Abstract
The hematopoietic system is well established as a paradigm for the study of cellular hierarchies, their disruption in disease and therapeutic use in regenerative medicine. Traditional approaches to study hematopoiesis involve purification of cell populations based on a small number of surface markers. However, such population-based analysis obscures underlying heterogeneity contained within any phenotypically defined cell population. This heterogeneity can only be resolved through single cell analysis. Recent advances in single cell techniques allow analysis of the genome, transcriptome, epigenome and proteome in single cells at an unprecedented scale. The application of these new single cell methods to investigate the hematopoietic system has led to paradigm shifts in our understanding of cellular heterogeneity in hematopoiesis and how this is disrupted in disease. In this review, we summarize how single cell techniques have been applied to the analysis of hematopoietic stem/progenitor cells in normal and malignant hematopoiesis, with a particular focus on recent advances in single-cell genomics, including how these might be utilized for clinical application.
Keywords: Hematopoietic stem cell, single cell RNA-seq, hematopoiesis, leukemia, genomics, heterogeneity
1. Introduction
The hematopoietic system is perhaps the best-defined model of cellular differentiation due to ease of access, readily identifiable mature blood lineages and plethora of in vitro and in vivo assays. Hematopoiesis is organized as a hierarchical process originating from a rare population of multipotent and self-renewing hematopoietic stem cells (HSCs) that provide a life-long supply of multiple different types of morphologically distinct mature blood cells, through a series of intermediary progenitor cells. Consequently, the hematopoietic system is well established as a paradigm for the study of cellular hierarchies and their disruption in disease [1, 2].
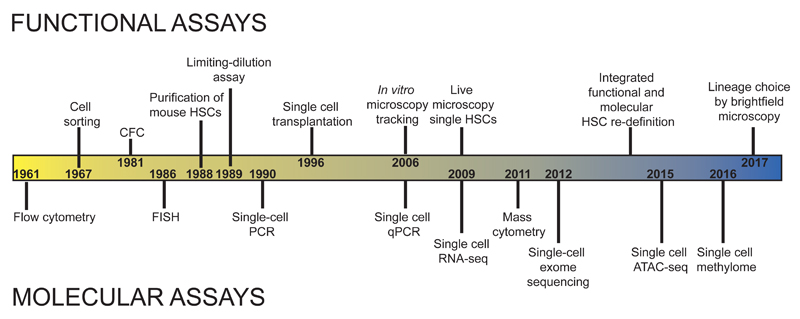
The regenerative capacity of cells within the hematopoietic system was first demonstrated through the rescue of lethally irradiated mice by transplantation of untreated bone marrow [3]. Following these initial experiments, HSC transplantation in patients was established as a routine treatment, and this remains by far the most widely used regenerative therapy in medicine [4]. The occurrence of macroscopic spleen colonies in early transplantation experiments also suggested the high proliferative capacity of some single cells within the hematopoietic system and the consequent need for single cell assays to study normal hematopoietic function. Subsequent experiments using marrow from aneuploidy mice confirmed the unicellular origin of transplant-derived spleen colonies [5]. Since these original observations, hematopoiesis has led the way in the development and application of a plethora of single cell phenotypic and functional analysis techniques to study blood cell development in vitro and in vivo (Figure 1). It is perhaps not surprising, therefore, that hematopoiesis has also emerged as a key developmental system to apply recent technical advances in single cell genomics. According to Sydney Brenner, “Progress in science depends on new techniques, new discoveries and new ideas, probably in that order”[6]. As predicted, the application of new single cell methods to investigate the hematopoietic system has led to paradigm shifts in our understanding of cellular heterogeneity in hematopoiesis and how this is disrupted in disease. In this review, we summarize how single cell approaches have been applied to the analysis of hematopoietic stem/progenitor cells (HSPC) in normal and malignant hematopoiesis, with a particular focus on recent single-cell genomics techniques.
Figure 1. Timeline illustrating key developments in the application of single-cell assays in hematopoiesis.
2. Single cell analysis and normal hematopoiesis
2.1. Limitations of phenotypically defined cell populations in hematopoiesis
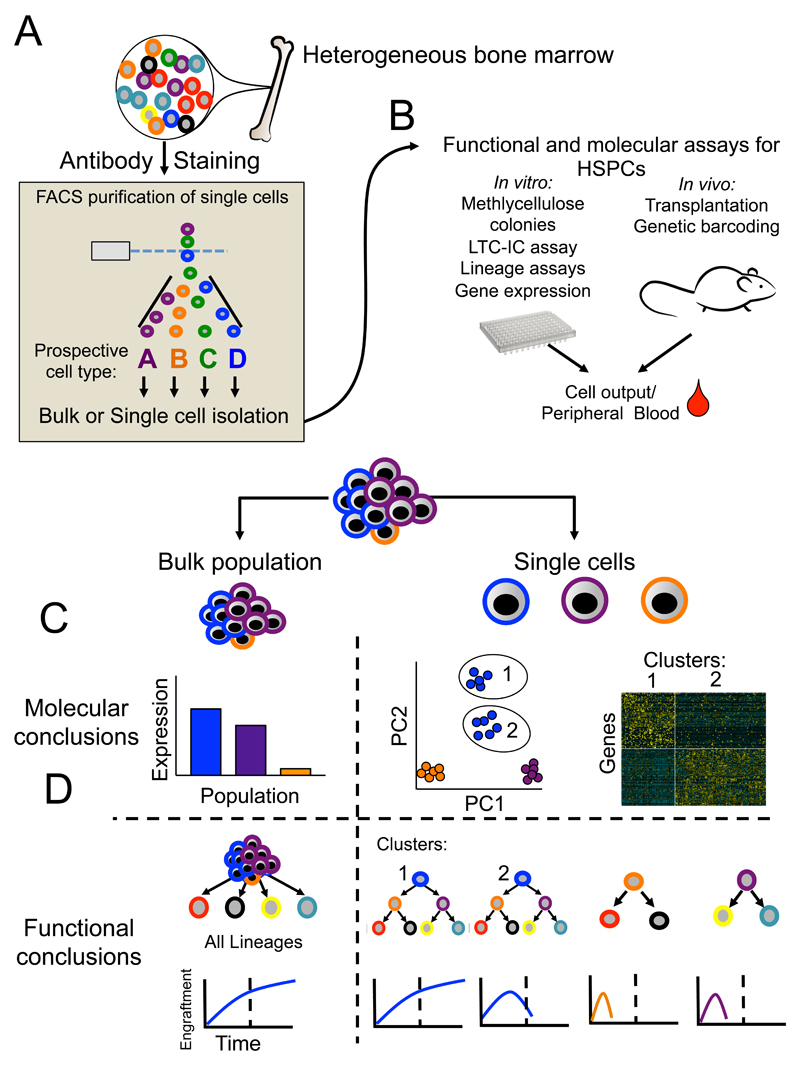
The ability to prospectively isolate immunophenotypic subsets of bone marrow was established through the use of monoclonal fluorescent antibodies and fluorescence-activated cell sorting (FACS, Figure 2A), pioneered by the Weissman laboratory. This single cell analysis method enabled the purification of a rare subset of bone marrow cells by excluding the cell surface markers for mature blood lineages (Lin-), and selecting for the cell surface markers Thy-1 and Sca-1 [7]. The long term repopulating capacity of bone marrow was also shown to be confined to this subset [8]. Subsequently, the phenotypic definition of HSCs has been further refined using a number of different markers, fluorescent dyes and/or transgenic mouse lines [9]. However, all methods to purify HSCs based on cell surface phenotype are limited by the same fundamental problem relating to heterogeneity within the phenotypically defined HSC compartment, including “contamination” by variable numbers of non-HSCs depending on the method used. Furthermore, purity of functional HSCs within the phenotypically-defined HSC compartment is affected, sometimes dramatically, by genetic background of mice, following perturbations such as 5-FU treatment and in disease models [9]. Heterogeneity within phenotypically defined stem/progenitor cell populations is particularly problematic in human hematopoiesis [10]. Ultimately, any phenotypically defined hematopoietic cell population will encompass a range of heterogeneous cell-types. Assays of stem cell function and lineage potential at the cell population level obscure this heterogeneity and can lead to false conclusions, highlighting the need for single-cell approaches to study hematopoiesis (Figure 2B-D).
