Abstract
Many studies have revealed pathways of epigenetic gene silencing by Polycomb Repressive Complex 2 (PRC2) in vivo, but understanding molecular mechanisms requires biochemistry. Here we analyze reconstituted human PRC2-nucleosome complexes. Histone modifications, the H3K27M cancer mutation, and inclusion of JARID2 or EZH1 in the PRC2 complex have unexpectedly minor effects on PRC2-nucleosome binding. Instead, protein-free linker DNA dominates the PRC2-nucleosome interaction. Specificity for CG-rich sequences is consistent with PRC2 occupying CG-rich DNA in vivo. Intriguingly, PRC2 preferentially binds methylated DNA via AEBP2, suggesting how DNA- and histone-methylation collaborate to repress chromatin. RNA is known to inhibit PRC2 activity. We find that RNA is not a methyltransferase inhibitor per se, but instead sequesters PRC2 from nucleosome substrates; this occurs because PRC2 binding requires linker DNA, and RNA and DNA binding are mutually exclusive. Together, we provide a model for PRC2 recruitment and a straightforward explanation of how actively transcribed portions of the genome bind PRC2 but escape silencing.
INTRODUCTION
Polycomb Repressive Complex 2 (PRC2) is a histone methyltransferase that specifically deposits methyl groups onto lysine 27 of histone H3. PRC2 is absolutely required for epigenetic silencing during embryonic development and cancer, and the importance of PRC2 for stem cell renewal and pluripotency is highlighted by the early embryonic lethality of mice with deletions of these genes1. In Drosophila, recruitment of Polycomb group proteins to target genes involves DNA sequences called Polycomb Responsive Elements (PREs)2, 3. However, how PRC2 is recruited to sites of action in mammalian cells has remained poorly understood.
Earlier work suggested that RNA could contribute to targeting PRC2 to specific genomic loci4 or holding it “poised but in check”5. PRC2 clearly interacts with RNA in vitro and in vivo, in both cases binding so many RNAs that its binding could be characterized as promiscuous6,7. Recently, our group demonstrated that PRC2 reads RNA motifs consisting of short repeats of consecutive guanines, which are ubiquitous in the human transcriptome8. While the hidden specificity in promiscuous RNA-binding has been revealed, the question of RNA functionality still remains: what role do RNA molecules play in the regulation of PRC2?
Two studies showed that RNA inhibits PRC2 catalytic activity on histone H3K275,9. Additional insight came from the Jenner lab, who concluded via pull-down experiments that purified nuclear RNA antagonizes PRC2 binding to chromatin10. Despite being an important initial observation, chromatin includes nucleosomes with different histone modifications, and whether such modifications would counteract RNA inhibition was not explored.
Pre-existing histone modifications have been proposed to be important determinants of PRC2 recruitment to genomic loci. Prior studies have shown that the PRC2 complex reads H3K27me3 marks of repressive chromatin and that binding to these histone modifications stimulates PRC2 enzymatic activity11. On the other hand, H3K4me3 and H3K36me3 marks of active chromatin are also recognized by PRC2, and such binding events inhibit PRC2 catalytic activity12. A histone-based perspective of PRC2 recruitment is epitomized by studies of pediatric high-grade gliomas, where it was suggested that a H3K27M missense mutation is able to sequester PRC2 and suppress its enzymatic function13. Although the equilibrium dissociation constants (Kd) for interaction of various modified histone peptides with PRC2 have been measured14 and the binding of modified histone peptides even visualized at atomic resolution15, the nucleosome-binding activity of PRC2 has remained thus far understudied16. In addition, knowing the affinity of PRC2 for nucleosomes vis-a-vis RNAs would inform models of PRC2-RNA functionality in mammals.
Here we undertake quantitative binding studies with recombinant PRC2 and reconstituted chromatin with and without RNA. We find that PRC2 binds protein-free nucleosome linkers and prefers GC-rich DNA, in agreement with bioinformatics showing PRC2 peaks at CG-rich DNA regions8, 17. Remarkably, the intrinsic properties of PRC2-nucleosome interactions provide a straightforward explanation for previously perplexing observations of PRC2 in vivo.
RESULTS
PRC2 binds longer nucleosome arrays with increased affinity
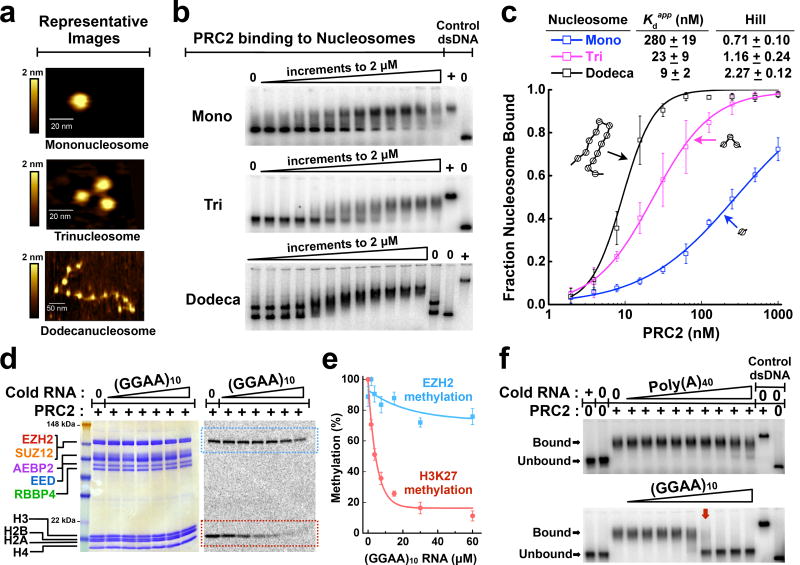
To quantify the binding affinities of human PRC2 with nucleosomes, we assembled 601-positioned mononucleosomes by standard salt gradient dialysis (Supplementary Fig. 1a,b). The 601-Widom sequence positions the nucleosome symmetrically with a DNA linker at each end18. EcoRI-digestion and atomic-force microscopy (AFM) verified that nucleosomes were neither under- nor over-saturated with histones (Fig. 1a and Supplementary Fig. 1c–e).
Figure 1.
RNA controls H3K27 methylation status by preventing PRC2 binding to nucleosomes. (a) Representative topographic images of mono-, tri-, and dodeca-nucleosomes as visualized by atomic-force microscopy. (b) PRC2 binding to 32P-radiolabeled mono-, tri- and dodeca-nucleosome substrates examined by EMSA. Data points represent 2-fold titrations ranging from 0–2 µM PRC2. (c) EMSA data fit with equilibrium binding curves.. Error bars give SD, n = 3 independent experiments. In this figure and others, several of the error bars are not visible because they are hidden by the size of the data point symbol. (d) Histone methyltransferase (HMTase) assays show that (GGAA)10 RNA inhibits H3K27 methylation. The persistence of EZH2 automethylation signal (blue-dashed box), but not H3K27 methylation signal (red-dashed box), indicates that RNA is not an active site inhibitor. 14C-SAM was used as a cofactor. PRC2 and histone proteins were visualized by Coomassie-staining (left-hand gel) and methylation levels were determined by 14C-autoradiography (right-hand gel). Data points represent 2-fold titrations ranging from 0–60 µM RNA. (e) Quantification of autoradiography gel in (d). Data fit with exponential decay curves. Error bars give SD, n = 3 independent experiments. (f) Pre-formed PRC2-trinucleosome complexes are disrupted by (GGAA)10 RNA, but not by negative control Poly(A)40 RNA. PRC2-trinucleosome complexes were fully disrupted (lane with red arrow) at a PRC2:RNA molar ratio of ~2:1. Data points represent 3-fold titrations ranging from 0–10 µM RNA. Uncropped gel images are shown in Supplementary Data Set 1.
Recombinant human PRC2 5-mer complex (EZH2−EED−SUZ12−RBBP4−AEBP2) was purified6 (Supplementary Fig. 1f,g). Binding of PRC2 to nucleosomes was measured by an Electrophoretic Mobility Shift Assay (EMSA), and the shifted bands were shown to contain PRC2 as well as nucleosomes (Supplementary Fig. 1h). The apparent dissociation constant (Kdapp) for mononucleosomes was 280 ± 19 nM (error defined in figure legend; Fig. 1b,c). Notably, this binding is dramatically stronger than the 40–400 µM Kdapp range measured for PRC2 binding to various H3 peptide substrates11, 14.
