Abstract
DNA is constantly exposed to a wide array of genotoxic agents, generating a variety of forms of DNA damage. DNA-protein crosslinks (DPCs) – the covalent linkage of proteins with a DNA strand – are one of the most deleterious and understudied forms of DNA damage, posing as steric blockades to transcription and replication. If not properly repaired, these lesions can lead to mutations, genomic instability, and cell death. DPCs can be induced endogenously or through environmental carcinogens and chemotherapeutic agents. Endogenously, DPCs are commonly derived through reactions with aldehydes, as well as through trapping of various enzymatic intermediates onto the DNA. Proteolytic cleavage of the protein moiety of a DPC is a general strategy for removing the lesion. This can be accomplished through a DPC-specific protease and and/or proteasome-mediated degradation. Nucleotide excision repair and homologous recombination are each involved in repairing DPCs, with their respective roles likely dependent on the nature and size of the adduct. The Fanconi anemia pathway may also have a role in processing DPC repair intermediates. In this review, we discuss how these lesions are formed, strategies and mechanisms for their removal, and diseases associated with defective DPC repair.
Keywords: DNA-protein crosslinks, nucleotide excision repair, SPRTN, Fanconi anemia
Introduction
The integrity of cellular DNA is constantly challenged by a wide variety of genotoxic agents, both environmentally and endogenously, leading to an array of mutational or deleterious lesions in the genome. To maintain genomic stability, cells must continually repair such damage in a timely manner [Lindahl, 1993]. Common forms of DNA damage include oxidation and alkylation of DNA bases, ultraviolet light-induced pyrimidine dimers, base mismatches, and single- and double-strand breaks. The structure and repair mechanisms for these forms of DNA damage have all been studied extensively. However, one category of DNA lesions that remains poorly understood is DNA crosslinks, of which there are two general types: DNA-DNA crosslinks and DNA-protein crosslinks. A DNA-DNA crosslink lesion entails a covalent bond linking either the same strand of DNA (DNA intrastrand crosslinks) or opposing strands of DNA (DNA interstrand crosslinks, ICLs). DNA-protein crosslinks (DPCs) are formed when a nucleotide residue on DNA forms a covalent bond with a protein (or a peptide, to form a DNA-peptide crosslink, DpC). Crosslinks are particularly hazardous, as they can effectively block gene transcription and DNA replication. Repair of crosslinks appears to involve complex repair mechanisms encompassing a large number of protein factors. While the overall pathway(s) of DPC repair have yet to be fully unraveled, several possible mechanisms have recently begun to emerge. In this article, we summarize current progress and postulate potential mechanisms of DPC repair.
I. Formation of DPCs
DPCs can form through a variety of different means, facilitated by naturally occurring and synthetic compounds. Such compounds can broadly be categorized as environmentally-induced, therapeutically-induced, and endogenously-induced (Figure 1).
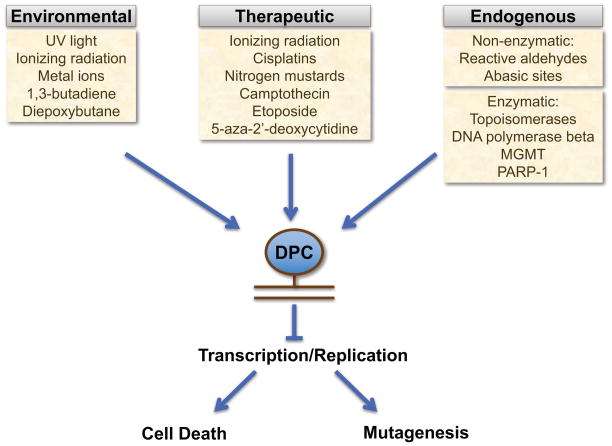
Figure 1. A variety of sources induce DPCs in cells.
DPCs can be formed through a variety of different means, including from environmental, therapeutic, and endogenously-generated agents. DPCs arrest transcription and replication, resulting in mutagenesis or cell death.
a. Environmentally-Induced DPCs
Several different DPC-inducing agents exist in our environments. DPCs can be induced upon exposure to ionizing radiation, ultraviolet light, and various transition metal ions, including chromium and nickel [Swenberg et al., 2011]. A number of other carcinogens are also known to induce DPCs, including bifunctional alkylating agents such as 1,3-butadiene, diepoxybutane, acrolein, and crotonaldehyde [Costa et al., 1997; Kurtz and Lloyd, 2003; Loeber et al., 2006; Minko et al., 2008].
b. Therapy-Induced DPCs
Ionizing radiation and chemical compounds that induce interstrand crosslinking or DNA-protein crosslinking are routinely used in chemotherapy regimens, either independently or in conjunction with other regimens [Huang and Li, 2013]. Each gray of ionizing radiation is thought to produce about 150 DPCs in the genome per cell [Barker et al., 2005]. Several cancer drugs induce DPCs, including nitrogen mustards, 5-aza-2′-deoxycitidine (5-azadC, also clinically known as decitabine), and platinum-based agents such as cisplatin- and transplatin-derivatives [Santi et al., 1983; Barker et al., 2005; Loeber et al., 2009; Ide et al., 2011; Stingele and Jentsch, 2015]. The chemotherapeutic drugs camptothecin and etoposide are used clinically to induce a specialized class of DPCs, as will be discussed later.
c. Endogenously-Induced DPCs
Endogenously-induced DPCs can be derived through enzymatic and non-enzymatic means. Certain DNA-interacting enzymes that otherwise form transient covalent complexes with DNA can become entrapped onto the DNA, forming DPCs. There are also multitudes of endogenously-produced metabolites and other species that lead to the formation of DPCs in cells through non-enzymatic means. Reactive aldehydes are a well-known class of DPC-inducing agent occurring endogenously in cells.
i. Non-Enzymatic
Assortments of reactive aldehydes are present in cells [Swenberg et al., 2011]. Reactive aldehydes are often generated as a result of various metabolic and regulatory processes in cells, such as amino acid metabolism and oxidative demethylation [Swenberg et al., 2011]. When the carbonyl carbon of an aldehyde is sufficiently electrophilic, it can react with nucleophiles in the surrounding environment [Dellarco, 1988]. When such an aldehyde is in the vicinity of chromatin, it can react with the primary amine of a DNA base, producing a methylol adduct; following a dehydration reaction to form a Schiff base, this intermediate adduct can then further react with a primary amine of another nearby DNA base (forming an ICL or intrastrand crosslink) or, alternatively, form a stable amide bond with a lysine or arginine residue of a nearby protein (forming a DPC). This mechanism of DPC formation has been validated experimentally, as NEIL1 has been shown to form this Schiff base intermediate with DNA [Bandaru et al., 2002].
Acetaldehyde, which is an intermediate in sugar metabolism as well as a key metabolite of ethanol, has also been demonstrated to react with the primary amines of DNA bases, yielding adducts of assorted chemical natures, including both DPCs and ICLs [Dellarco, 1988; Lorenti Garcia et al., 2009; Noguchi et al., 2017]. The nucleotide excision repair, homologous recombination, and Fanconi anemia pathways have each been shown to be important in the repair or prevention of DNA adducts formed by acetaldehyde, according to genetic studies in fission yeast [Noguchi et al., 2017].
The best-studied DPC-inducing aldehyde is formaldehyde. DPCs are the principal DNA lesion induced by formaldehyde in cells [Conaway et al., 1996; Quievryn and Zhitkovich, 2000]. For this reason, formaldehyde has commonly been used as a DPC-inducing agent in many studies in the field thus far. However, it is important to recognize that formaldehyde generates an assortment of other lesions in the DNA as well, including ICLs [Chaw et al., 1980]. Such heterogeneity of lesions can complicate readouts of experiments, as it is difficult to distinguish whether the readouts are a consequence of formaldehyde-induced DPCs versus other lesions such as ICLs.