Figure 2. Unique insights gained through single cell techniques in hematopoiesis.
The bone marrow is a highly heterogeneous mix of mature and immature blood cells as well as supportive niche (a). The identity of these cells can be ascertained through antibody staining. Using known cell surface markers FACS purification of prospective populations can be achieved for either bulk or single cell assays. (b) A wide variety of in vitro and in vivo assays for both bulk and single cells can be utilized for functional or molecular readouts. (c and d) An illustrative comparison of the results and conclusion from gene expression through bulk (left) or single cell (right) experiments used to analyse a theoretical phenotypic HSC population consisting of bona fide HSCs (blue cells, population 1), multipotent progenitors (blue cells, population 2), and two different bipotent progenitor cells (purple and orange). (c) At the molecular level, bulk analysis obscures considerable heterogeneity encompassed within this phenotypic HSC population, while single cell gene expression allows unsupervised clustering (illustrative PCA analysis) and identifies a sub-population structure associated with distinct gene expression. (d) At the functional level, bulk assays mask considerable heterogeneity that can be seen through single cell functional analysis (right, single cell transplantation uncovers heterogeneity in self-renewal and lineage potential in the phenotypically defined HSC populations). Importantly, this example demonstrates the functional difference in single cell transplantation from a population of HSCs previously assumed to be homogeneous.
2.2. Single-cell functional assays in hematopoiesis
Hematopoiesis has the considerable advantage of an array of in vitro and in vivo single-cell assays [9, 11]. Early colony assays provided evidence of heterogeneity in self-renewal capacity and lineage potential of individual HSPCs [12, 13]. Subsequently, methods were developed to purify and transplant phenotypically-defined single HSCs, yielding long term multilineage reconstitution [14]. This single cell transplantation approach has been used to characterize phenotypically defined HSCs, with ~15% to ~67% of transplanted cells yielding long-term engraftment with mouse HSCs [15] and up to ~20% with human HSCs [16]. Importantly, single cell transplantation has also identified a striking degree of functional heterogeneity within the phenotypic HSC compartment with individual stem cells demonstrating mature blood cell output biases or restrictions [17–20].
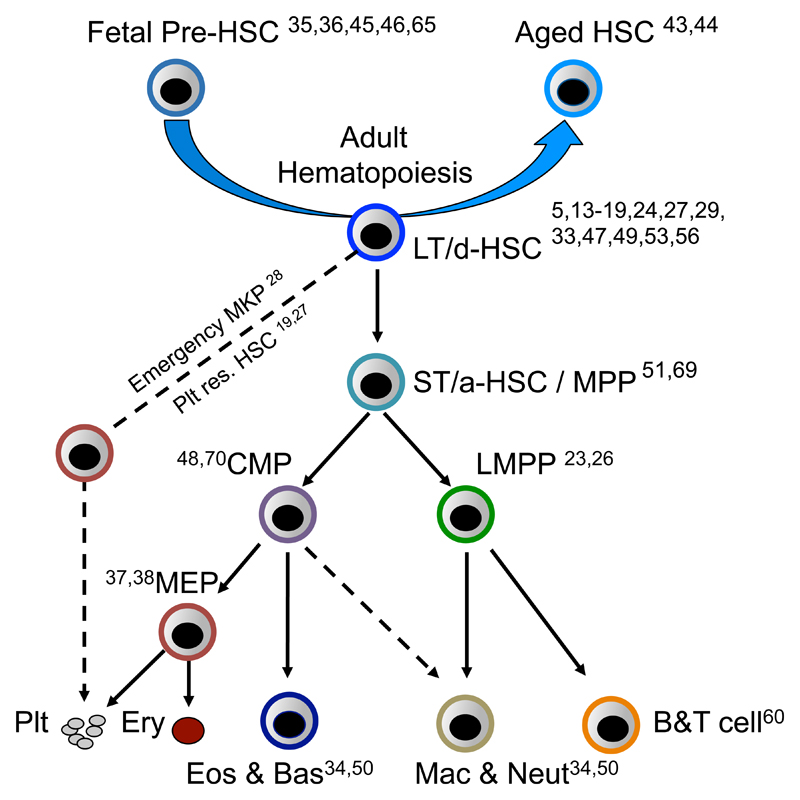
Similarly to the use of single cell in vivo experiments to resolve the phenotypic identity and functional heterogeneity of HSCs, single cell analysis has been instrumental in defining the major lineage branch points in the hematopoietic hierarchy. In the classical model of hematopoiesis, long-term HSCs differentiate through a series of increasingly lineage restricted progenitors concordantly with a loss of self-renewal [21]. By sorting single cells for in vitro culture and identifying their lineage output, the pathways of progenitor differentiation have been identified (Figure 3). As an illustrative example, in mouse hematopoiesis, this single cell analysis approach was used to revise classical models of hematopoietic differentiation through identification of the earliest lineage restriction step as a loss of megakaryocyte and erythroid potential in single cells sustaining other blood lineage developmental potentials; the so called lymphoid primed multipotent progenitor (LMPP) [22]. Lineage outputs from individual LMPPs in vivo have been shown to be more heterogeneous using cellular-barcoding approaches [23], however the sensitivity to detect lineage outputs of a particular clone in vivo using this approach remains to be established. Similarly, single cell in vitro studies have challenged classical models of human adult hematopoietic differentiation, raising the possibility of a two-tier hierarchy of multipotent and unipotent cells, without lineage restricted multipotent intermediaries [24]. Together, these observations from single cell in vivo and in vitro functional assays have provide evidence of considerable functional heterogeneity even within the most stringently defined HSPC populations. Ultimately, the self-renewal and lineage potentials of cells residing within a HSPC population can only be definitively established through single cell analysis, including both in vitro and in vivo functional assays, and hematopoiesis has led the way in the development of such single cell approaches (Figure 2D).
Figure 3. Single cell analysis applied to understand the hematopoietic roadmap.
A simplified hematopoietic hierarchy with citations highlighting key single cell analysis studies of the indicated HSPC population. Single cell analysis has resolved the emergence of HSCs in embryonic development (Fetal pre-HSCs), as well as each major branch point in adult hematopoiesis. LT/d-HSCs indicates long-term or dormant HSCs. ST/a-HSC/MPP indicates short-term/activated HSCs or multipotent progenitors. CMP indicates common myeloid progenitor. LMPP indicates lymphoid-primed multipotent progenitor. MEP indicates megakaryocyte erythroid progenitor. MKP indicates megakaryocyte progenitor.