Previous studies19,20 reported that PRC2 prefers to methylate polynucleosomes over mononucleosomes; here we tested whether this preference might occur at the level of binding. We assembled tri- and dodeca-nucleosome arrays, both with a linker DNA length of 60 base pairs. The Kdapp values of PRC2 for tri- and dodeca-nucleosomes were 23 ± 9 nM and 9 ± 2 nM, respectively, much higher affinities than measured for mononucleosomes (Fig. 1b,c). Notably, we did not adjust these Kdapp values for the number of binding sites on tri- and dodeca-nucleosomes, so the affinity per nucleosome increases from mono- to tri-nucleosomes but does not increase further for dodecanucleosomes. In addition, cooperativity increased substantially for PRC2 binding to arrays compared to mononucleosomes (Hill coefficient, Fig. 1c). Thus, PRC2 prefers binding to tandem nucleosome repeats over mononucleosomes, with enhanced affinity largely achieved with trinucleosomes, and the dodecanucleosomes binding most cooperatively.
For our EMSA experiments, controls for protein-free DNA and the PRC2-DNA complex (right-hand lanes of each gel in Fig. 1b) address some potential concerns about the binding studies. Namely, the nucleosomes do not dissociate at the sub-nanomolar concentrations used in the binding reaction, because protein-free DNA runs distinguishably from nucleosomes on the agarose gel, and no free DNA is observed in the experimental lanes. Alternatively, if nucleosomes unraveled and the released free DNA were then bound by PRC2, the resulting PRC2-DNA complex would have lower mobility than the PRC2-nucleosome complexes; no such PRC2-DNA species was observed in the experimental lanes. The exception is the dodecanucleosomes (bottom panel of Fig. 1b), where half of the DNA is fully assembled and the other half runs as under-saturated arrays. In this case, both the fully assembled and under-saturated arrays are bound by PRC2.
RNA is not an active site inhibitor of PRC2 methyltransferase
RNA has been previously shown to inhibit PRC2 catalytic activity5,9. To quantitatively measure RNA-mediated enzymatic inhibition, PRC2 and in vitro reconstituted mononucleosomes were incubated with radiolabeled S-adenosylmethionine (14C-SAM) methyl donor, and RNA was titrated into the reaction. For this analysis, (GGAA)10 RNA (which forms G-quadruplexes) was used due to its optimal binding, and Poly(A)40 provided a negative control RNA that does not bind PRC25,8.
In the absence of RNA, our histone methyltransferase (HMTase) assays revealed the expected methylation of histone H3 (dashed red box, Fig. 1d). We also observed automethylation of the EZH2 subunit, as has been previously reported by other groups21, 22 (dashed blue box, Fig. 1d). As seen in Fig. 1e, the presence of (GGAA)10 RNA in the HMTase assay dramatically inhibited H3K27 methylation but not EZH2 automethylation. Poly(A)40 RNA, which does not bind to PRC2, had no observable inhibitory effects (Supplementary Fig. 1i).
It is striking that RNA had only a small effect on EZH2 automethylation, even at the highest RNA concentration tested (60 µM). It is useful here to note that an active-site mutation in EZH2 abolishes both automethylation and H3K27 methylation (X. Wang, R. Paucek, Y. Long, A. Gooding and T.R. Cech, personal observations), indicating that the methylation of EZH2 is intrinsic and not due to a contaminating protein. Thus, the persistence of automethylation in the presence of RNA indicates that the RNA is not itself an active-site inhibitor, but interferes with H3K27 methylation by other means.
One obvious hypothesis for the mechanism of RNA inhibition is that RNA simply disrupts the association of PRC2 with nucleosomes. Therefore, we titrated unlabeled RNA with pre-formed complexes of PRC2 and radiolabeled trinucleosomes. As shown in the top panel of Fig. 1f, (GGAA)10 RNA stripped PRC2 from nucleosomes. Dissociation was substantially complete at a sub-stoichiometric RNA concentration, consistent with each RNA molecule possibly interacting with two PRC2 complexes. In contrast, Poly(A)40 failed to compete (bottom panel, Fig. 1f). Together, these data support the conclusion that RNA and nucleosomes share the same or mutually exclusive binding sites for PRC2. These findings are in agreement with a recent study suggesting that the interaction of PRC2 with RNA and chromatin is mutually antagonistic10.
Modified histones have small effects on PRC2 binding or RNA inhibition
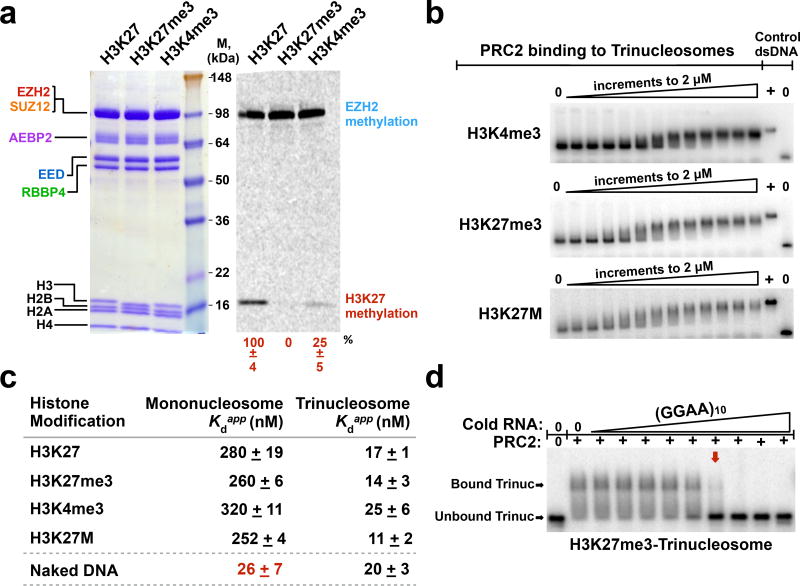
PRC2 is known to bind the N-terminal tail of H3 where it recognizes unmodified H3K27 via the EZH2 subunit, and it binds H3K4me3 and H3K27me3 at allosteric regulatory sites. Accordingly, we hypothesized that the binding of PRC2 to nucleosomes would be significantly affected if the nucleosomes carried these covalent modifications. More specifically, perhaps H3K27me3 histone modifications would counteract the inhibitory effects of RNA. We therefore reconstituted mono- and tri-nucleosomes containing unmodified H3, H3K4me3, or H3K27me3 modifications (Supplementary Fig. 2, 3 and 4a). We validated each of the modified nucleosomes with HMTase assays; PRC2 methylated the unmodified H3 nucleosomes, but not H3K27me3 nucleosomes (Fig. 2a). Furthermore, as expected, PRC2’s catalytic activity was diminished four-fold when nucleosomes contained H3K4me3 active marks (Fig. 2a). We also prepared nucleosomes harboring a H3K27M mutation, which is associated with pediatric glioma and thought to sequester PRC2 in vivo due to a tight binding interaction13.
Figure 2.
Histone modifications have small effects on PRC2 affinity for nucleosome substrates in vitro. (a) Validation of unmodified and modified (H3K27me3 and H3K4me3) nucleosome variants using HMTase assays. PRC2 and histone proteins were visualized by Coomassie-staining (left-hand gel); methylation levels were visualized by 14C-autoradiography (right-hand gel). Quantification of methylation signal relative to unmodified nucleosomes +SD, n = 3 independent experiments. (b) Representative EMSA gels for PRC2 binding to 32P-radiolabeled trinucleosomes with indicated histone modifications. 2-fold titrations ranged from 0–2 µM. (c) Quantifications of PRC2 binding affinity to mono- and tri-nucleosomes containing different modifications. SD was calculated from three replicates. Bottom row shows Kd values for PRC2 binding to protein-free DNA controls (207 bp mononucleosome template DNA and 621 bp trinucleosome template DNA). (d) (GGAA)10 RNA disrupts pre-formed complexes of PRC2-H3K27me3-trinucleosome. PRC2-H3K27me3-trinucleosome complexes were fully disrupted (red arrow) at a PRC2:RNA molar ratio of ~2:1. Data points represent 3-fold titrations ranging from 0–10 µM RNA. Uncropped gel images are shown in Supplementary Data Set 1.