Formaldehyde is particularly genotoxic, given that it is released as a byproduct during histone demethylation at nucleosomes [Walport et al., 2012]. Formaldehyde is also produced as an intermediate during enzymatic removal of methyl groups from DNA [Trewick et al., 2002]. To help mitigate the problem of having such a toxic and reactive chemical species being produced at such close proximity to DNA, cells have evolved a class of enzymes called aldehyde dehydrogenases. These enzymes metabolize various aldehydes such as formaldehyde and acetaldehyde, effectively detoxifying them [Vasiliou et al., 2004]. Formaldehyde dehydrogenase converts formaldehyde to formic acid, thereby reducing formaldehyde levels and likely diminishing the abundance of DPCs in the genome. However, evidence suggests that the formaldehyde concentration in human blood typically ranges between 2–3 mg/L (66–100 micromolar) [Zhang et al., 2009], and so DPC lesions still may be expected to arise on a regular basis. Whether other cellular and metabolic sources also contribute to the steady state levels of circulatory formaldehyde is unclear. Genes associated with DNA repair, DNA damage tolerance, and chromatin remodeling have been identified as notable contributors to ameliorating formaldehyde sensitivity, according to genetic evidence in budding yeast [de Graaf et al., 2009].
Abasic sites, frequent endogenous DNA lesions arising from hydrolysis of nitrogenous bases, are formed during base excision repair and upon spontaneous base loss. Abasic sites harbor reactive aldehyde groups as well, which are capable of covalent reactions with nearby proteins [Sczepanski et al., 2010]. Several in vitro studies have demonstrated that various metabolically-relevant peptides covalently bind with DNA, especially through aldehydic adducts and abasic sites, forming DpCs [Kuykendall and Bogdanffy, 1994; Kurtz et al., 2002; Kurtz and Lloyd, 2003; Minko et al., 2005]. For instance, DNA-glutathione crosslinks form after treatment with chromate [Zhitkovich et al., 1995; Voitkun et al., 1998]. As DpCs are smaller and often more efficiently repaired, they are predicted to be somewhat less genotoxic to cells than are DPCs.
ii. Enzymatic
DPCs can arise from normal enzymatic transactions with DNA. Various enzymes form transient complexes covalently linked to DNA as a reaction intermediate. Sometimes these transient covalent complexes can become trapped onto the DNA, giving rise to an enzymatically-derived DPC. The most frequent occurrence is with topoisomerases upon inhibitor treatment [Pommier et al., 2006]. Abasic sites can misalign the DNA strands, preventing DNA religation and trapping intermediate Topoisomerase 1 cleavage complexes (Top1ccs) onto the DNA [Pourquier et al., 1997]. These complexes are commonly formed in cells while topoisomerases interact with DNA to relieve torsional stress, but they are typically only transient intermediates rather than persisting structures. Several commonly used chemotherapeutic agents, such as camptothecin and etoposide, specifically trap these Top1ccs or Topoisomerase 2 cleavage complexes (Top2ccs), respectively, onto the DNA by inhibiting the religation reaction, thereby preventing replication and transcription [Mao et al 2001; Pommier et al., 2006].
DPCs can also form during base excision repair (BER). DNA bases are frequently subjected to hydrolysis, oxidation, and alkylation [Lindahl, 1993]. When a DNA glycosylase cleaves the damaged base, an abasic site is formed. If these abasic sites become oxidized by free radicals or reactive oxygen species, they can form structurally distinct derivatives. One such oxidized abasic site is 2-deoxyribonolactone. Repair by short-patch BER can induce covalent DPCs with this lesion, whereas long-patch BER removes the lesion directly, thereby avoiding DPC formation [Hashimoto et al., 2001; Sung et al., 2005]. The most abundant DPC species involved in such oxidized abasic sites is DNA polymerase β (Polβ), according to a study using site-specific 2-deoxyribonolactone in a cell-free extract system [Sung et al., 2005]. Polβ has also been shown to form DPCs with Ape1-processed 2-deoxyribonolactone [DeMott et al., 2002]. Covalent crosslinks between 2-deoxyribonolactone and bifunctional DNA glycosylase/lyase enzymes have also been observed [Faure et al., 2005].
Other DNA repair enzymes have also been found to form DPCs. O6-methylguanine-DNA methyltransferase (MGMT), an enzyme responsible for removing alkyl adducts from DNA, readily forms DPCs when cells are treated with nitrogen mustard or the carcinogen 1,2,3,4-diepoxybutane [Loeber et al., 2006; Loeber et al., 2008; Cheng et al., 2016]. Poly(ADP-ribose) polymerase-1 (PARP-1), which is implicated in several DNA repair, DNA damage detection, and chromatin remodeling processes, has been shown to covalently bond with abasic sites derived during BER, forming a DPC [Prasad et al., 2014].
The chemotherapeutic drug 5-azadC specifically induces DPCs, covalently trapping various DNA methyltransferases (DNMTs) onto the DNA [Santi et al., 1983]. 5-azadC is a nucleoside analog that is metabolized by cells, then phosphorylated to 5-azadCTP, and subsequently incorporated into the DNA, partially substituting for cytosine bases. When DNMTs attempt to methylate this nucleotide analog, they become covalently entrapped on the DNA, forming a DPC. Therefore, 5-azadC is also a potent inhibitor of DNA methylation [Juttermann et al., 1994; Christman, 2002; Palii et al., 2008].
II. Cytotoxicity of DPCs
ICLs and DPCs are highly deleterious to living cells, constituting steric blockades to the DNA replication and DNA transcription machineries, as well as interfering with accessibility of DNA repair and chromatin remodeling factors [Barker et al., 2005]. If not repaired in a timely manner, these lesions can lead to deleterious mutations and cell death [Barker et al., 2005; Fu et al., 2011; Nakano et al., 2012; Nakano et al., 2013]. DNA and RNA polymerases are unable to extend past DPCs and some DpCs in vitro [Chvalova et al., 2007; Yeo et al., 2014]. DPCs also prevent strand separation during replication and repair. In E. coli, a 16 kDa DPC prevented UvrD (DNA helicase II) from separating DNA strands [Kumari et al., 2010]. DPCs less than 14.1 kDa could be cleared by several helicases [Nakano et al., 2013]. Replication fork stalling by DPCs is readily observed in E. coli [Kuo et al., 2007]. Treatment with 5-azadC or its nucleoside analog 5-azacytidine (5-azaC) leads to γ-H2AX and 53BP1 foci, indicating the presence of DNA damage and double-strand breaks [Palii et al., 2008; Orta et al., 2013].
Failure to repair DPCs leads to a variety of genotoxic consequences, such as chromatid breaks, chromosomal aberrations, and mutations [Stingele and Jentsch, 2015]. It has been observed several decades ago that acetaldehyde exposure increases the incidence of mutations, sister chromatid exchanges, micronuclei, and aneuploidy in mammalian cells [Dellarco, 1988]. Replication fork collapse is known to occur from acetaldehyde treatment in fission yeast [Noguchi et al., 2017]. 5-azadC also leads to replication fork collapse in mammalian cells [Orta et al., 2013]. Much like mitomycin C, DPC-inducing reactive aldehydes also cause chromosomal aberrations [Speit et al., 2000; Mechilli et al., 2008; Lorenti Garcia et al., 2009]. It has also been observed that DPCs do not induce sister chromatid exchanges to the same extent as mitomycin C-induced DNA ICLs [Lorenti Garcia et al., 2009]. This observation suggests that chromosomal aberrations observed upon DPC induction may be the result of a DPC repair mechanism less favorable for homologous recombination [Lorenti Garcia et al., 2009]. Even the generally less deleterious DpC adducts have demonstrated mutagenicity. For instance, glutathione DNA-peptide crosslinks and a 10-mer Myc peptide incur mutagenic consequences in human fibroblasts and HEK 293T cells, respectively, particularly generating single base substitutions [Voitkun et al., 1998; Pande et al., 2017]. Translesion synthesis polymerases are important in bypassing smaller DpC lesions, thereby alleviating the replication block [Duxin et al., 2014; Yeo et al., 2014; Wickramaratne et al., 2016; Pande et al., 2017]. It is likely that proteolytic processing of larger DPCs and DpCs enables bypass by translesion synthesis polymerases as well.
III. Mechanistic Framework of Processing DPC Lesions
The protein moiety of a DPC poses a major steric hindrance to DNA repair factors, which require direct access to the affected bases. As a result, the general strategy for removing DPCs involves proteolytic size reduction of the protein adduct (Figure 2). This can be accomplished in one of two ways. Proteolytic cleavage by metalloproteases – including Wss1 (in S. cerevisiae), Dvc-1 (in C. elegans), and SPRTN (in metazoans) – degrades DPCs. Some evidence also suggests that proteasome-mediated degradation can achieve proteolytic size reduction of DPCs [Quievryn and Zhitkovich, 2000; Reardon and Sancar, 2006; Reardon et al., 2006; Baker et al., 2007; de Graaf et al., 2009] (Figure 2). Proteolytic size reduction is expected to produce a peptide remnant covalently linked to the DNA strand, allowing repair factors to access the damaged base to complete repair and restore the DNA to its original state. Alternatively, the small peptide remnant may allow lesion bypass polymerases to synthesize over the adducted base, alleviating replication fork blockage.
