glycol and flash frozen in liquid N2. 5.5 mg/ml C5-OmCI-RaCI1 was crystallized as above but mother liquor contained 10% PEG 6K, 0.1 M Bicine pH 9.0 and crystals were cryoprotected in mother liquor supplemented with 35% ethylene glycol. Data were collected on beamline I02 at the Diamond Light Source (Harwell, UK) as specified in Table 1. The Structure of C5-OmCI-RaCI3 was solved by molecular replacement using Phaser 46 within CCP4 47 with the structures of C5 (C345c domain removed, PDB ID 3cu7;19) and OmCI (PDB ID 2cm4; 26). The structure of RaCI3 was manually built into difference density and the model subjected to multiple rounds of manual rebuilding in Coot 48 and refinement in Phenix 49 . Once complete, this structure was used to phase the two other complexes with the sequence of the RaCI component set to poly alanine prior to molecular replacement, then the sequence rebuilt in the resulting difference density. Following manual rebuilding and refinement (as for the RaCI3 complex), the structures are characterized by the statistics shown in Table 1 with all three having less than 0.1% Ramachandran outliers and 95% of residues lying in the favorable regions of the Ramachandran plot. Structure factors and coordinates have been deposited in the PDB with the IDs 5HCC, 5HCD & 5HCE. Interactions between OmCI and C5, and RaCIs and C5 have been predicted by PISA 50.
Negative stain specimen preparation, electron microscopy and image processing
3 μL of quaternary complex C5-OmCI-RaCI-EcuFab solution with a concentration of ~0.1 mg/mL was applied to freshly glow-discharged carbon-coated 400-mesh Cu grids (Electron Microscopy Sciences). Following incubation for 30 s, excess solution was removed by blotting with a filter paper. The specimen was stained by application of 3 μL of 2% Uranyl Acetate solution. Following a few seconds incubation, excess staining solution was removed by blotting with a filter paper. The staining procedure was repeated twice. CCD images were recorded under low-dose conditions (~20e-/Å2) at 0.9-1.3 μm underfocus, using a FEI Tecnai T12 transmission electron microscope equipped with a LaB6 filament and operating at 120 kV acceleration voltage. Images were collected at a magnification that gave a sampling distance of 2.06 Å at the specimen level. The power spectra of the images were analyzed and images suffering from specimen drift during the exposure were excluded from further analysis. The images were low-pass filtered to the first zero crossing of the contrast transfer function (CTF) before manual windowing of 5,587 single-particle projections, using the particle windowing tool in the EMAN2 suite of programs 51. An ab initio 3D reconstruction was calculated using the PRIME algorithm 52, which is part of the SIMPLE suite of programs 53 , without using the X-ray model as a search reference at any point in the analysis. Fitting the C5 complex X-ray structures into the low resolution EM envelope validated the 3D reconstruction. To further assess the validity of the 3D reconstruction, reference-free 2D class averages were calculated using PRIME50. Next, a 16-Å-resolution EM density map, simulated from the X-ray structure using the pdb2mrc program in EMAN, was used as a search reference to align the reference-free 2D class averages in 3D. The simulated C5 complex EM density was re-projected in the orientations identified for the class averages in order to demonstrate the excellent agreement with the X-ray model and confirm the presence of the extra density due to bound EcuFab observed in 3D (see Fig. 5, main text). UCSF Chimera was used for automatic real-space rigid body docking 54. The reconstructed volume has been deposited in the EMDB (accession EMDB-8092)
Supplementary Material
Acknowledgements
We thank N. Darvill, T. Tang & L. Britton for experimental contributions, S. Jensen for technical support and M. Slovak (Institute of Zoology, Bratislava, Slovakia) for providing salivary glands. We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z and MRC Hub grant G0900747 91070) for the generation of the Sequencing data. This work was financially supported by a Rubicon grant from the Netherlands Organisation for Scientific Research to M.M.J. (825.11.030) and a Wellcome Investigator Award to S.M.L. (100298). We thank the staff at the Clive and Vera Ramaciotti Centre for Structural Cryo-Electron Microscopy at Monash University for assistance with EM image acquisition. We acknowledge Diamond Light Source for time on I02 under proposal MX9306. EM calculations were performed within the Multi-modal Australian ScienceS Imaging and Visualisation Environment (MASSIVE) (www.massive.org.au).