Unexpectedly, PRC2 bound all of these nucleosome variants with similar affinity (Fig. 2b,c). Unmodified mononucleosomes were bound with Kdapp = 280 nM, while H3K27me3-nucleosomes and H3K27M-nucleosomes were bound only slightly more tightly. H3K4me3-nucleosomes bound to PRC2 with an affinity that was only marginally weakened (Fig. 2c and Supplementary Fig. 4b). Together, these data lead to the unanticipated conclusion that PRC2 binds nucleosomes containing different marks with similar nanomolar affinity.
Given the similar binding by PRC2 measured with modified nucleosomes, we anticipated that RNA would inhibit PRC2 binding to all of these modified nucleosomes. Indeed, we observed that RNA disrupted complexes of PRC2 and H3K27me3-, H3K4me3-, and H3K27M-nucleosomes at similar stoichiometry as observed previously for unmodified nucleosomes (Fig. 2d and Supplementary Fig. 4c). Therefore, we conclude that histone marks contribute little to the targeting of PRC2 to nucleosomes and do not impact the ability of RNA to sequester PRC2 away from nucleosomes.
Histone-free linker DNA dictates PRC2 binding to nucleosomes
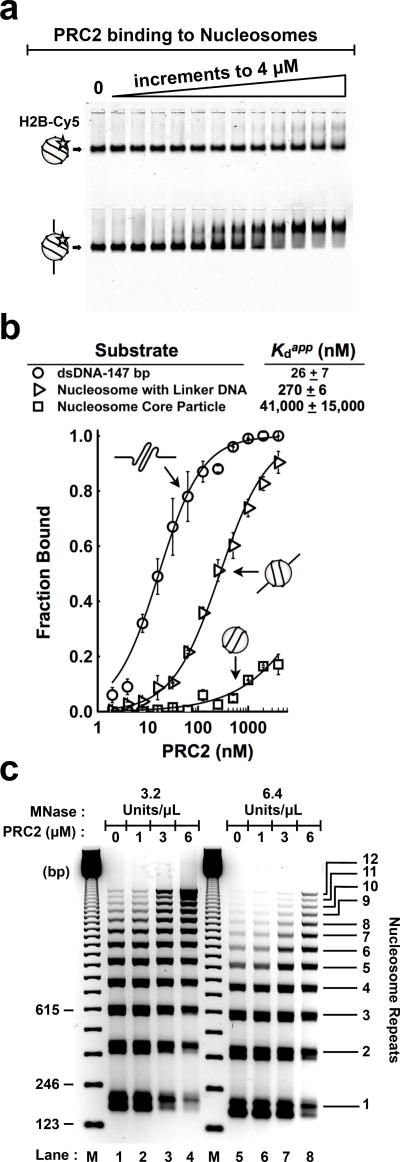
While investigating nucleosome-binding properties, we tested PRC2 binding to control naked DNA templates, expecting the affinities to the DNAs to be markedly weaker than their histone-coated counterparts. Surprisingly, the Kdapp values for PRC2 binding to the various naked DNA templates were all in the nanomolar range (Fig. 2c and Supplementary Fig. 4d). More remarkably, PRC2’s affinity for assembled mononucleosomes was 10-fold weaker than for the corresponding protein-free DNA.
The simplest explanation for this unexpected finding is that the histone octamer shields DNA surfaces that PRC2 would otherwise bind. In other words, we surmised that the protein-free linker regions of nucleosomes dictate PRC2 binding. To test this hypothesis, we reconstituted H2B-Cy5 labeled nucleosome core particles (NCPs) using 147 bp template DNA and mononucleosomes using 207 bp DNA (Supplementary Fig. 5a). Nucleosomes were purified by FPLC to assure homogeneity (Supplementary Fig. 5b). Astonishingly, with the linker DNA absent, PRC2 binding to nucleosomes was attenuated to a micromolar affinity of Kdapp = 40 ± 16 µM (Fig. 3a,b), consistent with the weak binding of PRC2 to histone tails14. Yet, robust PRC2 binding to nucleosomes was restored with the addition of linker DNA (Fig. 3a,b). And the strongest binding was observed for the protein-free 147 bp DNA (Fig. 3b and Supplementary Fig. 5c). These results with H2B-Cy5 fluorescently labeled nucleosomes were confirmed for nucleosomes labeled with 32P-DNA (Supplementary Fig. 5d).
Figure 3.
Nucleosome-free linker DNA dictates PRC2 binding to nucleosomes. (a) EMSA of PRC2 binding to H2B-Cy5 labeled nucleosome substrates. Nucleosome core particles lacking an accessible DNA linker (top gel) and mononucleosomes with 60 bp linker DNA (bottom gel), each at 2.5 nM. Binding reactions represent 2-fold titrations of PRC2 ranging from 0–4 µM. (b) PRC2 binds protein-free 147 bp dsDNA > mononucleosome with linker DNA >> nucleosome core particle. Error bars give SD, n = 3 independent experiments. (c) PRC2 protects linker regions of dodecanucleosome arrays from MNase-mediated digestion. M, 123-bp DNA ladder (1 µg per lane) size marker. Gel stained with ethidium bromide. Uncropped gel images are shown in Supplementary Data Set 1.
Because the lack of PRC2 binding to NCPs was unanticipated, we performed an additional test. We generated NCPs by treating 207 bp mononucleosomes with MNase-ExoIII23, which allowed the same nucleosome preparation to be tested with and without linker DNA. As shown in Supplementary Fig. 5e, MNase-ExoIII treatment successfully trimmed mononucleosomes to core particles. We found that binding to these trimmed NCPs was again attenuated to micromolar affinity (Supplementary Fig. 5f).
Given that PRC2 will rarely encounter isolated mononucleosomes in vivo, we proceeded to test whether PRC2 binds linker DNA in a chromatin array. Therefore, we subjected dodecanucleosome arrays to limited MNase digestion with or without PRC2. Typically, limited MNase digestion of nucleosome arrays produces double-stranded breaks within nucleosome linker regions. In the absence of PRC2, a ladder corresponding to the approximate positions of nucleosomes was revealed (Fig. 3c). However, as PRC2 was titrated into the reaction, PRC2 binding to DNA linker regions protected the dodecanucleosome arrays against MNase digestion. This is seen by the accumulation of higher molecular weight DNA fragments (lanes 4 and 8 of Fig. 3c and Supplementary Fig. 6a,b). Such a protection pattern has been observed for H1, a known linker histone24. Therefore, these data support the hypothesis that PRC2 binds linker regions of nucleosome arrays.