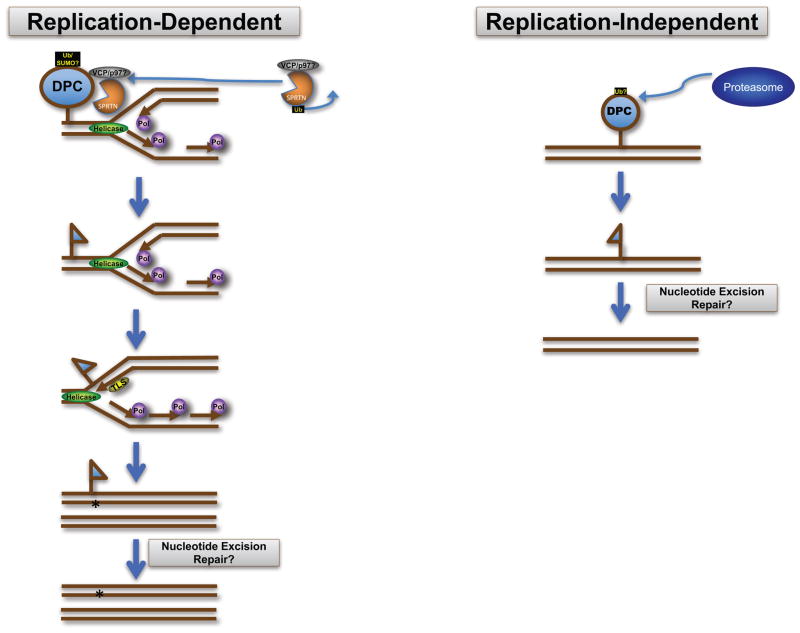
Figure 2. Proteolytic strategies for DPC removal.
In replication-dependent proteolytic size reduction, a leading-strand DPC prevents helicase-catalyzed strand separation. The DPC is cleaved by the protease SPRTN. The ubiquitin- and SUMO-dependent segregase VCP/p97/Cdc48 might assist DPC proteolysis. Following cleavage, the lesion remnant is bypassed by a translesion synthesis polymerase, possibly inducing a single base substitution. Nucleotide excision repair might excise the remaining lesion after replication has completed. In a possible replication-independent proteolytic size reduction, the proteasome directly degrades the protein portion of a DPC. Any remaining peptide remnant might be excised by the nucleotide excision repair pathway, restoring the damaged DNA to its original state.
a. Proteolytic Digestion of DPCs
Proteolytic cleavage of DPCs seems to be a highly conserved strategy to begin repair of these lesions, with DPC-specific proteases discovered in budding yeast, Xenopus, C. elegans, and mammalian systems. Processing of DPCs into smaller DpCs likely serves as an initial intermediate during the repair process [Quievryn and Zhitkovich, 2000].
The first proteolysis-based mechanism for DPC repair was characterized in budding yeast [Stingele et al., 2014]. In this system, the metalloprotease Wss1 promotes genome stability by specifically targeting DPC substrates, cleaving them into smaller adducts and enabling replication past the lesion [Stingele et al., 2014]. Yeast cells deficient in Wss1 are hypersensitive to formaldehyde-induced DPCs as well as drugs inducing Top1ccs on the DNA; these cells have elevated DPC levels and increased incidence of gross chromosomal rearrangements [Stingele et al., 2014].
Given that the size range of potential DPCs encompasses quite a wide spectrum, any protease that cleaves these lesions must exhibit broad substrate specificity. One way to achieve this broad specificity is to preferentially target Wss1 to sites of DNA damage, possibly via post-translational modification signaling. Because Wss1 bears two SUMO-interacting motifs, it is conceivable that DNA damage-associated SUMOylation might direct Wss1 to the appropriate sites [Stingele et al., 2014]. Supporting this conjecture, it has been observed that Top1ccs are SUMOylated, as are a number of proteins involved in the DNA damage response [Mao et al., 2000; Psakhye and Jentsch, 2012]. It will be interesting to investigate whether other DPCs are also targeted for SUMOylation, as well as whether this would indeed affect Wss1 recruitment to the lesion.
A study utilizing site-specific DPC-containing substrates processed in cell-free Xenopus egg extracts capable of DNA repair demonstrated further understanding of a replication-coupled DPC proteolysis mechanism [Duxin et al., 2014]. In this study, the eukaryotic replicative CMG helicase stalls upon encountering a DPC on the leading strand, effectively preventing further unwinding of the DNA duplex and arresting the replisome. The DPC is subsequently cleaved, and CMG navigates around the residual adduct. The leading strand then bypasses the cleaved DPC in a DNA Polymerase ζ (Polζ)-dependent manner. By comparison, DPCs on the lagging strand do not appear to cause major blockage to replication – lagging strand DPC adducts would only block extension of a single Okazaki fragment, which is approximately 150 nucleotides in length [Duxin et al., 2014]. In support of this difference, CMG was found to efficiently bypass streptavidin adducts situated only on the lagging strand [Fu et al., 2011].
Until recently, the identity of this replication-dependent protease in metazoans has remained elusive. Several studies have identified the metalloprotease SPRTN as a DPC-cleaving protease [Lopez-Mosqueda et al., 2016; Stingele et al., 2016; Vaz et al., 2016]. SPRTN is evolutionarily related to Wss1 in budding yeast, and they share similar domains in the C-terminus and other structural features. While Wss1 possesses a small ubiquitin-like modifier (SUMO)-binding domain, SPRTN harbors a ubiquitin-binding domain. Unlike Wss1, which binds chromatin directly [Stingele et al., 2014], SPRTN contains a PCNA-interacting peptide box that enables its recruitment to chromatin upon DNA damage [Centore et al., 2012]. Both Wss1 and SPRTN possess SHP-box domains that associate with the AAA ATPase VCP/p97/Cdc48, a ubiquitin- and SUMO-dependent segregase that plays several key roles in regulating the DNA damage response [Davis et al., 2012; Mosbech et al., 2012; Stingele et al., 2014; Balakirev et al., 2015]. Indeed, binding to this ATPase is required for in vivo function of Wss1 [Stingele et al., 2014]. It is conceivable that VCP/p97/Cdc48 might assist the proteolysis of DPC intermediates.
Prior to these studies, SPRTN was best known for its role in facilitating translesion synthesis [Centore et al., 2012; Davis et al., 2012; Ghosal et al., 2012; Juhasz et al., 2012; Machida et al., 2012; Mosbech et al., 2012]. Shortly thereafter, it was discovered that flies, mice, and human cells developed severe phenotypes that could not be explained by SPRTN’s role in translesion synthesis, indicating an additional function for SPRTN in maintaining genomic stability [Lessel et al., 2014; Maskey et al., 2014].
In mammalian cells, knockdown of SPRTN with siRNA resulted in hypersensitivity toward formaldehyde and the persistence of DPCs on DNA [Lopez-Mosqueda et al., 2016; Stingele et al., 2016; Vaz et al., 2016]. Consistently, replication-arrested worm larvae depleted of Dvc-1 exhibited severe sensitivity toward formaldehyde exposure [Stingele et al., 2016]. These results suggest that SPRTN is involved in DPC repair. SPRTN has several regulatory mechanisms to prevent rampant, uncontrolled proteolysis of proteins not involved in DPC lesions. The catalytic center of the protein is exposed to the solvent, making these regulatory mechanisms especially crucial to avoid excessive proteolysis on non-target proteins [Stingele et al., 2016]. The first of these mechanisms to be triggered involves a ubiquitin switch to control SPRTN activity. SPRTN in its monoubiquitinated state does not bind to chromatin; only when a DPC is detected does SPRTN become deubiquitinated, enabling the protein to bind to damaged chromatin sites. Therefore, the amount of SPRTN bound to chromatin is expected to be directly proportional to the abundance of DPC lesions [Stingele et al., 2016].