2.3. Single cell gene expression analysis applied to hematopoiesis
In parallel with single cell functional assays, early single cell gene expression analysis of HSPC provided evidence of transcriptional lineage-priming, preceding lineage specification [25]. This method, which allows analysis of combinatorial gene expression in single cells, is particularly informative when integrated with functional single cell analysis. For example, this approach provided evidence for hierarchical organization of transcriptional lineage programs, with down regulation of megakaryocyte and erythroid-associated gene expression in LMPPs, which sustained myeloid and lymphoid-associated gene expression [26], paralleling their in vitro functional capacity [22]. A similar approach was used to demonstrate megakaryocyte lineage-priming in platelet-biased HSCs and emergency megakaryopoiesis [27, 28]. By multiplexing assays for several genes encoding transcription factors, single cell analysis of HSPCs has also been employed to reveal novel transcription factor regulatory networks [29–31] and to explore transcriptional states associated with lineage commitment and loss of self-renewal [32]. This approach has also been used to uncover subtypes of HSCs based on their transcriptional signature and replicative history [15, 33], to detect heterogeneity in myeloid progenitor differentiation pathways [34] and to uncover the distinct transcriptional changes during early HSC specification [35, 36]. Targeted gene expression analysis has also been applied to study human hematopoiesis, identifying novel pathways of megakaryocyte commitment, questioning classical models of human erythro-megakaryopoiesis [37, 38].
Although a targeted gene expression approach is technically robust, it is also limited by the need to pre-select genes for interrogation. The first unbiased analysis of global gene expression in HSCs was conducted over ten years ago using microarrays [39]. This study, although limited to the analysis of just 12 HSCs, provided the first evidence of transcriptional heterogeneity in HSCs at the global level, including variation in key stem cell regulators such as Scl. Recent developments in single cell RNA-seq (scRNA-seq) has now enabled high-throughput, unbiased molecular analysis of single cells [40]. The development of single cell molecular analysis has been instrumental in the identification of previously unknown heterogeneity within pre-defined hematopoietic populations of presumed homogeneity (Figure 3). For example, scRNA-seq, incorporating index-FACS sorting in some cases (allowing integration of cell surface protein expression by FACS with gene expression in the same single cell), has helped to resolve heterogeneity within single, functional long-term reconstituting HSCs [15, 41], including changes in lineage-affiliated and cell cycle-associated gene expression occurring during aging [42, 43] and after gene deletion e.g. of Bcl11a [44]. Furthermore, scRNA-seq has also been used to resolve cellular pathways of emergence of HSCs during development, including generation of a transcriptional regulatory network model of HSC emergence and identification of key signaling pathways involved in this process [45, 46]. This approach has also been applied to hematopoietic progenitors to form a high-resolution molecular roadmap for hematopoiesis and to resolve previously unrecognized progenitor heterogeneity, and to measure transcription entropy of HSPCs [47–51]. For example, Paul et al analyzed HSPCs by scRNA-seq, identifying multiple previously unrecognized subpopulations within traditionally defined myeloid progenitor populations, each with distinct lineage priming towards specific lineage fates [48]. Olsson et al explored mixed lineage states in myeloid progenitors to delineate mechanisms governing neutrophil versus macrophage lineage specification in murine HSPCs, identifying interplay between Irf8 and Gfi1 as key components of this myeloid gene regulatory network [52]. In zebrafish, scRNA-seq has also been applied to reveal the continuous nature of thrombocyte lineage specification [53]. Similarly, two recent studies applying scRNA-seq to the study of human CD34+ HSPCs and HLR-DA+ cells have resolved a continuous and gradual lineage commitment of HSCs and novel dendritic cell lineages [54, 55]. Another recent study used scRNA-seq to help uncover a role for vitamin-A retinoic acid signaling in the regulation of HSC dormancy, demonstrating the power of this approach to discover novel regulators of HSC function [56]. This study highlights that although cell cycle can be viewed as a confounding variable in molecular analysis [57], the critical role of quiescence in adult HSC maintenance makes single cell analysis of cell cycle extremely important for hematopoiesis research in particular for the analysis of HSCs [56].
These single cell gene expression studies have challenged traditional views of the hematopoietic hierarchy, leading to major conceptual shifts in our understanding of the hematopoietic system. As techniques improve in relation to the number of cells that can be assayed, the capture of RNA molecules and the computational analysis of scRNA-seq data, these techniques will likely be increasingly useful for understanding hematopoietic heterogeneity and will move from primarily descriptive studies towards mechanistic insights.
2.4. Other single cell analysis approaches applied to hematopoiesis
Another exciting technical development in single cell analysis is the ability to measure multiple proteins expressed by single cells through mass cytometry. Unlike FACS analysis that is limited by the number of resolvable fluorescent molecules, mass cytometry allows for significantly higher number of targets that can be analyzed on single cells, including intracellular proteins. This technology has been applied to form a high-resolution map of HSPC response to cytokine stimulation [58, 59] and to investigate regulatory signaling during B-cell development [60]. This technique can also be combined with transcriptional measurements, and can be used as a highly-multiplexed imaging platform that could be applied to study the HSC niche [61, 62]. Indeed, advances in single-cell imaging techniques look set to provide an exciting new tool for hematopoiesis research, allowing reconstruction of spatial relationships between HSPC and neighboring cells which might reveal cell-extrinsic regulators of hematopoiesis [63, 64], and also allowing temporal tracking of individual cells, providing insights into HSC specification in development [65]. Finally, significant technical advancements have enabled single cell measurements of several epigenetic markers [66], including chromatin accessibility by ATAC or DNAse hypersensitivity [67], DNA methylation [68] and 5-hydroxymethlycytosine. These epigenetic techniques are likely to be of particular interest in the study of HSCs because epigenetic regulation appears to play a major role in the functional lineage biases of HSCs and have been studied extensively at the bulk population level [69]. However, a current limitation of these techniques, particularly single cell ATAC-seq, is the limited number of recoverable reads from single cells. Application of these approaches to resolve normal hematopoietic heterogeneity is early in development, but over the coming years looks set to provide new insights in relation to the role of genome regulation in HSPC heterogeneity; this may prove a powerful technique to resolve heterogeneity that is not apparent at the transcriptional level.
2.5. Integrating multiple single cell approaches in hematopoiesis
Linking together analysis of gene expression and cellular function at the single cell level is a potentially very powerful approach. However, most genomics techniques require the cell to be destroyed, precluding subsequent functional analysis. This limitation can be bypassed by studying small numbers of genes in HSPCs using transgenic fluorescent reporter mouse models. This allows for the combination of single cell functional and molecular analysis to dissect heterogeneity in cell populations. For example, using a Vwf-reporter, heterogeneity within the HSC compartment could be explored, leading to the identification of platelet-biased HSCs which reside at the apex of the hematopoietic hierarchy [27]. Single cell transcriptomics identified heterogeneity of Gata1 expression within myeloid progenitor cells, leading to the development of a Gata1-reporter line, which was used to establish a novel lineage bifurcation of Basophil/Eosinophils from neutrophil/monocyte lineage [50]. Fluorescent lineage reporters can also be combined with live cell imaging to model the interplay between the transcription factors Gata1 and Pu1 during lineage commitment of single HSCs [70]. Similarly to the use of gene reporters for lineage commitment, the refinement of phenotypic HSC purification has been advanced in recent years by the addition of reporters for HSC-specific genes including Fdg5 [71], Ctnna1 [72], Hoxb5 [73] and Pdzk1ip1 [74]. These models allow for the prospective isolation of single cells based on expression of markers not expressed at the cell surface. The ability to efficiently label single HSCs through gene reporters enables novel imaging of the HSC niche and robust lineage tracing. Together, these single cell gene expression reporter mouse models are an important tool in single cell biology and the hematopoietic field.