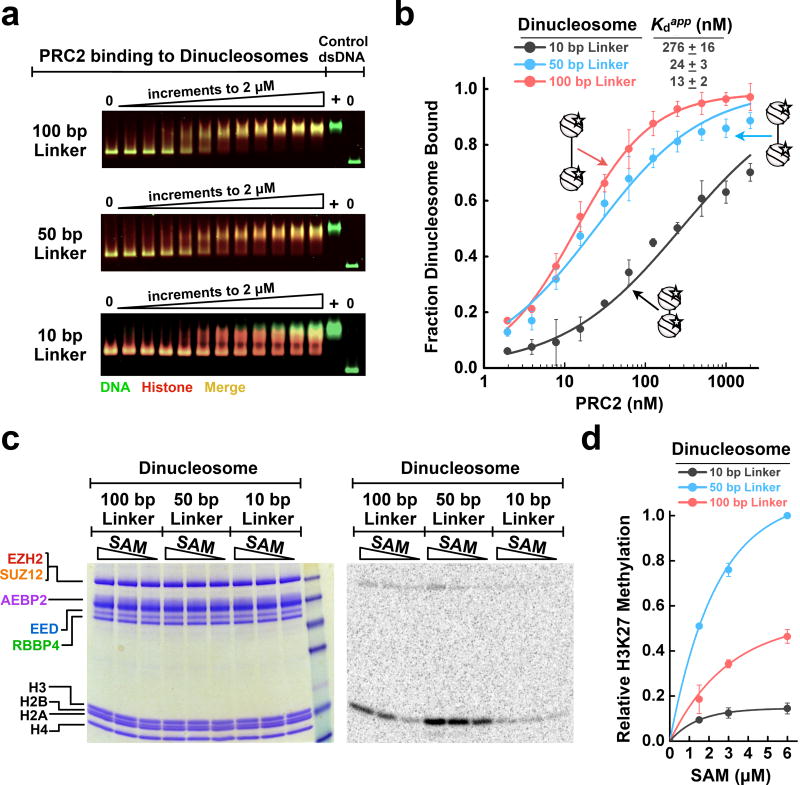
PRC2 prefers to bind and methylate long-linker dinucleosomes
PRC2 has been reported to occupy both promoter regions and inactive heterochromatin regions. However, these chromatin states may differ in nucleosome spacing and accessible linker DNA. Thus, we asked whether the spacing between two adjacent nucleosomes modulates PRC2 binding and activity. We reconstituted a fluorescent dinucleosome mimetic from a CpG island promoter of a PRC2-regulated gene17 (Supplementary Fig. 7a) by inserting a 100 bp linker between two 601-positioned nucleosomes. For comparison, we reconstituted dinucleosomes with a 50 or 10 bp linker, with the latter mimicking a gene-body dinucleosome25,26. Details of dinucleosome design are shown in Supplementary Fig. 7b,c. As shown by EMSA, PRC2 preferentially bound to long-linker dinucleosomes (Fig. 4a,b). Binding to dinucleosomes with a 10 bp linker was substantially diminished, yet PRC2 still bound with a nanomolar affinity of 276 ± 16 nM. Perhaps nucleosome breathing, the transient unwrapping of DNA from the histone core, could extend a short linker to allow binding of the entire PRC2 complex. Therefore, while PRC2 prefers binding to longer stretches of protein-free DNA, it is clear that PRC2 can still be targeted to more closely packed nucleosomes. Nevertheless, our conclusion still holds that a stretch of accessible DNA allows PRC2 to bind nucleosome substrates optimally.
Figure 4.
PRC2 binds and methylates long-linker dinucleosomes preferentially. (a) EMSA gels for PRC2 binding to fluorescent dinucleosomes with 10, 50, or 100 bp linker DNA joining the two nucleosomes. Histone proteins are site-specifically labeled with dye Cy5 on H2B (red channel); DNA was visualized by SYBR Green staining (green channel); assembled nucleosomes show an expected overlap of DNA and histones (yellow, merge). 2.5 nM of each nucleosome was incubated with 0–2 µM PRC2 in 2-fold increments. (b) EMSA data fit to binding curves. Error bars give SD, n = 3 independent experiments. (c) HMTase assays showing PRC2 enzymatic activity to dinucleosomes with 10, 50, or 100 bp linker DNA. Data points represent 6, 3, and 1.5 µM SAM. (d) Quantification of PRC2 HMTase. Error bars give SD, n = 3 independent experiments. Uncropped gel images are shown in Supplementary Data Set 1.
We hypothesized that dinucleosomes with a 100 bp linker length would have the greatest stimulation of HMTase due to optimal binding to PRC2. However, PRC2 activity did not increase monotonically with increasing dinucleosome spacing (Fig. 4c,d). Rather, there appears to be a “Goldilocks effect” with the 50 bp linker-dinucleosome stimulating PRC2 to a greater extent than either the 10 or 100 bp linker-dinucleosomes. Perhaps very long linker DNAs can position PRC2 too far from its H3 substrate for catalysis to occur.
PRC2 prefers CG-rich DNA and AEBP2 subunit confers mCpG specificity
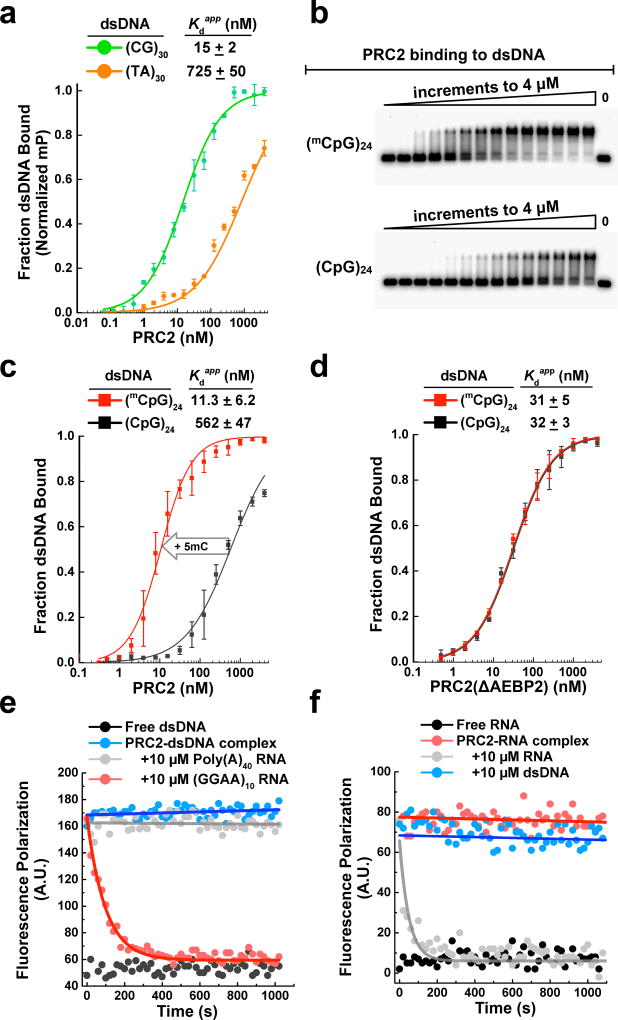
PRC2 has been broadly observed to localize with CpG islands (CGIs) in vivo, and it has been shown that there is a strong correlation between PRC2 occupancy and GC-rich sequences genome-wide8,17,27. It seemed likely to us that such targeting occurs via PRC2’s intrinsic DNA-binding activity. As shown in Fig. 5a, we tested PRC2 binding to (CG)30 and (TA)30 dsDNA substrates via fluorescence polarization. PRC2 bound the CG-rich substrate with a substantially lower Kdapp, in agreement with the previous in vivo observations.
Figure 5.
PRC2 preferentially binds CG-rich and CpG-methylated DNA. (a) PRC2 binding to (CG)30 and (TA)30 dsDNAs. Fluorescence polarization, mP = milli-polarization, 25 mM KCl. Error bars give SD, n = 3 independent experiments. (b) EMSA gels show that PRC2 recognizes mCpG dinucleotides in dsDNA. Top: (mCpG)24 dsDNA substrate; bottom: unmethylated (CpG)24 dsDNA substrate. Samples represent 2-fold titrations ranging from 0–4 µM PRC2. (c) EMSA data in (b) fit with equilibrium binding curves show that PRC2 has a substantial preference for binding methylated DNA. 100 mM KCl in binding buffer. Error bars give SD, n = 3 independent experiments. (d) Binding curves show that PRC2(ΔAEBP2) 4-mer complex does not discriminate between methylated and unmethylated DNA. Error bars give SD, n = 3 independent experiments. (e) Fluorescence polarization assays show pre-formed PRC2-dsDNA complex dissociation in the presence of RNA competitors. τ(GGAA)10 = 94.6 ± 4.4 s. Error is SD, n = 3 independent experiments. (f) Fluorescence polarization assays showing the lack of dissociation of pre-formed PRC2-RNA complex using dsDNA competitor. For all experiments, the binding buffer omitted yeast tRNA. Uncropped gel images are shown in Supplementary Data Set 1.
Given that 5-methyl-C (mC) is widespread throughout the mammalian genome and found on up to 80% of CpG dinucleotides28 and that methylation of CGIs near gene promoters is often associated with gene repression29, we hypothesized that PRC2 might preferentially bind mCpG sequences. As shown in Fig. 5b,c, we found that the methylated DNA, (mCpG)24, bound PRC2 with Kdapp = 11.3 ± 6.2 nM, which is about 50-fold stronger than the unmethylated (CpG)24 substrate. A similar strong preference for PRC2 binding to methylated DNA was found for TERT promoter DNA, which contained the eight mCpG dinucleotides found in vivo as determined by bisulfite sequencing analysis (J. Stern, R. Paucek, F. Huang, M. Ghandi, R. Nwumeh, J. Costello and T. Cech, personal communication).