Upon chromatin association, SPRTN is subjected to another regulatory mechanism, wherein the protein will only be capable of substrate proteolysis when bound to a region of single-stranded DNA. When bound to double-stranded DNA, SPRTN is only capable of catalyzing its own autocleavage. SPRTN’s ability to facilitate autocleavage represents a third means of controlling undesirable proteolysis [Stingele et al., 2016]. Because SPRTN must be bound to single-stranded DNA in order to exert its proteolysis activity, a single-stranded DNA intermediate must exist in close vicinity to a DPC during the repair process. This is consistent with the study in Xenopus egg extracts demonstrating that DPC proteolysis occurs on both the leading and lagging strand templates during DNA replication [Duxin et al., 2014].
SPRTN has only a low protein specificity when processing DPC substrates, which enables it to target a wide spectrum of DPCs for proteolysis. At the same time, SPRTN has low affinity to non-crosslinked proteins, ensuring that protein cleavage only occurs upon proper induction of the protease activity, a feature that minimizes rampant proteolysis [Stingele et al., 2016]. These seem to be evolutionarily advantageous features for such an enzyme, as it must perform proteolysis of proteins in an extremely precise setting.
Besides its role in replication-dependent DPC proteolysis, SPRTN is also believed to have a replication-independent function. In flies, SPRTN is apparently recruited to chromatin outside of S phase, independent of the replisome [Delabaere et al., 2014]. This suggests that DPCs might be proteolytically processed outside the replication context, which would prove especially useful for quiescent, non-dividing cells, and perhaps transcription-blocking DPCs.
b. Proteasome-Mediated Degradation of DPCs
Proteasome-mediated degradation may serve as either an alternative or complementary means of conferring DPC size reduction, thereby enabling nucleotide excision repair to remove the residual peptide adduct [Quievryn and Zhitkovich, 2000; Reardon and Sancar, 2006; Reardon et al., 2006; Baker et al., 2007; de Graaf et al., 2009]. However, the role of the proteasome in facilitating DPC repair must be better understood.
The protein portions of various enzymatic DPCs are suggested to be targets for degradation by the proteasome. For example, Top1ccs and Top2ccs are targeted for proteasomal degradation [Desai et al., 1997; Mao et al., 2001]. Following proteasome-mediated degradation, a covalently-bound DNA-peptide adduct remains. This peptide adduct is then processed by the enzyme tyrosyl-DNA-phosphodiesterase 1 or 2 (Tdp1, Tdp2), which hydrolyzes the covalent bond between the DNA and cleaves the topoisomerase [Pouliot et al., 1999; Pommier et al., 2006]. Moreover, Polβ DPCs are ubiquitinated and processed by the proteasome, as reflected by the observation that MG132 treatment causes nuclear accumulation of these ubiquitinated lesions [Quinones et al., 2015]. Proteasome activity also reduces sensitivity to 5-azadC treatment in mammalian cells, though interestingly FANCG-deficient cells are not further sensitized when MG132 is administered. This indicates that the proteasome and Fanconi anemia proteins function in the same pathway when coping with 5-azadC-induced damage [Orta et al., 2013].
Consistently, several reports showed that proteasome inhibition incurs sensitivity to and hinders the repair of assorted non-enzymatic DPCs. In mammalian cells, formaldehyde-induced DPCs were less efficiently repaired following proteasome inhibition [Quievryn and Zhitkovich, 2000; Baker et al., 2007]. Chromium(VI)-mediated DPC repair seemed to require proteasome function [Zecevic et al., 2010]. Proteasome inhibition increased sensitivity of human lung cells toward formaldehyde, suggesting a possible role for the proteasome in repairing formaldehyde-mediated DNA adducts [Ortega-Atienza et al., 2015]. Besides improving survival, proteasome activity was also important for replication recovery and cell cycle restoration after exposure to formaldehyde [Ortega-Atienza et al., 2015]. It is conceivable that the protein portion of certain DPCs can be degraded by the proteasome.
A number of other studies reported that proteasome inhibition does not significantly affect non-enzymatic DPC repair efficiency [Nakano et al., 2009; Duxin et al., 2014]. For instance, two different proteasome inhibitors added to Xenopus egg extracts did not affect DPC repair in vitro. However, the investigators did observe that depletion of ubiquitin blocked effective DPC repair, indicating that ubiquitination per se, but not proteasome-dependent degradation, is critical for DPC repair [Duxin et al., 2014]. Ubiquitin depletion affects a host of cellular processes; whether the coinciding inhibition of DPC repair is a direct or indirect consequence is not clear. The precise roles of ubiquitination in DPC repair have yet to be elucidated. Whether certain DPCs are in fact ubiquitinated prior to their cleavage by SPRTN remains to be fully demonstrated. Moreover, it is important to determine whether specific forms of lysine linkages (e.g., K48 versus K63) are associated with polyubiquitinated DPCs. The role of the proteasome in DPC repair necessitates combined genetic and biochemical studies.
c. Nucleotide Excision Repair and DPCs
Many studies suggested that nucleotide excision repair (NER) is an important pathway in the repair of DPCs. In general, the evidence actually suggests that the NER dependence of DPC and DpC removal may be influenced strongly by the model organism and experimental approaches used.
Reconstituted in vitro systems have been used to test the hypothesis that NER directly repairs DPCs and DpCs [Minko et al., 2002; Minko et al., 2005; Reardon and Sancar, 2006]. UvrABC, the bacterial NER complex, excised a site-specific 16 kDa protein covalently attached to DNA; however, this incision was observed at diminished kinetics relative to incision of smaller adducted proteins [Minko et al., 2002; Minko et al., 2005]. The UvrABC complex was able to excise a model DNA-peptide crosslink in a more efficient manner [Minko et al., 2005]. Similarly, E. coli NER proteins are equally efficient at incising other peptides covalently bound to abasic sites [Minko et al., 2005].
Compared to reconstituted bacterial NER, reconstituted mammalian NER proteins excised these same crosslinks with a greater disparity in efficiency, wherein DpCs were removed with high efficiency but repair of a 16 kDa protein adduct seemed unattainable [Reardon and Sancar, 2006]. Further in vitro experiments demonstrate that mammalian NER is capable of removing small DPCs wherein the protein size is below 8 kDa [Nakano et al., 2009]. Proteins above this approximate threshold were not effectively removed [Nakano et al., 2009]. These studies suggest that adduct size plays a critical role in determining the efficiency with which NER will be able to excise a DPC or DpC in vitro.
Generally, core and linker histone DPCs are likely the most abundant species of DPCs in cells [Solomon and Varshavsky, 1985], given their direct vicinity to DNA. Considering that histones are above 8 kDa in size, it is unlikely that the NER mechanism is capable of independently removing DPCs in the absence of prior size reduction by proteolytic activities.
Using defined DPC substrates, the bacterial NER system was capable of repairing DPCs below 12–14 kDa [Nakano et al., 2007; Ide et al., 2011]. NER mutants were also hypersensitive to formaldehyde treatment, but not 5-azaC treatment [Nakano et al., 2007]. 5-azaC, like its nucleoside analog 5-azadC, specifically induces DPCs instead of a mix of different lesions when incorporated into the DNA [Christman, 2002]. Because 5-azaC traps DNMTs onto DNA [Santi et al., 1983], the adducted protein moiety – 53 kDa, in the case of Dcm methylase in E. coli – is substantially larger than the range with which bacteria can excise DPCs by NER in vivo. Thus, it stands to reason that these DPCs may be repaired in E. coli through other mechanisms, such as homologous recombination [Nakano et al., 2007]. Intriguingly, 5-azadC incurs sensitivity to BER-deficient mammalian cells. Specifically, loss of XRCC1 results in an accumulation of single- and double-strand breaks, as well as persistence of DNMT DPCs, indicating that BER may play a role in removing these types of DPC lesions [Orta et al., 2014].
NER-deficient mammalian cells have yielded conflicting results with regards to the NER pathway’s involvement in DPC repair [Fornace and Seres, 1982; Quievryn and Zhitkovich, 2000; Speit et al., 2000; Nakano et al., 2009]. While NER-deficient human cells were shown to be capable of removing formaldehyde-induced DPCs [Quievryn and Zhitkovich, 2000; Speit et al., 2000; Zecevic et al., 2010], NER-deficient fibroblasts were defective in repairing crosslinks induced by another DPC-inducing compound, transplatin, suggesting a role for NER in repairing specific forms of DPC lesions [Fornace and Seres, 1982]. Cells deficient in NER exhibited a higher level of chromosomal aberrations and micronuclei following formaldehyde treatment [Speit et al., 2000]. Furthermore, mammalian NER was unable to excise even small formaldehyde-induced DPCs of 7.4 or 8.0 kDa in vivo, conflicting with the in vitro experiments using mammalian NER, and also in contrast with the DPC sizes excised in bacterial in vivo experiments [Nakano et al., 2007; Nakano et al., 2009].