Although single cell transplantation experiments allow definitive assessment of the lineage output of a single HSC, these experiments are also cumbersome and technically demanding. An alternative approach for in vivo lineage tracing of multiple individual hematopoietic clones in one experiment is through the use of cellular barcoding [23, 75–78]. This method involves the viral introduction of unique DNA barcodes into HSCs ex vivo followed by transplantation. By sequencing the nucleated mature blood cells post transplantation for the introduced barcodes, individual clones can be traced. This allows analysis of the lineage output of multiple single cells in vivo over time, in order to understand the clonal dynamics of individual HSPCs. A similar approach has been developed to allow the study of hematopoietic clones during steady-state, unperturbed (non-transplant) conditions. By using an inducible Sleeping Beauty transposon researchers were able to label single HSPCs and follow their output in vivo without transplantation [79]. Surprisingly, using this method little mature blood cell production from HSCs under non-transplant conditions was observed. Conflicting results using HSC-specific inducible fate mapping [74] highlights the need for further studies to fully establish the sensitivity of cellular barcoding approaches to detect outputs of single HSC clones. This method also does not allow the quantification of enucleated erythrocytes and platelets. Another innovative approach utilizes a modified version of the Confetti multicolor fluorescent reporter [80]. This model system allows for clonal tracing of up to 16 distinct single cell HSC-origin clones through flow cytomety. Transplantation of defined in vivo clones into new mice demonstrated a high degree of retained clonal characteristics. This system also allows for clonal tracing of non-nucleated erythrocytes. Together, these single cell barcoding and labeling approaches are providing an extremely valuable window into the function of normal hematopoiesis not obtainable through bulk methods. Integration of such approaches with functional genomic analysis at the single cell level would be an innovative way to link transcriptional or epigenetic heterogeneity of HSPCs with their lineage output in vivo.
3. Single cell approaches in malignant hematopoiesis
In the era of personalized medicine, intratumoral heterogeneity is increasingly recognized as a critical factor underlying a tumor’s ability to evolve and evade therapy. It is therefore crucial to understand such heterogeneity in order to aid the development of new therapeutic approaches [81]. Intratumoral heterogeneity takes many guises, from hierarchical organization of some tumors, including the presence of cancer stem cells (CSCs) [82], to genetically distinct subclones, which includes not only heterogeneity associated with the presence of somatic mutations [83] but also heterogeneity of transcriptional or epigenetic states [84]. Methods to fully characterize these multiple layers of heterogeneity have remained largely elusive until the introduction of single cell genomics. Once again, hematopoiesis research has led the way in the application of new single cell approaches to begin unraveling intratumoral heterogeneity in cancers.
3.1. Analysis of intratumoral heterogeneity with single cell analysis
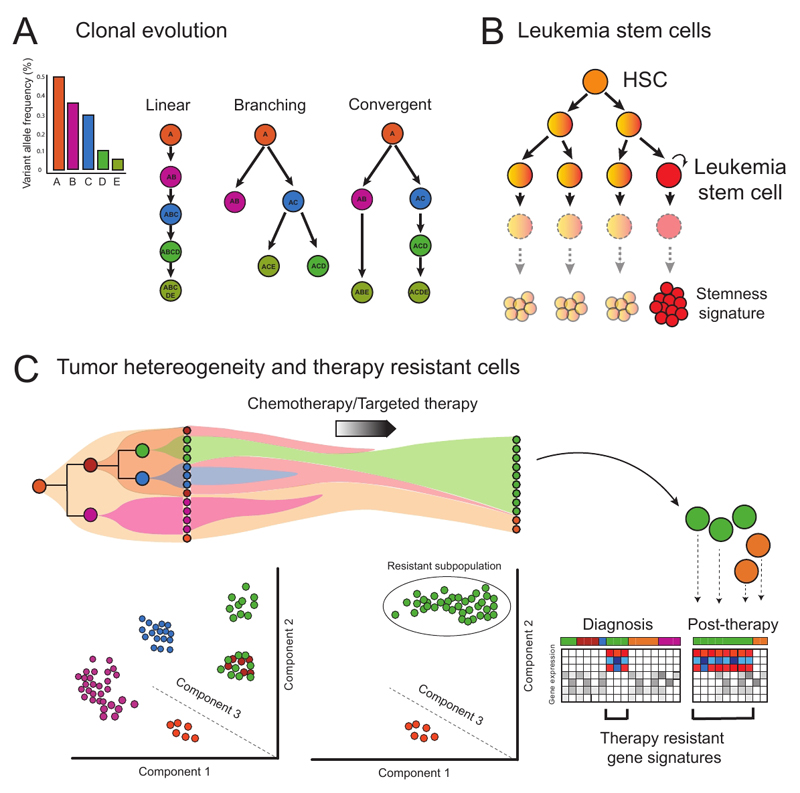
Solid tumors are characterized by the serial acquisition of consecutive genetic lesions, sometimes in a linear fashion, but more often leading to a complex subclonal structure which is generated through branched and convergent patterns of evolution (Figure 4A) [81]. Although the same principles govern the development of hematopoietic tumors, in many cases blood cancers are characterized by relatively few genetic lesions [85], making these diseases an ideal tractable model for understanding of the interplay of different somatic mutations during clonal evolution.
Figure 4. Concepts and applications of single cell genomics in resolving heterogeneity during malignant hematopoiesis.
(a) Single-cell sequencing is able to resolve different patterns of clonal evolution during tumour progression, providing valuable insights into pathways of cancer evolution. Each letter indicates a specific mutation. (b) Aberrant self-renewal in progenitor cell populations establishes leukemia stem cells. (c) Clonal dynamics and transcriptomic/epigenetic heterogeneity of leukemic populations can be revealed through single cell genomic techniques. The analysis of patient samples at diagnosis and after therapy provides crucial insights into different subpopulations and molecular pathways driving drug resistance and relapse. In the figure, a pre-leukemic population (orange) that acquires further mutations gives rise to a variety of leukemic subclones (purple, brown, green and blue). Single cell analysis can be used to identify these different subclones, importantly, also identifying additional heterogeneity within the green subclone, which is only uncovered by PCA (Principal Component Analysis). The pre-leukemic cells and one leukemia subclone (green) are resistant to therapy and the green subclone expands after treatment. The other subclones are eradicated, including a subpopulation of green cells. The molecular characterization of this persistent cell subpopulation can provide insights into mechanisms of therapy resistance that would not be obtained through analysis at the cell population level.