To determine which subunit could be responsible for recognizing methylated CpG, we focused on AEBP2, which contains three well-conserved C2H2 zinc finger motifs. These motifs have been suggested to specifically recognize mCpG dinucleotides in other proteins30. We purified PRC2(ΔAEBP2) 4-mer complex and repeated the binding experiments with the methylated and unmethylated DNA substrates. PRC2(ΔAEBP2) lost binding preference for methylated DNA (Fig. 5d). To further corroborate this result, we introduced point mutations into each of AEBP2’s three conserved C2H2 zinc finger motifs (Supplementary Fig. 8a), and found that this mutant PRC2-AEBP2 5-mer complex also lost its preference for methylated DNA (Supplementary Fig. 8b). Together, these data suggest that AEBP2 is a regulator that helps the PRC2 complex read DNA epigenetic marks and that DNA methylation and histone lysine methylation might be mutually reinforcing.
RNA and DNA binding to PRC2 are mutually exclusive
Understanding the importance of linker DNA binding for PRC2-nucleosome interaction led us to hypothesize that RNA was not just inhibiting PRC2 binding to nucleosomes, but more precisely, was antagonizing PRC2 binding to protein-free linker DNA. Thus we speculated that RNA and DNA binding are competitive. As a test, we performed fluorescence polarization competition experiments where RNAs were titrated into reactions of pre-formed PRC2-A488-dsDNA complexes. Competitor RNA substrates were (GGAA)10 and a negative control Poly(A)40. In the absence of PRC2, free dsDNA gave a fluorescence polarization signal of about 55 A.U. (Fig. 5e). When PRC2-dsDNA complexes were formed, the fluorescence polarization signal rose to ~170 A.U. PRC2 was effectively competed from DNA using excess (GGAA)10 RNA with a half-life of 91 ± 5 s (Fig. 5e). Decreasing the RNA competitor concentration 10-fold did not change the half-life substantially (Supplementary Fig. 8c), indicating that the competitor RNA traps PRC2 upon dissociation from DNA rather than actively invading the PRC2-DNA complex. (If the competitor RNA actively invaded the PRC2-DNA complex, the measured half-life would decrease with increasing RNA concentration.) Notably, Poly(A)40 was inert and did not sequester PRC2. We also performed a gel-based competition where RNAs were titrated into reactions of pre-formed PRC2-dsDNA complexes (Supplementary Fig. 8d) and observed similar results.
Next, we reversed the competition experiment by titrating (CG)30 competitor DNA into reactions of pre-formed PRC2-A488-RNA complexes. In the absence of PRC2, free RNA gave a fluorescence polarization signal of ~9 A.U. (Fig. 5f). When PRC2-RNA complexes were formed, the signal rose to ~80 A.U. We found that 10 µM DNA competitor failed to compete PRC2 from RNA. Note, in the absence of tRNA competitor, the Kdapp values of RNA were even much tighter, in the picomolar range. These data collectively support the key conclusion that RNA and DNA have mutually exclusive binding to PRC2, with RNA binding with higher affinity.
JARID2 and EZH1 do not alter PRC2-nucleosome binding or RNA inhibition
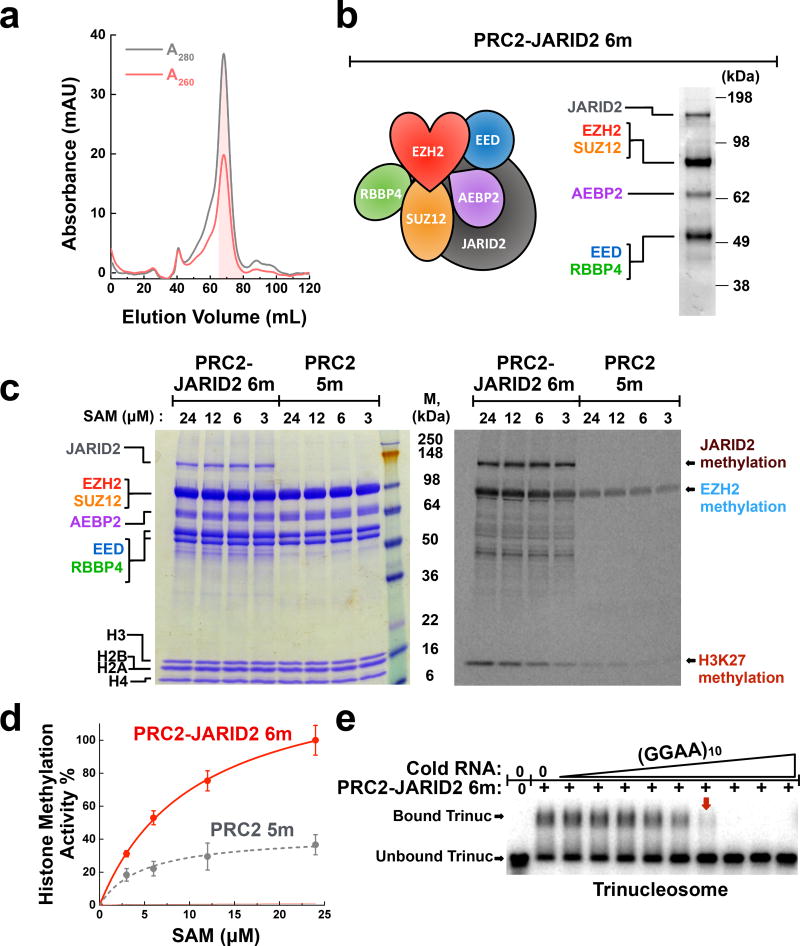
JARID2 and EZH1 are PRC2 components that have both been suggested to “load” PRC2 onto nucleosomes31. Therefore, we hypothesized that PRC2 affinity for nucleosomes would be enhanced upon the inclusion of JARID2 or upon substitution of EZH2 for EZH1. PRC2−JARID2 6-mer and PRC2−-EZH1 5-mer complexes were expressed and purified (see Methods). In both cases, a multi-step chromatography approach was used to ensure the purity of the complex, and the final size-exclusion column chromatogram showed a mono-dispersed peak (PRC2−JARID2 6-mer shown in Fig. 6a,b).
Figure 6.
JARID2 enhances PRC2 histone methyltransferase activity but does not prevent eviction by RNA. (a) Fractionation of PRC2−JARID2 6-mer over Sephacryl S-400 sizing column shows that the complex is monodisperse. A260/A280 ratio <0.7 indicates the lack of nucleic acid contamination. Shaded fractions were collected. (b) Typical SDS-PAGE gel showing the purity of recombinant PRC2-JARID2 6-mer complex. (c) A comparison of histone methyltransferase activity between PRC2−JARID2 6-mer and PRC2 5-mer using mononucleosome substrates. PRC2 and histone proteins visualized by Coomassie-staining (left-hand gel); methylation signal detected by 14C-autoradiography (right-hand gel). (d) Quantification of H3 methyltransferase activity. Error bars give SD, n = 3 independent experiments. (e) (GGAA)10 RNA disrupts pre-formed complexes of PRC2−JARID2-trinucleosome. Complexes were fully disrupted (red box arrow) at a PRC2:RNA molar ratio of ~2:1. Samples represent 3-fold titrations ranging from 0–10 µM RNA. Uncropped gel images are shown in Supplementary Data Set 1.
We first evaluated the enzymatic properties of each protein complex using nucleosome substrates. In our HMTase assay (Fig. 6c), the PRC2−JARID2 6-mer complex auto-methylated JARID2, as has been reported22. The JARID2 automethylation signal provides some additional evidence that our purification scheme preserves the integrity of JARID2. Furthermore, H3K27 methylation of nucleosomes by the reconstituted PRC2−JARID2 6-mer complex was ~3-fold improved when compared to PRC2 5-mer complex (Fig. 6c,d), consistent with published studies where JARID2 protein was titrated into PRC2-H3 reactions5,32. In the case of the PRC2−EZH1 5-mer complex, the enzymatic activity was substantially decreased (Supplementary Fig. 9a,b), in agreement with an in vivo study suggesting that EZH1 maintains basal H3K27 mono-methylation activity33.