Given that DPCs exhibit a high degree of structural diversity, it is conceivable that the NER pathway can remove a subset of DPCs with certain structural characteristics. Indeed, a study in Chinese hamster ovary (CHO) cells demonstrated that the transcription-coupled repair/NER pathways are involved in repairing formaldehyde-induced DPCs, but not to the same degree with acetaldehyde-induced DPCs [Lorenti Garcia et al., 2009].
Taken together, the steric hindrance of protein adducts necessitates that proteolytic cleavage provide a means to achieve the necessary size reduction to process the lesion into intermediates that can be effective NER substrates. Thus, a combined proteasome/protease-NER mechanism is a viable mechanism for the removal of DPCs.
d. Homologous Recombination and DPCs
Several studies suggested homologous recombination (HR) as a critical pathway for the repair of DPCs. In E. coli, HR can be utilized when the DPC is too large for direct NER processing [Nakano et al., 2007]. In bacteria, DPCs above 12–14 kDa depends on RecBCD-mediated HR, as such DPCs appear too large to be efficiently excised by the NER complex [Nakano et al., 2007; Ide et al., 2011]. HR in bacteria is able to participate in the repair of DPCs independent of adduct size [Nakano et al., 2007]. In mammalian cells, HR appears to play a key role in tolerance of all formaldehyde-mediated DPCs, even those as small as 7.4 kDa [Nakano et al., 2009]. It has been reported that HR-deficient cells also exhibit sensitivity toward acetaldehyde, suggesting that recombination is involved in the repair of various aldehyde-induced DPCs [Mechilli et al., 2008]. DNMT DPCs induced by 5-azadC yielded increased RAD51 foci and sister chromatid exchanges, indicating that HR is critical to repair this form of damage [Orta et al., 2013].
Yeast mutants in HR were particularly sensitive toward low-dose, chronic formaldehyde exposure [de Graaf et al., 2009]. Yeast NER mutants, on the other hand, displayed mild sensitivity when presented with a similar formaldehyde exposure regime; however, acute treatment with a high dose of formaldehyde required NER proteins to a much higher extent than HR proteins. In support of this data, acute formaldehyde treatment involves NER-dependent single-strand breaks in yeast, without formation of double-strand breaks, even when HR genes are deleted [de Graaf et al., 2009]. A report in hamster cells confirms the finding that formaldehyde treatment does not induce detectable double-strand breaks [Speit et al., 2000]. Moreover, double-strand break formation is not observed during the replication-dependent repair of DPCs in Xenopus egg extracts [Duxin et al., 2014]. This is in stark contrast with studies in E. coli and chicken cells demonstrating the need for double-strand break intermediates in processing DPCs [Nakano et al., 2007; Ridpath et al., 2007]. RAD52 and Wss1 contribute to the repair of formaldehyde-induced DPCs in a non-epistatic manner in yeast, indicating that HR and proteolysis may be separate strategies for enabling DPC processing [Stingele et al., 2014]. Peculiarly, yeast strains that were depleted of HR proteins Mre11 and RAD52 were less sensitive to the cytotoxic effects of acute, high-dosage formaldehyde treatment than were wild-type strains. This suggests that HR in some contexts can be detrimental in response to formaldehyde-induced DPCs, possibly disrupting preferable repair processes such as NER [de Graaf et al., 2009]. Thus, the involvement of HR and NER in DPC repair may depict a pathway choice governed by several different factors, such as protein adduct size, levels of DPC damage, context of exposure to DPC-inducing agents, and the replication status of the cell.
e. Fanconi Anemia Pathway and DPC Repair
Fanconi anemia (FA) proteins are involved in the DNA damage response upon recognition of ICLs [Kottemann and Smogorzewska, 2013]. While the precise roles of FA proteins in recognizing and facilitating the repair of ICLs has been heavily pursued, it remains unclear whether these proteins have a direct role in repairing DPC lesions [Duxin and Walter, 2015].
Several studies implicate various FA proteins in the repair of DPCs [Ridpath et al., 2007; Langevin et al., 2011; Rosado et al., 2011]. FA-deficient chicken and mammalian cell lines exhibited hypersensitivity toward formaldehyde and 5-azadC, respectively [Ridpath et al., 2007; Rosado et al., 2011; Orta et al., 2013]. CHO cells depleted in FANCG were unable to engage in HR to repair DNMT DPCs incurred by 5-azadC, resulting in increased chromatid breaks and radial fusions [Orta et al., 2013]. This same FANCG-depleted CHO cell line exhibited high sensitivity toward acetaldehyde [Mechilli et al., 2008], though formaldehyde treatment did not incur the same level of sensitivity [Lorenti Garcia et al., 2009]. FANCD2-depleted cells, however, did exhibit sensitivity toward formaldehyde [Karanja et al., 2014; Vaz et al., 2016]. Intake of acetaldehyde in human cells resulted in FANCD2 monoubiquitination [Marietta et al., 2009; Abraham et al., 2011]. Upon acetaldehyde exposure, cells depleted of FANCQ/XPF and FANCG also had an elevated incidence of chromosomal aberrations [Mechilli et al., 2008; Lorenti Garcia et al., 2009]. Collectively, these studies suggest roles for FA proteins in repairing lesions incurred by reactive aldehydes and 5-azadC.
Other studies have found that FA proteins are not required for repairing DPC lesions. Immunodepletion of FANCI-FANCD2 from Xenopus egg extracts only inhibited ICL repair, but not DPC repair/bypass [Duxin et al., 2014]. FANCD2 depletion in C. elegans larvae did not affect formaldehyde sensitivity, even in the absence of Dvc-1 [Stingele et al., 2016]. Consistently, mouse embryonic fibroblasts deficient in FANCD2 were not defective in formaldehyde-induced DPC repair [Stingele et al., 2016]. FANCD2 depletion also had no effect on DPC accumulation as measured by DPC detection from chromatin [Vaz et al., 2016]. While FANCD2-depleted cells did exhibit sensitivity toward formaldehyde in HeLa cells, camptothecin treatment had no such effect [Vaz et al., 2016], suggesting that sensitivity toward formaldehyde in these cells might be due primarily to the ratio of ICL lesions formaldehyde is known to induce [Chaw et al., 1980]. However, it is important to note that these results do not preclude the possibility that the FA pathway plays a role in DPC repair, given that Tdp1 can process the Top1ccs induced by camptothecin.
So far, results from different groups remain conflicting with regards to the precise role of the FA pathway in DPC repair. It is possible that DPCs encountered during replication are processed by the FA pathway, rendering its function as indirect. Perhaps some DPC structures require FA-mediated nucleolytic incisions. Given that the established FA pathway function is to repair ICLs, further studies are needed to improve the understanding of FA mechanisms in DPC repair by using agents that specifically induce DPCs and/or defined DPC lesions.
IV. Diseases Associated with DPC Repair Defects
Deficiencies in DPC repair mechanisms are related to various diseases, including Ruijs-Aalfa syndrome and perhaps Fanconi anemia.
Formaldehyde has been shown to be tumorigenic in rodent models [Monticello et al., 1996]. Exposure to formaldehyde significantly increases risks of nasopharyngeal cancer, squamous cell carcinomas, and leukemia in mammals [Schwilk et al., 2010; Swenberg et al., 2011]. However, it is unclear whether it is formaldehyde-induced DPCs that cause this increased susceptibility to leukemia, as rats inhaling formaldehyde did not develop detectable formaldehyde-DNA adducts in their bone marrow [Lu et al., 2010].
SPRTN-knockout mice are embryonically lethal, demonstrating the critical importance of this protein. However, hypomorphic mice exhibit genomic instability and premature aging [Maskey et al., 2014], similar to phenotypes observed in human patients with SPRTN mutations. Three patients have been identified with germline mutations in SPRTN that lead to Ruijs-Aalfa syndrome, a disease associated with genomic instability, accelerated aging, and early-onset hepatocellular carcinoma [Ruijs et al., 2003; Lessel et al., 2014; Maskey et al., 2014]. Indeed, the DPC repair defect of SPRTN mutants in these patients likely causes this disease [Lopez-Mosqueda et al., 2016; Stingele et al., 2016; Vaz et al., 2016]. This might explain why Ruijs-Aalfa syndrome is associated with early-onset hepatocellular carcinoma, as reactive aldehydes are produced extensively as a byproduct of detoxification of metabolites and alcohol. Therefore, it stands to reason that DPCs may be substantially more abundant in the liver than other tissues. However, because most liver cells exist in a non-replicative state, DPCs may need to be processed by SPRTN in a replication-independent manner, as described earlier.