Next Generation Sequencing (NGS) has revolutionized our understanding of the genetics events that drive cancer development [86]. Although NGS data derived from bulk tumor samples can provide important information about the phylogenetic tree of the tumor [83], the resolution to fully reconstruct clonal hierarchies and identify which mutations are present in each subclone, is further enhanced through mutational analysis in single cells [40] (Figure 4A). Indeed, assessment of clonal evolution in hematological cancers at the single cell level is already in routine clinical use through application of cytogenetic analysis, including fluorescent in-situ hybridization (FISH). More recently, single cell mutation analysis can also be carried out either by targeted next generation sequencing of previously characterized mutations [87–90] or single cell whole exome [91, 92] and single cell whole genome sequencing [40, 93]. One of the first examples of single cell exome sequencing in human cancer was a case of JAK2-mutation negative myeloproliferative neoplasm (MPN), where 58 cells were sequenced, revealing monoclonal evolution of the disease in this patient [91]. Whole genome sequencing techniques such as MDA (Multiple Displacement Amplification) have also been used for the analysis of clonal dynamics in childhood Acute Lymphoblastic Leukemia (ALL) [94] and secondary Acute Myeloid Leukemia (AML) [95]. Each approach has advantages and also limitations in terms of the spectrum of mutations that can be analyzed and the sensitivity of mutation detection. Whole genome and whole exome techniques are discovery-type approaches that allow the characterization of new genetic events, whereas targeted techniques rely on mutations previously identified by other methods. However, targeted mutational analysis is a higher throughput and more cost effective approach than whole exome or genome sequencing of single cells. The sensitivity and specificity of mutation detection using a targeted approach is superior in comparison with whole genome techniques which suffer from technical problems, such as high rates of allelic dropout [40]. To help address such technical problems, a wide range of bioinformatics tools to resolve the branches of the clonal tree have been developed [96–98].
Analysis of hematopoietic cancers has illustrated the power of single cell analysis to resolve the genetic architecture in cancer. Pioneering studies by Mel Greaves and colleagues examined the clonal architecture of childhood acute lymphoblastic leukemia through the use of multiplexed, single cell FISH, enabling the detection of up to eight genetic abnormalities in individual cells [99]. This analysis revealed a pattern of convergent evolution in some leukemias, with the same copy number alteration being independently acquired in distinct subclones. In other cancers such as myelodysplastic syndromes, colonies derived from single cells were genotyped, revealing that mutations were acquired in a linear, rather than branching manner in most cases [100]. In both cases this single cell mutation analysis allowed the likely initiating genetic event to be established [99, 100]. Furthermore, analysis of clones derived from single cells from patients with JAK2 and TET2 co-mutated MPN revealed differences in disease phenotype and response to targeted therapy that were contingent upon the order of acquisition of these mutations [101]. The integration of single cell genotyping with FACS purification of specific cell populations allows mutations to be mapped to distinct phenotypically defined cell types. For example, analysis of clones derived from single cells helped to identify pre-leukemic HSCs which reside in the phenotypic HSC compartment and carry only a subset of the mutations found in the bulk leukemia, and also have a phenotype that is distinct from leukemia stem cells (LSCs) in patients with acute myeloid leukemia [89]. These single cell studies of hematopoietic cancers have provided valuable insights into pathways of evolution in cancer, impact of mutation order and cellular hierarchies during disease evolution that could not be resolved through bulk analysis.
3.2. Resolving transcriptional and epigenetic heterogeneity
Tumor heterogeneity is not restricted to different genetic subclones, as molecular heterogeneity in cancer also reflects different transcriptional and epigenomic states [102]. To explore this layer of heterogeneity, analysis of the transcriptional and epigenetic landscape of individual cells is required to establish the cellular composition of a cancer, and integration of these data with mutational status is essential. Gene expression analysis at the single cell level either using a targeted strategy or with scRNA-seq is an ideal approach to identify how specific mutations disrupt transcription in distinct subpopulations of cells [40, 103, 104]. This necessitates integration of transcriptional and mutational readouts. However, obtaining mutational information from scRNA-seq datasets remains challenging, as they usually lack sufficient coverage due to both technical dropouts and stochastic gene expression at the single cell level [105]. Until recently, this integration of mutation status of single cells with their transcriptional profile has remained elusive. Using chronic myeloid leukemia (CML) as the disease model, we have recently demonstrated that by combining a targeted mutation analysis with scRNA-seq, it is possible to detect mutations with high sensitivity, uniquely allowing characterization of LSC before and after therapy [105]. This approach revealed previously unrecognized heterogeneity in CML stem cells and, importantly, allowed analysis of non-leukemic HSCs from CML patients, also providing insights into microenvironmental disruption in CML, which was associated with clinical outcome.
Single-cell epigenomic profiling of HSPCs has been applied to assess heterogeneity of DNA methylation [68] and chromatin accessibility [67] in healthy human hematopoiesis. A single pioneering study to date has assessed the degree to which this heterogeneity contributes to malignant hematopoiesis and how it is disrupted to give rise to malignant phenotypes in single cells [67]. Interestingly, this study found that epigenomic profiling of different stem and progenitor cells better discriminates between previously known cell types as compared with transcriptomic analyses, suggesting that chromatin accessibility captures cell identity more accurately. Moreover, using enhancer footprints, this study also showed that AML LSCs and blasts are heterogeneous populations, containing cells with enhancer signatures corresponding to various stages of hematopoietic differentiation. This is likely to be a critical factor to understand the molecular mechanisms of leukemogenesis considering that mutations affecting epigenetic modifiers are commonly early events during disease evolution [106]. New methods now allow epigenetic and transcriptome information to be obtained from the same single cell [107–110]. Although these approaches have not been used in the hematopoietic system to date, they look set to further enhance understanding of the relationship between transcriptional and epigenetic heterogeneity and how this correlates with malignant phenotypes.
3.3. Hierarchical organization of hematopoietic tumors
There is a growing body of evidence suggesting that some tumors are organized in a cellular hierarchy, with cancer stem cells (CSC), which are the cells responsible for initiating and propagating the disease, residing at the apex of that hierarchy [82, 111] (Figure 4B). Therefore, conceptually, the elimination of CSC is not only necessary, but also potentially sufficient to result in the complete eradication of a tumor and subsequent cure. As a consequence, the identification and characterization of CSC has obvious implications for the development of efficient and durable treatments [112]. Myeloid neoplasms are an excellent model of CSC as candidate LSC have been identified in the majority of myeloid tumors [113–116]. In chronic myeloid malignancies, such as CML, Myelodysplastic Syndromes or Myeloproliferative Neoplasms, the mutation of the founding clone can be traced to the phenotypic HSC compartment [113, 116]. In the case of CML, although LSCs reside in the phenotypic HSC compartment, scRNA-seq has demonstrated that CML-LSCs are mostly distinct from normal HSCs [105], allowing putative therapeutic targets that might differentially target LSCs to be identified.
In the case of AML, multiple different myeloid progenitor populations have been shown to act as LSCs [117–120]. Single-cell technologies offer an unprecedented opportunity to resolve such heterogeneity of cancer propagating populations as well as to characterize their transcriptional program, which may correlate with clinical outcome, as demonstrated at the bulk level [121]. Given that no single surface marker to date can purify leukemic stem cells, single-cell RNA-seq could also be used to identify putative LSC populations on the basis of “stemness” signatures, using available data [121], or to predict de novo stem cell identities from scRNA-seq data using recently developed algorithms [49]. Indeed, the principle of identifying LSCs using single cell gene expression analysis has already been shown in model systems [122]. Single cell analysis could not only identify LSCs but also provide functional insights, including proteomic characteristics [123], which in turn will help to guide new therapeutic strategies towards their elimination.