We thought that these differences in activity might foreshadow differences in nucleosome-binding activity. However, neither the PRC2−JARID2 6-mer complex nor the PRC2−EZH1 5-mer complex had differences in nucleosome-binding when compared to the typical PRC2 5-mer complex (Supplementary Fig. 9c). In addition, both complexes showed trends similar to PRC2 5-mer in binding to RNA (Supplementary Fig. 9d) and to DNA (Supplementary Fig. 9e), and in RNA-mediated disruption of PRC2-nucleosome binding (Fig. 6e).
DISCUSSION
Numerous studies have established the importance of PRC2 for epigenetic gene silencing in mammals and mapped the genomic sites of H3K27me3 deposition in multiple cell types. Yet fundamental mechanistic questions remained. How does PRC2 bind chromatin? How is PRC2 recruited to particular sites in the genome? Does RNA binding to PRC2 regulate these events, and how? Here we use quantitative biochemistry with purified PRC2 and reconstituted nucleosomes to address those questions. Although in vivo there will undoubtedly be numerous exceptions to these “rules” established in vitro, these exceptions should stimulate the search for additional factors (e.g., non-histone chromosomal proteins or post-translational modifications) that explain why events in cells might diverge from the biochemical paradigm.
How does PRC2 bind chromatin?
Candidates for PRC2 recruitment to chromatin have included histone tails, DNA, non-histone proteins, and RNA1,34,35. Binding of PRC2 by various N-terminal tails of histone H3 is well established and has been directly observed by X-ray crystallography15,36. Yet, do such interactions provide sufficient binding energy, given measured affinities of histone peptide-PRC2 in the 40–400 µM range? Here we found that different histone marks make minor contributions to net PRC2 binding of nucleosomes in vitro (see below). Instead, the histone-free linker DNA has a central and direct role in the recruitment of PRC2 to an adjoining nucleosome core particle, with an observed binding affinity in the nanomolar range. In the absence of an available DNA linker, such as with a nucleosome core particle, PRC2 targeting to nucleosomes is substantially weakened in vitro. The groups of Beat Fierz and Juerg Mueller have also found that PRC2 binding to DNA provides the main contribution to its affinity for chromatin (Choi et al., 2017; accompanying manuscript), in agreement with our conclusion. Their use of single-molecule TIRF microscopy provides data complementary to the ensemble assays used in our study.
What in vivo data relate to our findings? ChIP-seq studies find PRC2 peaking near transcriptional start sites, which are typically nucleosome-free regions15,37; however, the resolution of current ChIP-seq datasets does not allow for a definitive comparison of PRC2 binding sites and nucleosome positioning. Nevertheless, one pioneering study has provided evidence of mutual exclusion between SUZ12 binding and nucleosome density on targeted CpG islands38. Moreover, our biochemical findings are in agreement with a recent study that concluded that H3K27M is not involved in the recruitment or sequestration of PRC2 in vivo39.
Another conclusion from our investigation is that the core PRC2 complex (EZH2−EED− SUZ12) has robust DNA-binding activity. This offers an alternative perspective to the standing assumption that core PRC2 subunits lack the ability to bind DNA40. Because DNA and RNA binding are competitive, the implication is that the same core PRC2 components that bind RNA8 also possess intrinsic DNA-binding activity through still uncharacterized domains.
Thus, our new perspective is that nucleosome-free regions of chromatin and nucleosome linkers recruit PRC2, after which it will bind histone tails in its vicinity. PRC2 HMTase will then be stimulated by existing H3K27me3 or repressed by binding H3K4me3, as in the canonical model. Inherent to this new model is that PRC2 enrichment at a target site can be explained by chromatin binding in hierachical fashion, with nanomolar affinity DNA-binding components cooperating with micromolar affinity histone tail-binding modules. This engagement with histone tails might re-orient the enzyme on chromatin, leading to activation of HMTase activity, or inhibition in the case of H3K27M. Conceivably, there might be an offsetting energetic penalty associated with simultaneous engagement of PRC2 with DNA and histones tails, explaining why the net effect of H3K27M on overall binding is observed to be modest (i.e., lower than simple binding additivity would predict).
What defines a gene target for PRC2-mediated epigenetic silencing?
In search of a recruiting DNA element, our in vitro analysis showed that PRC2 prefers binding to CG-rich sequences. This finding is consistent with multiple studies showing that PRC2 target genes contain CG-rich sequences in vivo8,17,27,41. Past studies have also proposed that CpG islands could be novel recruiters for PRC2 in mammals. Interestingly, CpG island and CG content have been shown to be key factors that promote nucleosome instability and depletion42,43; this may further contribute to PRC2 binding CG-rich regions in vivo.
Our finding that PRC2 5-mer complexes have an intrinsic preference for mCpG-DNA is surprising, given that genome-wide data suggest cytosine methylation antagonizes PRC2 binding35,44–46. On the other hand, PRC2 has been suggested to be directly associated with DNA methylation and loss of DNA methylation can lead to loss of H3K27me3 and PRC2, in agreement with our in vitro results showing that methylated DNA binds optimally47,48. The discrepancy between the various studies could come from different compositions of PRC2 subcomplexes. We have found that the AEBP2 subunit of PRC2 acts as an effector of specificity for methylated DNA. PRC2 complexes lacking AEBP2 bind DNA with low-nM affinity, regardless of CpG methylation status. Yet, the incorporation of AEBP2 into the PRC2 complex reconfigures the binding properties of PRC2 to greatly reduce its affinity for unmethylated DNA. The targeting to methylated DNA by AEBP2 is dependent on its C2H2 zinc-finger domains. In fact, one recent study reports that AEBP2 plays a role in defining the mutually exclusive composition of PRC2 subcomplexes49. AEBP2 may not be the only epigenetic regulator that associates with the PRC2 core complex. Indeed, other accessory subunits in PRC2 subcomplexes may promote exclusion from methylated CpG islands, and mCpG-binding proteins could in some cases hide mCpG from PRC2. Future studies are needed to test such hypotheses.
Drosophila has PREs, which recruit proteins such as PHO and other DNA-binding factors50,51. Such factors then recruit Drosophila PRC2 (dPRC2) in hierarchical fashion. Even though the sequence specificity of dPRC2 for PREs is not fully understood, this DNA-based recruitment mechanism is widely accepted. In mammals, no homologs of PREs have been identified, and neither have orthologs of the dPRC2-recruiting proteins52. However, our study demonstrates that human PRC2 has in fact robust DNA-binding activity (with nanomolar Kdapp). In addition, nucleosome-kinetic studies have suggested PREs are areas of histone replacement and reduced nucleosome occupancy53. And our findings indicate that the intrinsic DNA-binding activity of human PRC2 is important for binding to chromatin and promoting activity. Therefore, PRC2 recruitment in mammals and Drosophila may be more similar than previously thought.
JARID2
JARID2 has been identified and recognized as an important regulator of PRC2. Initial studies concluded that JARID2 negatively regulates PRC2 activity, but subsequent studies have reported that JARID2 is an activator54, 55. This contradiction could simply arise from different substrates used in activity assays (i.e., unfolded histone H3 vs. histone octamer vs. nucleosome), or it could be due to difficulties during protein purification5. We reconstituted PRC2−JARID2 6-mer complex by co-expressing JARID2 together with the other five subunits, and we then used well-assembled nucleosomes as our substrate in activity assays. We found that the incorporation of JARID2 substantially stimulates EZH2 catalytic activity, as indicated by an increase in H3K27 methylation and EZH2 automethylation. This finding supports the conclusion that JARID2 may allosterically regulate PRC2, in agreement with previous findings from mixing experiments using PRC2 and JARID25,31. However, we did not observe increased nucleosome-binding affinity as a previous study suggested31. This discrepancy could be due to the previous use of truncated JARID2 proteins to test nucleosome-binding affinity, rather than full-length JARID2 in complex with PRC2 as used here. Furthermore, although JARID2 has been proposed to have an N-terminal noncoding RNA-binding region (RBR)56, we found that the inclusion of JARID2 did not alter PRC2 RNA-binding activity (Supplementary Fig. 9d). However, our RNA crosslinking analysis showed that JARID2 may contribute a surface that contacts RNA when it is in complex with PRC2 (Supplementary Fig 9f,g).