DPCs may also contribute to the Fanconi anemia phenotype. Fanconi anemia is a rare, recessive hereditary disease characterized by chromosomal instability and hypersensitivity to DNA ICLs. Patients with Fanconi anemia eventually develop bone marrow failure and have a substantially heightened risk of acute myeloid leukemia and solid tumors [Kottemann and Smogorzewska, 2013]. Studies in mice have shown synthetic phenotypes between FA gene knockouts and aldehyde dehydrogenases such that these mice develop anemia and leukemia in response to ethanol exposure [Langevin et al., 2011; Rosado et al., 2011; Garaycoechea et al., 2012]. Aldehyde dehydrogenases act as aldehyde-detoxifying enzymes, metabolizing reactive aldehydes into more benign chemical species [Vasiliou et al., 2004]. Thus, deficiency in aldehyde dehydrogenases allows for the accumulation of reactive aldehydes that may in turn form deleterious DNA crosslinks, including, and perhaps predominantly, DPCs. Given that the mice also share phenotypic manifestations of Fanconi anemia, it is plausible that endogenous aldehydes cause or exacerbate the phenotype of this disease. Thus, the FA pathway may have roles in repairing DPC lesions that are yet to be fully defined.
Concluding Remarks
DNA-protein crosslinks present a major risk to genomic stability, as they constitute potent blocks to essential DNA functions, including transcription and replication. The challenge in removing DPCs may come from their excessive bulkiness, which hinders accessibility of canonical DNA repair factors. DPCs arise from numerous sources, with a wide spectrum of protein moieties, allowing an extremely diverse array of structures. Recent studies describe the proteolytic processing by DPC-specific proteases Wss1 and SPRTN. While progress has been made in detailing the mechanisms of regulating SPRTN activity, many questions still remain. For instance, little is known about its recruitment to DPC lesions. What is the nature of the ubiquitination critical for replication-coupled DPC proteolysis? Does its p97-interacting motif, which is thought to play a role in SPRTN’s involvement in regulating translesion synthesis, also have a role in modulating its proteolytic activity? SPRTN is an essential gene in mammals, and so its absence would disable a crucial means of removing DPCs. In this light, the hurdles posed to cells by these lesions may be even more profound than initially thought. Precisely how abundant DPCs are and how often they arise in the genome remains unclear. Finally, what is the role of the Fanconi anemia pathway, NER pathway, and homologous recombination in resolving DPCs? An enhanced understanding of these cellular processes will almost certainly provide insightful and exciting avenues for improving chemotherapy and disease treatment.
Acknowledgments
We thank Erica Lynn for helpful comments on the manuscript. This work was supported by the National Institutes of Health (CA179441, CA193124-Project 3 to Lei Li) and the Oliver Stringer Endowed Professorship (Lei Li).
Footnotes
COMPLIANCE AND ETHICS
The authors declare that they have no conflict of interest.
References
- Abraham J, Balbo S, Crabb D, Brooks PJ. Alcohol metabolism in human cells causes DNA damage and activates the Fanconi anemia-breast cancer susceptibility (FA-BRCA) DNA damage response network. Alcoholism, clinical and experimental research. 2011;35:2113–2120. doi: 10.1111/j.1530-0277.2011.01563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wuenschell G, Xia L, Termini J, Bates SE, Riggs AD, O’Connor TR. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. The Journal of biological chemistry. 2007;282:22592–22604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- Balakirev MY, Mullally JE, Favier A, Assard N, Sulpice E, Lindsey DF, Rulina AV, Gidrol X, Wilkinson KD. Wss1 metalloprotease partners with Cdc48/Doa1 in processing genotoxic SUMO conjugates. eLife. 2015:4. doi: 10.7554/eLife.06763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA repair. 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutation research. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Centore RC, Yazinski SA, Tse A, Zou L. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Molecular cell. 2012;46:625–635. doi: 10.1016/j.molcel.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw YF, Crane LE, Lange P, Shapiro R. Isolation and identification of cross-links from formaldehyde-treated nucleic acids. Biochemistry. 1980;19:5525–5531. doi: 10.1021/bi00565a010. [DOI] [PubMed] [Google Scholar]
- Cheng J, Ye F, Dan G, Zhao Y, Wang B, Zhao J, Sai Y, Zou Z. Bifunctional alkylating agent-mediated MGMT-DNA cross-linking and its proteolytic cleavage in 16HBE cells. Toxicology and applied pharmacology. 2016;305:267–273. doi: 10.1016/j.taap.2016.06.022. [DOI] [PubMed] [Google Scholar]
- Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Chvalova K, Brabec V, Kasparkova J. Mechanism of the formation of DNA-protein cross-links by antitumor cisplatin. Nucleic acids research. 2007;35:1812–1821. doi: 10.1093/nar/gkm032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway CC, Whysner J, Verna LK, Williams GM. Formaldehyde mechanistic data and risk assessment: endogenous protection from DNA adduct formation. Pharmacology & therapeutics. 1996;71:29–55. doi: 10.1016/0163-7258(96)00061-7. [DOI] [PubMed] [Google Scholar]
- Costa M, Zhitkovich A, Harris M, Paustenbach D, Gargas M. DNA-protein cross-links produced by various chemicals in cultured human lymphoma cells. Journal of toxicology and environmental health. 1997;50:433–449. doi: 10.1080/00984109708984000. [DOI] [PubMed] [Google Scholar]
- Davis EJ, Lachaud C, Appleton P, Macartney TJ, Nathke I, Rouse J. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nature structural & molecular biology. 2012;19:1093–1100. doi: 10.1038/nsmb.2394. [DOI] [PubMed] [Google Scholar]
- de Graaf B, Clore A, McCullough AK. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA repair. 2009;8:1207–1214. doi: 10.1016/j.dnarep.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delabaere L, Orsi GA, Sapey-Triomphe L, Horard B, Couble P, Loppin B. The Spartan ortholog maternal haploid is required for paternal chromosome integrity in the Drosophila zygote. Current biology: CB. 2014;24:2281–2287. doi: 10.1016/j.cub.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Dellarco VL. A mutagenicity assessment of acetaldehyde. Mutation research. 1988;195:1–20. doi: 10.1016/0165-1110(88)90013-9. [DOI] [PubMed] [Google Scholar]
- DeMott MS, Beyret E, Wong D, Bales BC, Hwang JT, Greenberg MM, Demple B. Covalent trapping of human DNA polymerase beta by the oxidative DNA lesion 2-deoxyribonolactone. The Journal of biological chemistry. 2002;277:7637–7640. doi: 10.1074/jbc.C100577200. [DOI] [PubMed] [Google Scholar]
- Desai SD, Liu LF, Vazquez-Abad D, D’Arpa P. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. The Journal of biological chemistry. 1997;272:24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- Duxin JP, Dewar JM, Yardimci H, Walter JC. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell. 2014;159:346–357. doi: 10.1016/j.cell.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Walter JC. What is the DNA repair defect underlying Fanconi anemia? Current opinion in cell biology. 2015;37:49–60. doi: 10.1016/j.ceb.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure V, Saparbaev M, Dumy P, Constant JF. Action of multiple base excision repair enzymes on the 2′-deoxyribonolactone. Biochemical and biophysical research communications. 2005;328:1188–1195. doi: 10.1016/j.bbrc.2005.01.082. [DOI] [PubMed] [Google Scholar]
- Fornace AJ, Jr, Seres DS. Repair of trans-Pt(II) diamminedichloride DNA-protein crosslinks in normal and excision-deficient human cells. Mutation research. 1982;94:277–284. doi: 10.1016/0027-5107(82)90291-3. [DOI] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- Ghosal G, Leung JW, Nair BC, Fong KW, Chen J. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. The Journal of biological chemistry. 2012;287:34225–34233. doi: 10.1074/jbc.M112.400135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Greenberg MM, Kow YW, Hwang JT, Cunningham RP. The 2-deoxyribonolactone lesion produced in DNA by neocarzinostatin and other damaging agents forms cross-links with the base-excision repair enzyme endonuclease III. Journal of the American Chemical Society. 2001;123:3161–3162. doi: 10.1021/ja003354z. [DOI] [PubMed] [Google Scholar]