Footnotes
Author Contributions.
S.M.L. & M.A.N. initially conceived the work. S.M.L. & M.M.J. designed the biochemical experiments and M.M.J. performed them with the assistance of N.M.B. M.M.J. crystallized the complexes and the structures were solved by S.M.L. & S.J. M.M.J., S.M.L., S.J. analyzed the structures. D.S. designed, collected and solved the NMR structure. H.E. designed the EM experiments and together with S.M.L. collected and analyzed the data. Y.I.L. processed the HiSeq data and assembled the transcriptome. All authors were involved in discussion of results and preparation of the manuscript.
Competing financial interests
M.M.J., M.A.N & S.M.L. hold IP in the area of tick complement inhibitors.
Accession Codes
Structure factors and coordinates for X-ray structures have been deposited in the Protein Data Bank (PDB) with accession codes 5HCC, 5HCD & 5HCE. Coordinates for the NMR structure have also been deposited in the PDB with accession code 5IEC and experimental restraints have been deposited in the BMRB (30024). The EM volume has been deposited in the EMDB (EMDB-8092). The Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GEFJ00000000. The version described in this paper is the first version, GEFJ01000000. RaCI protein sequences have been deposited in GenBank with accession codes KU533794 - KU533797 and BK009406 - BK009408.
References
- 1.Kemper C, Kohl J. Novel roles for complement receptors in T cell regulation and beyond. Mol Immunol. 2013;56(3):181–190. doi: 10.1016/j.molimm.2013.05.223. [DOI] [PubMed] [Google Scholar]
- 2.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11(9):785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Law SK, Dodds AW. The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 1997;6(2):263–274. doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulthard LG, Woodruff TM. Is the complement activation product C3a a proinflammatory molecule? Re-evaluating the evidence and the myth. J Immunol. 2015;194(8):3542–3548. doi: 10.4049/jimmunol.1403068. [DOI] [PubMed] [Google Scholar]
- 5.Pangburn MK, Rawal N. Structure and function of complement C5 convertase enzymes. Biochem Soc Trans. 2002;30(Pt 6):1006–1010. doi: 10.1042/bst0301006. [DOI] [PubMed] [Google Scholar]
- 6.Rawal N, Pangburn M. Formation of high-affinity C5 convertases of the alternative pathway of complement. J Immunol. 2001;166(4):2635–2642. doi: 10.4049/jimmunol.166.4.2635. [DOI] [PubMed] [Google Scholar]
- 7.Rawal N, Pangburn MK. Formation of high affinity C5 convertase of the classical pathway of complement. J Biol Chem. 2003;278(40):38476–38483. doi: 10.1074/jbc.M307017200. [DOI] [PubMed] [Google Scholar]
- 8.Daha MR, Fearon DT, Austen KF. C3 requirements for formation of alternative pathway C5 convertase. J Immunol. 1976;117(2):630–634. [PubMed] [Google Scholar]
- 9.Medicus RG, Schreiber RD, Gotze O, Muller-Eberhard HJ. A molecular concept of the properdin pathway. Proc Natl Acad Sci U S A. 1976;73(2):612–616. doi: 10.1073/pnas.73.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodruff TM, Nandakumar KS, Tedesco F. Inhibiting the C5-C5a receptor axis. Mol Immunol. 2011;48(14):1631–1642. doi: 10.1016/j.molimm.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Morgan BP. The membrane attack complex as an inflammatory trigger. Immunobiology. 2015 doi: 10.1016/j.imbio.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Markiewski MM, Lambris JD. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am J Pathol. 2007;171(3):715–727. doi: 10.2353/ajpath.2007.070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricklin D, Lambris JD. Complement in immune and inflammatory disorders: pathophysiological mechanisms. J Immunol. 2013;190(8):3831–3838. doi: 10.4049/jimmunol.1203487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mollnes TE, Kirschfink M. Strategies of therapeutic complement inhibition. Mol Immunol. 2006;43(1-2):107–121. doi: 10.1016/j.molimm.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Nunn MA, Sharma A, Paesen GC, Adamson S, Lissina O, Willis AC, et al. Complement inhibitor of C5 activation from the soft tick Ornithodoros moubata. J Immunol. 2005;174(4):2084–2091. doi: 10.4049/jimmunol.174.4.2084. [DOI] [PubMed] [Google Scholar]