How does RNA binding regulate PRC2?
Why is the PRC2 gene-silencing complex recruited to actively transcribed genes? Why does PRC2 not suppress transcriptional activity at those genes when bound? These perplexing observations can now be explained by the intrinsic properties of PRC2-RNA-nucleosome interactions. In highly expressed genes, PRC2 may be bound by RNA emerging from active genes but it is not able to promote silencing, because the RNA shuttles PRC2 away from chromatin. RNA inhibition of PRC2-chromatin binding was previously observed in non-equilibrium pull-down assays10. Our work now provides an unexpectedly simple mechanism for this eviction: (1) RNA and DNA bind PRC2 competitively, with RNA having higher affinity. (2) DNA binding is necessary for PRC2 to bind nucleosomes. (3) Thus, PRC2 cannot bind nucleosomes in the presence of a saturating amount of RNA. Furthermore, these results provide mechanistic understanding for how RNA can hold PRC2 “poised but in check”5. RNA is not an active-site inhibitor of PRC2 methyltransferase activity, as evidenced by the unperturbed automethylation activity, but instead it prevents PRC2 from binding its nucleosome substrate.
In conclusion
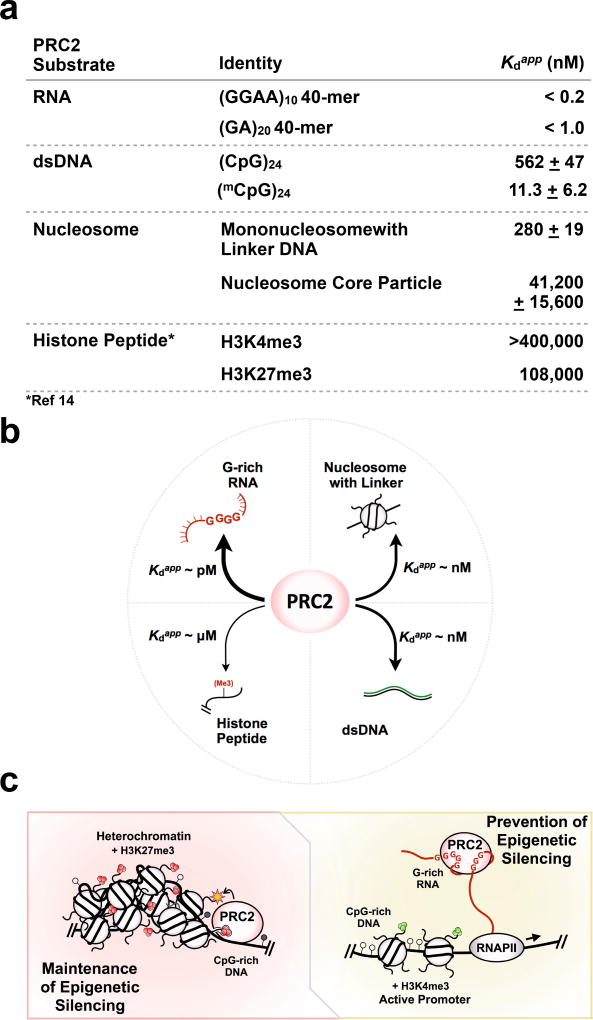
PRC2 recruitment to target sites is perhaps one of the most enigmatic aspects of its function. As a gene silencer with an intrinsic ability to propagate its own heritable enzymatic product, PRC2 must possess a repertoire of biochemical properties suitable to its biological function. Together, our findings reveal new insights into how PRC2 associates with nucleosomes and on the interplay of PRC2 binding to chromatin and RNA. As shown in Figure 7a,b, PRC2 binds RNA >> nucleosomes with linker DNA ≈ DNA >> histone tails. Based on these relative affinities, as well as the passive role of RNA in dissociating PRC2-nucleosomes complexes, we propose the following model (Figure 7c): the PRC2 complex is recruited to target genes by binding to DNA, with contributions in specificity conferred by CG-rich sequences or CpG islands. This DNA binding by PRC2 could promote scanning of nearby chromatin and the recognition of histone marks. Most of the affinity of PRC2 binding comes from its binding to DNA, not histones; its binding to histone tails is of course necessary for H3K27-trimethylation and for regulation, but it makes a minor contribution to affinity. In the case of active transcription units, on the other hand, the nascent pre-mRNA or nearby lncRNA transcripts bind PRC2 and prevent its deposition to chromatin; thus, the active state is maintained. These models will no doubt prove to be oversimplified in particular biological situations, where the presence of other proteins or RNPs (either PRC2-bound or chromatin-bound) may overcome or compete with the intrinsic properties of the PRC2 complex. Yet understanding the fundamental properties of the PRC2-nucleosome interaction provides a framework for interpretation of specific instances where epigenetic gene silencing is maintained or is switched on or off.
Figure 7.
A model for PRC2-chromatin-RNA interactions and regulation of epigenetic silencing. (a) A summary of relative PRC2 binding to RNA, DNA, nucleosomes, and histone peptides. All Kdapp values were determined by EMSA under the same conditions, in the absence of tRNA competitor, with the exception of the literature Kd values for histone peptide binding14 which were determined by ITC. (b) A comparison of binding affinity of PRC2 to various substrates reveals that PRC2 binds RNA >> nucleosomes with linker DNA ≈ DNA >> histone tails. (c) Model includes (left) maintenance of epigenetic silencing, where high-affinity DNA-binding modules of PRC2 allow the complex to preferentially bind nucleosome-free regions and methylate an adjacent nucleosome, and (right) prevention of epigenetic silencing, where RNA binding suppresses H3K27 methylation by interfering with the DNA binding required by PRC2 to bind chromatin. Heterochromatin is characterized by H3K27me3 marks and dense nucleosome packing. Active promoters feature H3K4me3 marks. CpG islands featuring unmethylated (open circles) and methylated (filled circles) CpG dinucleotides in gene promoter regions may serve as a conduit for PRC2 targeting. Even though the model may be an oversimplification, it provides an interpretation of how epigenetic silencing could be switched on-and-off under certain chromatin states, as deduced from our quantitative study of PRC2 substrate binding.
ONLINE METHODS
Protein expression and purification
Human PRC2-JARID2 6-mer complexes were expressed in insect cells similarly to PRC2 5-mer as previously described6,8. In brief, sequences encoding human EZH2, SUZ12, EED, RBBP4, AEBP2, and JARID2 (UniProtDB entry isoform sequences Q15910-2, Q15022, O75530-1, Q09028-1, Q6ZN18-1, and Q92833-1, respectively) were cloned into the pfast-bac1 expression vector (Invitrogen) with PreScission-cleavable N-terminal hexahistidine-MBP tags. JARID2 had an additional 3×FLAG-tag before the PreScission cutting site. Standard Bac-to-Bac baculovirus expression system (Invitrogen) was used to generate baculovirus stocks according to manufacturer’s protocol. The titer of each baculovirus stock was measured by gp64 detection (Expression Systems). Equal amounts of baculovirus of each subunit were used to infect sf9 cells (Invitrogen) at a density of 2.0 × 106 cells/ml. Following infection, the cells were incubated for 72 h (27°C, 130 rpm) before they were harvested. The harvested cells were snap-frozen with liquid nitrogen for later purification.