3.4. Single cell analysis of therapy resistant mechanisms in malignant hematopoiesis
Intratumoral heterogeneity is closely linked to therapy resistance [81, 102], raising the possibility that single cell analysis could be applied to identify and characterize subpopulations of cells that are selectively resistant to the treatment (Figure 4C). This approach has been applied in AML by carrying out single cell targeted mutation analysis following treatment with the FLT3 inhibitor quizartinib [124], demonstrating marked clonal complexity associated with therapy resistance that was mediated by multiple subclones carrying both on- and off-target mutations.
Single cell gene expression analysis has recently been applied to identify therapy resistant LSCs in CML patients receiving tyrosine kinase inhibitor (TKI) therapy, either using a targeted gene expression analysis [125], or by scRNA-seq [105]. LSCs which persisted during TKI therapy could be identified at the single cell level through detection of the BCR-ABL fusion gene, the defining genetic lesion in CML and the target of TKI therapy in CML [126]. LSC that persisted during TKI therapy showed quiescence-associated gene expression. Whole transcriptome analysis by scRNA-seq has the advantage that molecular signatures associated with selective resistance to TKI in LSCs could be analyzed, revealing distinct molecular signatures associated with therapy resistance in LSCs, including increased TNFα and TGFβ-associated gene expression [105]. This study serves as a proof of concept for the application of scRNA-seq to uncover cellular and molecular mechanisms of therapy resistance in LSCs, an approach that could be more broadly applied to analyze any cancer.
4. The Future: Towards Clinical application of single-cell analysis in hematology
Single cell analysis techniques are already firmly established in laboratory-based hematological diagnostic pathways, from flow cytometry to cytogenetic/FISH analysis, both of which currently complement routine analysis of the blood count and morphological analysis of the blood and bone marrow. As scRNA-seq and similar single cell technologies become more routinely available, cost-effective and high-throughput, such analysis could be integrated into diagnostic pathways to definitively establish the cellular composition of the blood or bone marrow and aid disease classification, including analysis of tissue sections by in situ sequencing [127]. Although this may appear to be “science fiction”, not so long ago the possibility of routine clinical use of whole genome sequencing would have seemed far-fetched, yet this is now being integrated into diagnostic algorithms on a large scale [128].
The more obvious and immediate clinical application of single cell analysis techniques, however, is in cancer diagnostics, for example, to identify new biomarkers for prediction of prognosis or response to candidate therapies in order to help refine personalized medicine. For example, single cell analysis could allow measurement of intratumoral heterogeneity, with derivation of a “diversity index”, identifying clonally complex and evolving cancers which may be associated with adverse prognosis [129]. Using single cell approaches to analyse non-clonally involved cells can also be informative for prognosis; we recently demonstrated the potential prognostic utility of scRNA-seq in patients with CML through analysis of inflammatory signatures in the non-leukemic HSC compartment [105]. Single cell gene expression analysis has also been shown to predict response to certain therapies in multiple myeloma [130].
Another possible application of single cell techniques includes use of single cell approaches for minimal residual disease (MRD) detection. Indeed, flow-cytometry based MRD analysis (a single cell method) is already in clinical use for leukemia monitoring [131]. In a recent proof of concept study, massively-parallel RNA-sequencing has also been used to establish the percentage of host versus donor chimerism in bone marrow mononuclear cells of two patients who underwent hematopoietic stem cell transplantation [132]. This study illustrates how single-cell technologies can aid treatment monitoring and at the same time provide biological insights into the characteristics of these residual cells. This principle could also be applied to cancer patients receiving therapy, with single-cell analysis applied to monitor for the presence of therapy-resistant cells at earlier stages allowing early clinical intervention.
Whilst considerable hurdles remain before new single cell genomics techniques can be applied directly in the clinic for patient benefit, the next few years are likely to see extensive efforts towards translation of this new technology towards personalized medicine, including biomarker identification to predict response to specific therapies, prognostic risk stratification, MRD detection and therapeutic target discovery.
Acknowledgments
This work was funded by a Medical Research Council Senior Clinical Fellowship (MR/L006340/1), the MRC Molecular Haematology Unit core award (A.J.M.; MC_UU_12009/5), the MRC funded Oxford Consortium for Single-cell Biology (MR/M00919X/1), a CRUK DPhil Prize Studentship (A.R.M) and the Oxford NIHR Biomedical Centre based at Oxford University Hospitals NHS Trust and University of Oxford (131/030). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or the NIH.
Bibliography
- 1.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–44. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ema H, Morita Y, Suda T. Heterogeneity and hierarchy of hematopoietic stem cells. Exp Hematol. 2014;42(2):74–82 e2. doi: 10.1016/j.exphem.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Till JE, M EA. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res. 1961;14:213–22. [PubMed] [Google Scholar]
- 4.Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med. 2006;354(17):1813–26. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- 5.Becker AJ, McC E, Till JE. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature. 1963;197:452–4. doi: 10.1038/197452a0. [DOI] [PubMed] [Google Scholar]
- 6.Brenner S. Life sentences: Hunters and gatherers. Genome Biology. 2002;3(2) comment1003.1-comment1003.2. [Google Scholar]
- 7.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241(4861):58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 8.Uchida N, Weissman IL. Searching for hematopoietic stem cells: evidence that Thy-1.1lo Lin- Sca-1+ cells are the only stem cells in C57BL/Ka-Thy-1.1 bone marrow. J Exp Med. 1992;175(1):175–84. doi: 10.1084/jem.175.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purton LE, Scadden DT. Limiting factors in murine hematopoietic stem cell assays. Cell Stem Cell. 2007;1(3):263–70. doi: 10.1016/j.stem.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Doulatov S, et al. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10(2):120–36. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Ema H, et al. Adult mouse hematopoietic stem cells: purification and single-cell assays. Nat Protoc. 2006;1(6):2979–87. doi: 10.1038/nprot.2006.447. [DOI] [PubMed] [Google Scholar]
- 12.Humphries RK, Eaves AC, Eaves CJ. Self-renewal of hemopoietic stem cells during mixed colony formation in vitro. Proc Natl Acad Sci U S A. 1981;78(6):3629–33. doi: 10.1073/pnas.78.6.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suda T, Suda J, Ogawa M. Single-cell origin of mouse hemopoietic colonies expressing multiple lineages in variable combinations. Proc Natl Acad Sci U S A. 1983;80(21):6689–93. doi: 10.1073/pnas.80.21.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osawa M, et al. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273(5272):242–5. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 15.Wilson NK, et al. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell. 2015;16(6):712–24. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Notta F, et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333(6039):218–21. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 17.Dykstra B, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1(2):218–29. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Sieburg HB, et al. The hematopoietic stem compartment consists of a limited number of discrete stem cell subsets. Blood. 2006;107(6):2311–6. doi: 10.1182/blood-2005-07-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto R, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154(5):1112–26. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Copley MR, Beer PA, Eaves CJ. Hematopoietic stem cell heterogeneity takes center stage. Cell Stem Cell. 2012;10(6):690–7. doi: 10.1016/j.stem.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Babovic S, Eaves CJ. Hierarchical organization of fetal and adult hematopoietic stem cells. Exp Cell Res. 2014;329(2):185–91. doi: 10.1016/j.yexcr.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Naik SH, et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496(7444):229–32. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- 24.Notta F, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016;351(6269):aab2116. doi: 10.1126/science.aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu M, et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11(6):774–85. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 26.Mansson R, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26(4):407–19. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Sanjuan-Pla A, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502(7470):232–6. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- 28.Haas S, et al. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell. 2015;17(4):422–34. doi: 10.1016/j.stem.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Moignard V, et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat Cell Biol. 2013;15(4):363–72. doi: 10.1038/ncb2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schutte J, et al. An experimentally validated network of nine haematopoietic transcription factors reveals mechanisms of cell state stability. Elife. 2016;5:e11469. doi: 10.7554/eLife.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamey FK, et al. Reconstructing blood stem cell regulatory network models from single-cell molecular profiles. Proc Natl Acad Sci U S A. 2017;114(23):5822–5829. doi: 10.1073/pnas.1610609114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pina C, et al. Inferring rules of lineage commitment in haematopoiesis. Nat Cell Biol. 2012;14(3):287–94. doi: 10.1038/ncb2442. [DOI] [PubMed] [Google Scholar]
- 33.Sawen P, et al. Mitotic History Reveals Distinct Stem Cell Populations and Their Contributions to Hematopoiesis. Cell Rep. 2016;14(12):2809–18. doi: 10.1016/j.celrep.2016.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franco CB, et al. Distinguishing mast cell and granulocyte differentiation at the single-cell level. Cell Stem Cell. 2010;6(4):361–8. doi: 10.1016/j.stem.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swiers G, et al. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat Commun. 2013;4:2924. doi: 10.1038/ncomms3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson AC, et al. Single-cell analyses of regulatory network perturbations using enhancer-targeting TALEs suggest novel roles for PU.1 during haematopoietic specification. Development. 2014;141(20):4018–30. doi: 10.1242/dev.115709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Psaila B, et al. Single-cell profiling of human megakaryocyte-erythroid progenitors identifies distinct megakaryocyte and erythroid differentiation pathways. Genome Biol. 2016;17:83. doi: 10.1186/s13059-016-0939-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyawaki K, et al. Identification of unipotent megakaryocyte progenitors in human hematopoiesis. Blood. 2017;129(25):3332–3343. doi: 10.1182/blood-2016-09-741611. [DOI] [PubMed] [Google Scholar]
- 39.Ramos CA, et al. Evidence for diversity in transcriptional profiles of single hematopoietic stem cells. PLoS Genet. 2006;2(9):e159. doi: 10.1371/journal.pgen.0020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wills QF, Mead AJ. Application of single-cell genomics in cancer: promise and challenges. Hum Mol Genet. 2015;24(R1):R74–84. doi: 10.1093/hmg/ddv235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulte R, et al. Index sorting resolves heterogeneous murine hematopoietic stem cell populations. Exp Hematol. 2015;43(9):803–11. doi: 10.1016/j.exphem.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grover A, et al. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat Commun. 2016;7:11075. doi: 10.1038/ncomms11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kowalczyk MS, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25(12):1860–72. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsang JC, et al. Single-cell transcriptomic reconstruction reveals cell cycle and multi-lineage differentiation defects in Bcl11a-deficient hematopoietic stem cells. Genome Biol. 2015;16:178. doi: 10.1186/s13059-015-0739-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou F, et al. Tracing haematopoietic stem cell formation at single-cell resolution. Nature. 2016;533(7604):487–92. doi: 10.1038/nature17997. [DOI] [PubMed] [Google Scholar]
- 46.Moignard V, et al. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotechnol. 2015;33(3):269–76. doi: 10.1038/nbt.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nestorowa S, et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128(8):e20–31. doi: 10.1182/blood-2016-05-716480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paul F, et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell. 2015;163(7):1663–77. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 49.Grun D, et al. De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell. 2016;19(2):266–77. doi: 10.1016/j.stem.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drissen R, et al. Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat Immunol. 2016;17(6):666–76. doi: 10.1038/ni.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pietras EM, et al. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell. 2015;17(1):35–46. doi: 10.1016/j.stem.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olsson A, et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537(7622):698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macaulay IC, et al. Single-Cell RNA-Sequencing Reveals a Continuous Spectrum of Differentiation in Hematopoietic Cells. Cell Rep. 2016;14(4):966–77. doi: 10.1016/j.celrep.2015.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villani AC, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356(6335) doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Velten L, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol. 2017;19(4):271–281. doi: 10.1038/ncb3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cabezas-Wallscheid N, et al. Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell. 2017 doi: 10.1016/j.cell.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 57.Buettner F, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33(2):155–60. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 58.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–96. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knapp DJ, et al. Distinct signaling programs control human hematopoietic stem cell survival and proliferation. Blood. 2017;129(3):307–318. doi: 10.1182/blood-2016-09-740654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bendall SC, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157(3):714–25. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frei AP, et al. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Methods. 2016;13(3):269–75. doi: 10.1038/nmeth.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–22. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 63.Silberstein L, et al. Proximity-Based Differential Single-Cell Analysis of the Niche to Identify Stem/Progenitor Cell Regulators. Cell Stem Cell. 2016;19(4):530–543. doi: 10.1016/j.stem.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee JH, et al. Highly Multiplexed Subcellular RNA Sequencing in Situ. Science. 2014;343(6177):1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eilken HM, Nishikawa S, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457(7231):896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- 66.Wills QF, Higgs DR, Mead AJ. Studying epigenomics in single cells: what is feasible and what can we learn? Epigenomics. 2015;7(8):1231–4. doi: 10.2217/epi.15.93. [DOI] [PubMed] [Google Scholar]
- 67.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48(10):1193–203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farlik M, et al. DNA Methylation Dynamics of Human Hematopoietic Stem Cell Differentiation. Cell Stem Cell. 2016;19(6):808–822. doi: 10.1016/j.stem.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cabezas-Wallscheid N, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15(4):507–22. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 70.Hoppe PS, et al. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature. 2016;535(7611):299–302. doi: 10.1038/nature18320. [DOI] [PubMed] [Google Scholar]
- 71.Gazit R, et al. Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J Exp Med. 2014;211(7):1315–31. doi: 10.1084/jem.20130428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Acar M, et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 2015;526(7571):126–30. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen JY, et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature. 2016;530(7589):223–7. doi: 10.1038/nature16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sawai CM, et al. Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity. 2016;45(3):597–609. doi: 10.1016/j.immuni.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerrits A, et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 2010;115(13):2610–8. doi: 10.1182/blood-2009-06-229757. [DOI] [PubMed] [Google Scholar]