- 16.Nene V, Lee D, Kang'a S, Skilton R, Shah T, de Villiers E, et al. Genes transcribed in the salivary glands of female Rhipicephalus appendiculatus ticks infected with Theileria parva. Insect Biochem Mol Biol. 2004;34(10):1117–1128. doi: 10.1016/j.ibmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Barratt-Due A, Thorgersen EB, Lindstad JK, Pharo A, Lissina O, Lambris JD, et al. Ornithodoros moubata complement inhibitor is an equally effective C5 inhibitor in pigs and humans. J Immunol. 2011;187(9):4913–4919. doi: 10.4049/jimmunol.1101000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hepburn NJ, Chamberlain-Banoub JL, Williams AS, Morgan BP, Harris CL. Prevention of experimental autoimmune myasthenia gravis by rat Crry-Ig: A model agent for long-term complement inhibition in vivo. Mol Immunol. 2008;45(2):395–405. doi: 10.1016/j.molimm.2007.06.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fredslund F, Laursen NS, Roversi P, Jenner L, Oliveira CL, Pedersen JS, et al. Structure of and influence of a tick complement inhibitor on human complement component 5. Nat Immunol. 2008;9(7):753–760. doi: 10.1038/ni.1625. [DOI] [PubMed] [Google Scholar]
- 20.Laursen NS, Andersen KR, Braren I, Spillner E, Sottrup-Jensen L, Andersen GR. Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex. EMBO J. 2011;30(3):606–616. doi: 10.1038/emboj.2010.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laursen NS, Gordon N, Hermans S, Lorenz N, Jackson N, Wines B, et al. Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus. Proc Natl Acad Sci U S A. 2010;107(8):3681–3686. doi: 10.1073/pnas.0910565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Boyar W, Galakatos N, Gonnella NC. Solution structure of a unique C5a semi-synthetic antagonist: implications in receptor binding. Protein Sci. 1997;6(1):65–72. doi: 10.1002/pro.5560060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kini RM, Doley R. Structure, function and evolution of three-finger toxins: mini proteins with multiple targets. Toxicon. 2010;56(6):855–867. doi: 10.1016/j.toxicon.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Mans BJ, Ribeiro JM. Function, mechanism and evolution of the moubatin-clade of soft tick lipocalins. Insect Biochem Mol Biol. 2008;38(9):841–852. doi: 10.1016/j.ibmb.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roversi P, Ryffel B, Togbe D, Maillet I, Teixeira M, Ahmat N, et al. Bifunctional lipocalin ameliorates murine immune complex-induced acute lung injury. J Biol Chem. 2013;288(26):18789–18802. doi: 10.1074/jbc.M112.420331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roversi P, Lissina O, Johnson S, Ahmat N, Paesen GC, Ploss K, et al. The structure of OMCI, a novel lipocalin inhibitor of the complement system. J Mol Biol. 2007;369(3):784–793. doi: 10.1016/j.jmb.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimura J, Yamamoto M, Hayashi S, Ohyashiki K, Ando K, Brodsky AL, et al. Genetic variants in C5 and poor response to eculizumab. N Engl J Med. 2014;370(7):632–639. doi: 10.1056/NEJMoa1311084. [DOI] [PubMed] [Google Scholar]
- 28.Thomas TC, Rollins SA, Rother RP, Giannoni MA, Hartman SL, Elliott EA, et al. Inhibition of complement activity by humanized anti-C5 antibody and single-chain Fv. Mol Immunol. 1996;33(17-18):1389–1401. doi: 10.1016/s0161-5890(96)00078-8. [DOI] [PubMed] [Google Scholar]
- 29.Elmlund D, Elmlund H. Cryogenic electron microscopy and single-particle analysis. Annu Rev Biochem. 2015;84:499–517. doi: 10.1146/annurev-biochem-060614-034226. [DOI] [PubMed] [Google Scholar]