- Huang Y, Li L. DNA crosslinking damage and cancer - a tale of friend and foe. Translational cancer research. 2013;2:144–154. doi: 10.3978/j.issn.2218-676X.2013.03.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide H, Shoulkamy MI, Nakano T, Miyamoto-Matsubara M, Salem AM. Repair and biochemical effects of DNA-protein crosslinks. Mutation research. 2011;711:113–122. doi: 10.1016/j.mrfmmm.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Juhasz S, Balogh D, Hajdu I, Burkovics P, Villamil MA, Zhuang Z, Haracska L. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic acids research. 2012;40:10795–10808. doi: 10.1093/nar/gks850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanja KK, Lee EH, Hendrickson EA, Campbell JL. Preventing over-resection by DNA2 helicase/nuclease suppresses repair defects in Fanconi anemia cells. Cell cycle. 2014;13:1540–1550. doi: 10.4161/cc.28476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A, Minko IG, Smith RL, Lloyd RS, McCullough AK. Modulation of UvrD helicase activity by covalent DNA-protein cross-links. The Journal of biological chemistry. 2010;285:21313–21322. doi: 10.1074/jbc.M109.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HK, Griffith JD, Kreuzer KN. 5-Azacytidine induced methyltransferase- DNA adducts block DNA replication in vivo. Cancer research. 2007;67:8248–8254. doi: 10.1158/0008-5472.CAN-07-1038. [DOI] [PubMed] [Google Scholar]
- Kurtz AJ, Dodson ML, Lloyd RS. Evidence for multiple imino intermediates and identification of reactive nucleophiles in peptide-catalyzed beta-elimination at abasic sites. Biochemistry. 2002;41:7054–7064. doi: 10.1021/bi020026y. [DOI] [PubMed] [Google Scholar]
- Kurtz AJ, Lloyd RS. 1,N2-deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. The Journal of biological chemistry. 2003;278:5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- Kuykendall JR, Bogdanffy MS. Formation and stability of acetaldehyde-induced crosslinks between poly-lysine and poly-deoxyguanosine. Mutation research. 1994;311:49–56. doi: 10.1016/0027-5107(94)90072-8. [DOI] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- Lessel D, Vaz B, Halder S, Lockhart PJ, Marinovic-Terzic I, Lopez-Mosqueda J, Philipp M, Sim JC, Smith KR, Oehler J, Cabrera E, Freire R, Pope K, Nahid A, Norris F, Leventer RJ, Delatycki MB, Barbi G, von Ameln S, Hogel J, Degoricija M, Fertig R, Burkhalter MD, Hofmann K, Thiele H, Altmuller J, Nurnberg G, Nurnberg P, Bahlo M, Martin GM, Aalfs CM, Oshima J, Terzic J, Amor DJ, Dikic I, Ramadan K, Kubisch C. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nature genetics. 2014;46:1239–1244. doi: 10.1038/ng.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Loeber R, Michaelson E, Fang Q, Campbell C, Pegg AE, Tretyakova N. Cross-linking of the DNA repair protein Omicron6-alkylguanine DNA alkyltransferase to DNA in the presence of antitumor nitrogen mustards. Chemical research in toxicology. 2008;21:787–795. doi: 10.1021/tx7004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeber R, Rajesh M, Fang Q, Pegg AE, Tretyakova N. Cross-linking of the human DNA repair protein O6-alkylguanine DNA alkyltransferase to DNA in the presence of 1,2,3,4-diepoxybutane. Chemical research in toxicology. 2006;19:645–654. doi: 10.1021/tx0600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeber RL, Michaelson-Richie ED, Codreanu SG, Liebler DC, Campbell CR, Tretyakova NY. Proteomic analysis of DNA-protein cross-linking by antitumor nitrogen mustards. Chemical research in toxicology. 2009;22:1151–1162. doi: 10.1021/tx900078y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maddi K, Prgomet S, Kalayil S, Marinovic-Terzic I, Terzic J, Dikic I. SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks. eLife. 2016:5. doi: 10.7554/eLife.21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenti Garcia C, Mechilli M, Proietti De Santis L, Schinoppi A, Kobos K, Palitti F. Relationship between DNA lesions, DNA repair and chromosomal damage induced by acetaldehyde. Mutation research. 2009;662:3–9. doi: 10.1016/j.mrfmmm.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Lu K, Collins LB, Ru H, Bermudez E, Swenberg JA. Distribution of DNA adducts caused by inhaled formaldehyde is consistent with induction of nasal carcinoma but not leukemia. Toxicological sciences: an official journal of the Society of Toxicology. 2010;116:441–451. doi: 10.1093/toxsci/kfq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y, Kim MS, Machida YJ. Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell cycle. 2012;11:3395–3402. doi: 10.4161/cc.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. The Journal of biological chemistry. 2001;276:40652–40658. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marietta C, Thompson LH, Lamerdin JE, Brooks PJ. Acetaldehyde stimulates FANCD2 monoubiquitination, H2AX phosphorylation, and BRCA1 phosphorylation in human cells in vitro: implications for alcohol-related carcinogenesis. Mutation research. 2009;664:77–83. doi: 10.1016/j.mrfmmm.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskey RS, Kim MS, Baker DJ, Childs B, Malureanu LA, Jeganathan KB, Machida Y, van Deursen JM, Machida YJ. Spartan deficiency causes genomic instability and progeroid phenotypes. Nature communications. 2014;5:5744. doi: 10.1038/ncomms6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechilli M, Schinoppi A, Kobos K, Natarajan AT, Palitti F. DNA repair deficiency and acetaldehyde-induced chromosomal alterations in CHO cells. Mutagenesis. 2008;23:51–56. doi: 10.1093/mutage/gem042. [DOI] [PubMed] [Google Scholar]
- Minko IG, Kozekov ID, Kozekova A, Harris TM, Rizzo CJ, Lloyd RS. Mutagenic potential of DNA-peptide crosslinks mediated by acrolein-derived DNA adducts. Mutation research. 2008;637:161–172. doi: 10.1016/j.mrfmmm.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko IG, Kurtz AJ, Croteau DL, Van Houten B, Harris TM, Lloyd RS. Initiation of repair of DNA-polypeptide cross-links by the UvrABC nuclease. Biochemistry. 2005;44:3000–3009. doi: 10.1021/bi0478805. [DOI] [PubMed] [Google Scholar]
- Minko IG, Zou Y, Lloyd RS. Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1905–1909. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticello TM, Swenberg JA, Gross EA, Leininger JR, Kimbell JS, Seilkop S, Starr TB, Gibson JE, Morgan KT. Correlation of regional and nonlinear formaldehyde-induced nasal cancer with proliferating populations of cells. Cancer research. 1996;56:1012–1022. [PubMed] [Google Scholar]
- Mosbech A, Gibbs-Seymour I, Kagias K, Thorslund T, Beli P, Povlsen L, Nielsen SV, Smedegaard S, Sedgwick G, Lukas C, Hartmann-Petersen R, Lukas J, Choudhary C, Pocock R, Bekker-Jensen S, Mailand N. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nature structural & molecular biology. 2012;19:1084–1092. doi: 10.1038/nsmb.2395. [DOI] [PubMed] [Google Scholar]
- Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, Van Houten B, Iijima K, Tauchi H, Ide H. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. The Journal of biological chemistry. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Miyamoto-Matsubara M, Shoulkamy MI, Salem AM, Pack SP, Ishimi Y, Ide H. Translocation and stability of replicative DNA helicases upon encountering DNA-protein cross-links. The Journal of biological chemistry. 2013;288:4649–4658. doi: 10.1074/jbc.M112.419358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AM, Izumi S, Pack SP, Makino K, Ide H. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Molecular cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Nakano T, Ouchi R, Kawazoe J, Pack SP, Makino K, Ide H. T7 RNA polymerases backed up by covalently trapped proteins catalyze highly error prone transcription. The Journal of biological chemistry. 2012;287:6562–6572. doi: 10.1074/jbc.M111.318410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi C, Grothusen G, Anandarajan V, Martinez-Lage Garcia M, Terlecky D, Corzo K, Tanaka K, Nakagawa H, Noguchi E. Genetic controls of DNA damage avoidance in response to acetaldehyde in fission yeast. Cell cycle. 2017;16:45–58. doi: 10.1080/15384101.2016.1237326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orta ML, Calderon-Montano JM, Dominguez I, Pastor N, Burgos-Moron E, Lopez-Lazaro M, Cortes F, Mateos S, Helleday T. 5-Aza-2′-deoxycytidine causes replication lesions that require Fanconi anemia-dependent homologous recombination for repair. Nucleic acids research. 2013;41:5827–5836. doi: 10.1093/nar/gkt270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orta ML, Hoglund A, Calderon-Montano JM, Dominguez I, Burgos-Moron E, Visnes T, Pastor N, Strom C, Lopez-lazaro M, Helleday T. The PARP inhibitor Olaparib disrupts base excision repair of 5-aza-2′-deoxycytidine lesions. Nucleic acids research. 2014;42:9108–9120. doi: 10.1093/nar/gku638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Atienza S, Green SE, Zhitkovich A. Proteasome activity is important for replication recovery, CHK1 phosphorylation and prevention of G2 arrest after low-dose formaldehyde. Toxicology and applied pharmacology. 2015;286:135–141. doi: 10.1016/j.taap.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Molecular and cellular biology. 2008;28:752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande P, Ji S, Mukherjee S, Scharer OD, Tretyakova NY, Basu AK. Mutagenicity of a Model DNA-Peptide Cross-Link in Human Cells: Roles of Translesion Synthesis DNA Polymerases. Chemical research in toxicology. 2017;30:669–677. doi: 10.1021/acs.chemrestox.6b00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, Agama K, Nitiss JL, Redon C. Repair of topoisomerase I-mediated DNA damage. Progress in nucleic acid research and molecular biology. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- Pourquier P, Ueng LM, Kohlhagen G, Mazumder A, Gupta M, Kohn KW, Pommier Y. Effects of uracil incorporation, DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I. The Journal of biological chemistry. 1997;272:7792–7796. doi: 10.1074/jbc.272.12.7792. [DOI] [PubMed] [Google Scholar]
- Prasad R, Horton JK, Chastain PD, 2nd, Gassman NR, Freudenthal BD, Hou EW, Wilson SH. Suicidal cross-linking of PARP-1 to AP site intermediates in cells undergoing base excision repair. Nucleic acids research. 2014;42:6337–6351. doi: 10.1093/nar/gku288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Quievryn G, Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis. 2000;21:1573–1580. [PubMed] [Google Scholar]
- Quinones JL, Thapar U, Yu K, Fang Q, Sobol RW, Demple B. Enzyme mechanism-based, oxidative DNA-protein cross-links formed with DNA polymerase beta in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:8602–8607. doi: 10.1073/pnas.1501101112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon JT, Cheng Y, Sancar A. Repair of DNA-protein cross-links in mammalian cells. Cell cycle. 2006;5:1366–1370. doi: 10.4161/cc.5.13.2892. [DOI] [PubMed] [Google Scholar]
- Reardon JT, Sancar A. Repair of DNA-polypeptide crosslinks by human excision nuclease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4056–4061. doi: 10.1073/pnas.0600538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, Buerstedde JM, Gillespie DA, Sale JE, Yamazoe M, Bishop DK, Takata M, Takeda S, Watanabe M, Swenberg JA, Nakamura J. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer research. 2007;67:11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nature structural & molecular biology. 2011;18:1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- Ruijs MW, van Andel RN, Oshima J, Madan K, Nieuwint AW, Aalfs CM. Atypical progeroid syndrome: an unknown helicase gene defect? American journal of medical genetics Part A. 2003;116A:295–299. doi: 10.1002/ajmg.a.10730. [DOI] [PubMed] [Google Scholar]
- Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- Schwilk E, Zhang L, Smith MT, Smith AH, Steinmaus C. Formaldehyde and leukemia: an updated meta-analysis and evaluation of bias. Journal of occupational and environmental medicine. 2010;52:878–886. doi: 10.1097/JOM.0b013e3181ef7e31. [DOI] [PubMed] [Google Scholar]
- Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speit G, Schutz P, Merk O. Induction and repair of formaldehyde-induced DNA-protein crosslinks in repair-deficient human cell lines. Mutagenesis. 2000;15:85–90. doi: 10.1093/mutage/15.1.85. [DOI] [PubMed] [Google Scholar]
- Stingele J, Bellelli R, Alte F, Hewitt G, Sarek G, Maslen SL, Tsutakawa SE, Borg A, Kjaer S, Tainer JA, Skehel JM, Groll M, Boulton SJ. Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN. Molecular cell. 2016;64:688–703. doi: 10.1016/j.molcel.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele J, Jentsch S. DNA-protein crosslink repair. Nature reviews Molecular cell biology. 2015;16:455–460. doi: 10.1038/nrm4015. [DOI] [PubMed] [Google Scholar]
- Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell. 2014;158:327–338. doi: 10.1016/j.cell.2014.04.053. [DOI] [PubMed] [Google Scholar]
- Sung JS, DeMott MS, Demple B. Long-patch base excision DNA repair of 2-deoxyribonolactone prevents the formation of DNA-protein cross-links with DNA polymerase beta. The Journal of biological chemistry. 2005;280:39095–39103. doi: 10.1074/jbc.M506480200. [DOI] [PubMed] [Google Scholar]
- Swenberg JA, Lu K, Moeller BC, Gao L, Upton PB, Nakamura J, Starr TB. Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicological sciences: an official journal of the Society of Toxicology. 2011;120(Suppl 1):S130–145. doi: 10.1093/toxsci/kfq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug metabolism reviews. 2004;36:279–299. doi: 10.1081/dmr-120034001. [DOI] [PubMed] [Google Scholar]
- Vaz B, Popovic M, Newman JA, Fielden J, Aitkenhead H, Halder S, Singh AN, Vendrell I, Fischer R, Torrecilla I, Drobnitzky N, Freire R, Amor DJ, Lockhart PJ, Kessler BM, McKenna GW, Gileadi O, Ramadan K. Metalloprotease SPRTN/DVC1 Orchestrates Replication-Coupled DNA-Protein Crosslink Repair. Molecular cell. 2016;64:704–719. doi: 10.1016/j.molcel.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voitkun V, Zhitkovich A. Analysis of DNA-protein crosslinking activity of malondialdehyde in vitro. Mutation research. 1999;424:97–106. doi: 10.1016/s0027-5107(99)00011-1. [DOI] [PubMed] [Google Scholar]
- Voitkun V, Zhitkovich A, Costa M. Cr(III)-mediated crosslinks of glutathione or amino acids to the DNA phosphate backbone are mutagenic in human cells. Nucleic acids research. 1998;26:2024–2030. doi: 10.1093/nar/26.8.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walport LJ, Hopkinson RJ, Schofield CJ. Mechanisms of human histone and nucleic acid demethylases. Current opinion in chemical biology. 2012;16:525–534. doi: 10.1016/j.cbpa.2012.09.015. [DOI] [PubMed] [Google Scholar]
- Wickramaratne S, Ji S, Mukherjee S, Su Y, Pence MG, Lior-Hoffmann L, Fu I, Broyde S, Guengerich FP, Distefano M, Scharer OD, Sham YY, Tretyakova N. Bypass of DNA-Protein Cross-links Conjugated to the 7-Deazaguanine Position of DNA by Translesion Synthesis Polymerases. The Journal of biological chemistry. 2016;291:23589–23603. doi: 10.1074/jbc.M116.745257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo JE, Wickramaratne S, Khatwani S, Wang YC, Vervacke J, Distefano MD, Tretyakova NY. Synthesis of site-specific DNA-protein conjugates and their effects on DNA replication. ACS chemical biology. 2014;9:1860–1868. doi: 10.1021/cb5001795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecevic A, Hagan E, Reynolds M, Poage G, Johnston T, Zhitkovich A. XPA impacts formation but not proteasome-sensitive repair of DNA-protein cross-links induced by chromate. Mutagenesis. 2010;25:381–388. doi: 10.1093/mutage/geq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Steinmaus C, Eastmond DA, Xin XK, Smith MT. Formaldehyde exposure and leukemia: a new meta-analysis and potential mechanisms. Mutation research. 2009;681:150–168. doi: 10.1016/j.mrrev.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Zhitkovich A, Voitkun V, Costa M. Glutathione and free amino acids form stable complexes with DNA following exposure of intact mammalian cells to chromate. Carcinogenesis. 1995;16:907–913. doi: 10.1093/carcin/16.4.907. [DOI] [PubMed] [Google Scholar]