- 76.Lu R, et al. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol. 2011;29(10):928–33. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim S, et al. Dynamics of HSPC repopulation in nonhuman primates revealed by a decade-long clonal-tracking study. Cell Stem Cell. 2014;14(4):473–85. doi: 10.1016/j.stem.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Biasco L, et al. In Vivo Tracking of Human Hematopoiesis Reveals Patterns of Clonal Dynamics during Early and Steady-State Reconstitution Phases. Cell Stem Cell. 2016;19(1):107–19. doi: 10.1016/j.stem.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun J, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514(7522):322–7. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu VW, et al. Epigenetic Memory Underlies Cell-Autonomous Heterogeneous Behavior of Hematopoietic Stem Cells. Cell. 2016;167(5):1310–1322 e17. doi: 10.1016/j.cell.2016.10.045. [DOI] [PubMed] [Google Scholar]
- 81.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27(1):15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 82.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21(3):283–96. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13(11):795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mazor T, et al. Intratumoral Heterogeneity of the Epigenome. Cancer Cell. 2016;29(4):440–51. doi: 10.1016/j.ccell.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349(6255):1483–9. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 87.Potter NE, et al. Single-cell mutational profiling and clonal phylogeny in cancer. Genome Res. 2013;23(12):2115–25. doi: 10.1101/gr.159913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Papaemmanuil E, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46(2):116–25. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jan, et al. Clonal Evolution of Preleukemic Hematopoietic Stem Cells Precedes Human Acute Myeloid Leukemia. Leukemia. 2012 doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paguirigan Amy L, S J, Meshinchi Soheil, Carroll Martin, Maley Carlo, Radich Jerald P. Single cell genotyping demonstrates complex clonal diversity in acute myeloid leukemia. Science Translational Medicine. 7 doi: 10.1126/scitranslmed.aaa0763. 281re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148(5):873–85. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 92.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148(5):886–95. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gawad C, Koh W, Quake SR. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(50):17947–17952. doi: 10.1073/pnas.1420822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hughes AE, et al. Clonal architecture of secondary acute myeloid leukemia defined by single-cell sequencing. PLoS Genet. 2014;10(7):e1004462. doi: 10.1371/journal.pgen.1004462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Roth A, et al. Clonal genotype and population structure inference from single-cell tumor sequencing. Nat Methods. 2016;13(7):573–6. doi: 10.1038/nmeth.3867. [DOI] [PubMed] [Google Scholar]
- 97.Ross EM, Markowetz F. OncoNEM: inferring tumor evolution from single-cell sequencing data. Genome Biol. 2016;17:69. doi: 10.1186/s13059-016-0929-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Salehi Sohrab, S A, Roth Andrew, Aparicio Samuel, Bouchard-Côté Alexandre, Shah Sohrab P. ddClone: joint statistical inference of clonal populations from single cell and bulk tumour sequencing data. Genome Biol. 2017 doi: 10.1186/s13059-017-1169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–61. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 100.Woll PS, et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell. 2014;25(6):794–808. doi: 10.1016/j.ccr.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 101.Ortmann CA, et al. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med. 2015;372(7):601–12. doi: 10.1056/NEJMoa1412098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alizadeh AA, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21(8):846–53. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Patel Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014 doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang L, et al. Integrated single-cell genetic and transcriptional analysis suggests novel drivers of chronic lymphocytic leukemia. Genome Res. 2017;27(8):1300–1311. doi: 10.1101/gr.217331.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Giustacchini A, et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat Med. 2017 doi: 10.1038/nm.4336. [DOI] [PubMed] [Google Scholar]
- 106.Levine RL, A-W O. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood. 2013;121(18) doi: 10.1182/blood-2013-01-451781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Macaulay IC, et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12(6):519–22. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 108.Angermueller C, et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods. 2016;13(3):229–32. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cheow LF, et al. Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat Methods. 2016;13(10):833–6. doi: 10.1038/nmeth.3961. [DOI] [PubMed] [Google Scholar]
- 110.Clark SJ, et al. Joint Profiling Of Chromatin Accessibility, DNA Methylation And Transcription In Single Cells. bioRxiv. 2017 doi: 10.1038/s41467-018-03149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 112.Eppert K, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17(9):1086–93. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 113.Mullally A, M AJ. Myeloproliferative neoplasm stem cells. Blood. 2017;129(12) doi: 10.1182/blood-2016-10-696005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Majeti R, T D. Biology and relevance of human acute myeloid leukemia stem cells. Blood. 2017;129(12) doi: 10.1182/blood-2016-10-696054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shastri Aditi, W B, Steidl Ulrich, Verma Amit. Stem and progenitor cell alterations in myelodysplastic syndromes. Blood. 2017;129(12) doi: 10.1182/blood-2016-10-696062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vetrie D, H TL. The chronic myeloid leukemia stem cell: stemming the tide of persistence. Blood. 2017;129(12) doi: 10.1182/blood-2016-09-696013. [DOI] [PubMed] [Google Scholar]
- 117.Goardon N, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19(1):138–52. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 118.Quek L, et al. Genetically distinct leukemic stem cells in human CD34-acute myeloid leukemia are arrested at a hemopoietic precursor-like stage. J Exp Med. 2016;213(8):1513–35. doi: 10.1084/jem.20151775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Taussig David C, M-M F, Anjos-Afonso Fernando, Pearce Daniel J, Allen Kirsty, Ridler Christopher, Lillington Debra, Oakervee Heather, Cavenagh Jamie, Agrawal Samir G, Lister T Andrew, et al. Anti-CD38 antibody–mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood. 2008;112(3) doi: 10.1182/blood-2007-10-118331. [DOI] [PubMed] [Google Scholar]
- 120.Taussig David C, V J, Miraki-Moud Farideh, Griessinger Emmanuel, Sharrock Kirsty, Luke Tina, Lillington Debra, Oakervee Heather, Cavenagh Jamie, Agrawal Samir G, Lister T Andrew, et al. Leukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34 fraction. Blood. 2010;115(10) doi: 10.1182/blood-2009-02-206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ng SW, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433–437. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 122.Guo G, et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell. 2013;13(4):492–505. doi: 10.1016/j.stem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Levine JH, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162(1):184–97. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Smith CC, et al. Heterogeneous resistance to quizartinib in acute myeloid leukemia revealed by single-cell analysis. Blood. 2017;130(1):48–58. doi: 10.1182/blood-2016-04-711820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Warfvinge R, et al. Single-cell molecular analysis defines therapy response and immunophenotype of stem cell subpopulations in CML. Blood. 2017;129(17):2384–2394. doi: 10.1182/blood-2016-07-728873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Longo DL. Imatinib Changed Everything. N Engl J Med. 2017;376(10):982–983. doi: 10.1056/NEJMe1700833. [DOI] [PubMed] [Google Scholar]
- 127.Chen KH, et al. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348(6233):aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Siva N. UK gears up to decode 100,000 genomes from NHS patients. Lancet. 2015;385(9963):103–4. doi: 10.1016/S0140-6736(14)62453-3. [DOI] [PubMed] [Google Scholar]
- 129.Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell. 2015;58(4):598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mitra AK, et al. Single-cell analysis of targeted transcriptome predicts drug sensitivity of single cells within human myeloma tumors. Leukemia. 2016;30(5):1094–102. doi: 10.1038/leu.2015.361. [DOI] [PubMed] [Google Scholar]
- 131.Thompson PA, Wierda WG. Eliminating minimal residual disease as a therapeutic end point: working toward cure for patients with CLL. Blood. 2016;127(3):279–86. doi: 10.1182/blood-2015-08-634816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]