- 30.Janssen BJ, Gomes L, Koning RI, Svergun DI, Koster AJ, Fritzinger DC, et al. Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex. EMBO J. 2009;28(16):2469–2478. doi: 10.1038/emboj.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortensen S, Kidmose RT, Petersen SV, Szilagyi A, Prohaszka Z, Andersen GR. Structural Basis for the Function of Complement Component C4 within the Classical and Lectin Pathways of Complement. J Immunol. 2015;194(11):5488–5496. doi: 10.4049/jimmunol.1500087. [DOI] [PubMed] [Google Scholar]
- 32.Rooijakkers SH, Wu J, Ruyken M, van Domselaar R, Planken KL, Tzekou A, et al. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat Immunol. 2009;10(7):721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444(7116):213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 34.Reis ES, Mastellos DC, Yancopoulou D, Risitano AM, Ricklin D, Lambris JD. Applying complement therapeutics to rare diseases. Clin Immunol. 2015;161(2):225–240. doi: 10.1016/j.clim.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogt W, Schmidt G, Von Buttlar B, Dieminger L. A new function of the activated third component of complement: binding to C5, an essential step for C5 activation. Immunology. 1978;34(1):29–40. [PMC free article] [PubMed] [Google Scholar]
- 36.Kinoshita T, Takata Y, Kozono H, Takeda J, Hong KS, Inoue K. C5 convertase of the alternative complement pathway: covalent linkage between two C3b molecules within the trimolecular complex enzyme. J Immunol. 1988;141(11):3895–3901. [PubMed] [Google Scholar]
- 37.Takata Y, Kinoshita T, Kozono H, Takeda J, Tanaka E, Hong K, et al. Covalent association of C3b with C4b within C5 convertase of the classical complement pathway. J Exp Med. 1987;165(6):1494–1507. doi: 10.1084/jem.165.6.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berends ET, Gorham RD, Jr, Ruyken M, Soppe JA, Orhan H, Aerts PC, et al. Molecular insights into the surface-specific arrangement of complement C5 convertase enzymes. BMC Biol. 2015;13:93. doi: 10.1186/s12915-015-0203-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rawal N, Pangburn MK. Functional role of the noncatalytic subunit of complement C5 convertase. J Immunol. 2000;164(3):1379–1385. doi: 10.4049/jimmunol.164.3.1379. [DOI] [PubMed] [Google Scholar]
- 40.Sandoval A, Ai R, Ostresh JM, Ogata RT. Distal recognition site for classical pathway convertase located in the C345C/netrin module of complement component C5. J Immunol. 2000;165:1066–1073. doi: 10.4049/jimmunol.165.2.1066. [DOI] [PubMed] [Google Scholar]
- 41.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roos A, Bouwman LH, Munoz J, Zuiverloon T, Faber-Krol MC, Fallaux-van den Houten FC, et al. Functional characterization of the lectin pathway of complement in human serum. Mol Immunol. 2003;39(11):655–668. doi: 10.1016/s0161-5890(02)00254-7. [DOI] [PubMed] [Google Scholar]
- 43.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 45.Tian Y, Opella SJ, Marassi FM. Improved chemical shift prediction by Rosetta conformational sampling. J Biomol NMR. 2012;54(3):237–243. doi: 10.1007/s10858-012-9677-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, Baker D, Brunger AT, Das R, DiMaio F, Read RJ, et al. Advances, interactions, and future developments in the CNS, Phenix, and Rosetta structural biology software systems. Annu Rev Biophys. 2013;42:265–287. doi: 10.1146/annurev-biophys-083012-130253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 51.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 52.Elmlund H, Elmlund D, Bengio S. PRIME: probabilistic initial 3D model generation for single-particle cryo-electron microscopy. Structure. 2013;21(8):1299–1306. doi: 10.1016/j.str.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Elmlund D, Elmlund H. SIMPLE: Software for ab initio reconstruction of heterogeneous single-particles. J Struct Biol. 2012;180(3):420–427. doi: 10.1016/j.jsb.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 54.Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, et al. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol. 2012;179(3):269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.