PRC2 5-mer complex was purified as previously described8 via a three-column purification scheme. For PRC2 6-mer complex, an additional FLAG-tag purification was incorporated, as follows. Cell extract was incubated with the amylose resin and washed thoroughly, followed by elution with 10 mM maltose. The elutate was concentrated to ~15 mg/ml (Amicon Ultra-15 Centrifugal Filter Unit, 30 kDa MWCO, Millipore cat # UFC903024). Then, the concentrated eluate was incubated with equilibrated Anti-FLAG G1 affinity resin (GenScript L00432-25) in the cold room with slow agitation for 2–3 h. Beads were washed with 20 C.V. of FLAG-buffer (10 mM Tris, pH 7.5 at RT and 150 mM NaCl) and eluted with FLAG-buffer supplemented with 3×FLAG peptide to 0.2 mg/ml ApexBio A6001). Each of three elutions proceeded with slow agitation for 40–50 min in the cold room. The TCEP concentration was adjusted to 1 mM and the eluate was concentrated with Amicon Ultra-0.5 Centrifugal Filter Unit with 30-kDa cutoff (Millipore UFC503096), followed by digestion with PreScission protease at a mass ratio of 1:50 protease:protein. After completion of cleavage, protein complex was chromatographed on a 5 ml Hi-Trap Heparin column (GE, 17-0407-03), followed by fractionation over a HiPrep 16/60 Sephacryl S-400 HR sizing column. The 6-mer peak fractions were identified using SDS-PAGE, pooled, and concentrated as above. Final protein concentration was measured by absorbance at 280 nm, and the ratio of absorbance at 260 nm/280 nm <0.7 provided an indication of no nucleic acid contamination.
Nucleosome reconstitution
Histone octamer was assembled using equal molar amount of each histone in 2 M NaCl. Octamer was purified via a sizing column (GE, HiLoad 16/60 Superdex 200 HR). Puc19 plasmid containing 601-Widom positioning sequence was purified using GigaPrep (Qiagen 12191) and cut with EcoRV (NEB R3195M). Efficiency of cutting was determined using a 1% agarose gel. Then, the DNA was adjusted to ~1 mg/ml and purified via Mono Q column (GE 17-5167-01). Fractions containing the nucleosome template were identified by agarose gel, pooled, and concentrated by ethanol precipitation. DNA was then dissolved in TE buffer.
Nucleosome reconstitutions were performed by standard protocol using salt dialysis18. In brief, nucleosome particles and arrays were reconstituted at a small scale from 1 to 6 µM in a volume of 25–50 µl. Widom-601 DNA (1-, 3-, or 12-repeats) were mixed with histone octamers and assembled by dialysis from RB-High buffer (2 M KCl, 10 mM Tris-HCl pH 7.5, 0.1 mM EDTA, 1 mM DTT) to RB-Low buffer (0.25 M KCl, 10 mM Tris-HCl pH 7.5, 1 mM EDTA, 1 mM DTT). The reactions were dialyzed gradually from RB-High buffer to RB-Low buffer at a flow-rate of 1.45 ml/min using a peristaltic pump (BioRad Model EP-1 Econo Pump). After dialysis for about 24 h, buffer was exchanged into TCS buffer (20 mM Tris-HCl pH 7.5, 1 mM EDTA, 1 mM DTT) and allowed to dialyze for 1–6 h. Nucleosomes were then transferred to LoBind tubes and stored at 4°C. After reconstitution, a portion of each sample was cut by EcoRI-HF (NEB R3101T) and analyzed by 5% native-PAGE on a 1% agarose gel. Based on ethidium bromide staining, the histone-saturation level of reconstituted nucleosomes could be assessed. In addition, each sample was subjected to AFM to further examine whether each reconstituted nucleosome batch was of good quality.
Atomic-force microscopy
For surface deposition, nucleosome samples were diluted with TCS buffer to a final concentration of 2 nM. 50 µL of diluted samples were pipetted onto APTES-modified mica (Grade V, SPI). Samples were then incubated at room temperature for 5 min. The mica discs were then rinsed with purified 18.2-MΩ deionized water. For samples intended for AFM imaging in air, the washed mica disks were then dried using a gentle N2 gas flow, perpendicular to the mica surface. For samples intended for imaging in liquid, the rinsed mica disks were quickly exchanged into imaging buffer (10 mM Tris-HCl pH 7.5, 3 mM NiCl2). All samples were imaged by AFM immediately following preparation. Data acquisition was performed using a MFP-3D AFM or a Cypher ES Environmental Atomic-Force Microscope (Asylum Research). The samples were imaged in tapping mode using a commercial silicon cantilever with a spring constant of 46 N/m. For air imaging, Asylum AC240 cantilevers were used; for liquid imaging, Bruker SNL-10 cantilevers were used. Images were captured at 512×512 pixels in the trace direction, at a scan size of 2 µm and a scan rate of 1.0 Hz. Image processing was carried out using publicly available software (Gwyddion).
Electrophoretic mobility shift assay (EMSA)
Reconstituted nucleosomes were radiolabeled in the cold room for 1 h using T4 PNK (NEB M0201L) by standard protocol, with the modification of reducing the concentration of MgCl2 in the T4 PNK reaction buffer to 0.5 mM. After labeling, excess [γ-32P]-ATP in the reaction was removed by running the samples over a G50 Sephadex column (Roche 11 273 949 001). The column was pre-equilibrated with TCS buffer. The counts of nucleosomes were determined by liquid scintillation counting. Radiolabeled nucleosomes were diluted with binding buffer (50 mM Tris-HCl pH 7.5 at 25°C, 100 mM KCl, 2.5 mM MgCl2, 0.1 mM ZnCl2, 2 mM 2-mercaptoethanol, 0.05% v/v NP-40, 0.1 mg/ml bovine serum albumin, 5% v/v glycerol). Next, stock PRC2 was diluted with binding buffer and added to radiolabeled nucleosomes. Binding was carried out at 30°C for 30 min, followed by loading samples onto non-denaturing 0.7% agarose gel (Fisher BP160-100) buffered with 1×TBE at 4°C. Gel electrophoresis was carried out for 90 min at 66 V in an ice box within a 4°C cold room. A Hybond N+membrane (Amersham, Fisher Scientific 45-000-927) and two sheets of Whatman 3 mm chromatography paper were put underneath the gel, which then was vacuum dried for 60 min at 80°C. Dried gels were exposed to phosphorimaging plates, which were scanned using a Typhoon Trio phosphorimager (GE Healthcare) for signal acquisition. Gel analysis was carried out with ImageQuant software (GE Healthcare) and data fitted to a sigmoidal binding curve using custom written code in MATLAB (MathWorks). Note, yeast tRNA is typically used as a competitor to eliminate nonspecific binding in RNA binding studies8. However, in our nucleosome and DNA binding studies, we did not include yeast tRNA in the binding buffer because it competes off DNA from PRC2.
Semisynthesis of H3 containing trimethyl-lysine (H2K4Kme3 and H3K27me3)
Semisynthetic histones were generated essentially as previously described57 with some minor changes. Briefly, synthetic peptides were synthesized as C-terminal hydrazides by Fmoc-SPPS and converted into the corresponding thioesters by oxidation using NaNO2 in the presence of thiols. Native chemical ligation was then performed with the requisite truncated H3 protein in ligation buffer (200 mM phosphate buffer, 6 M guanidine hydrochloride, pH 7.5) with 3% 2,2,2-trifluoroethanethiol (TFET). The cysteine-containing ligation product was subjected to radical-desulfurization to give the native H3 sequence bearing the Kme3 PTM at the desired position. Following purification by HPLC, the proteins were characterized by HPLC and ESI-MS (Supplementary Fig. 3 and Supplementary Fig. 4).
In vitro histone methyltransferase assay
Unless indicated otherwise, each 10 µl reaction contained 600 nM PRC2, 600 nM mononucleosome, and 12 µM S-[methyl-14C]-adenosylmethionine (PerkinElmer NEC363050UC) in Covfefe buffer (50 mM Tris-HCl pH 8.0 at 30°C, 100 mM KCl, 2.5 mM MgCl2, 0.1 mM ZnCl2, 2 mM 2-mercaptoethanol, 0.1 mg/ml bovine serum albumin, 5% v/v glycerol). Reactions were incubated for 1 h at 30°C and stopped by adding 4× loading dye. Each reaction was then heated at 95°C for 5 min and loaded onto either 4–12% Bis-Tris gel (ThermoFisher NP0322BOX) or 10–20% Tris-Glycine gel (ThermoFisher XP10202BOX). Gels were first stained by Coomassie and scanned, then vacuum dried for 60 min at 80°C. Signal was acquired with a Typhoon Trio phosphorimager (GE Healthcare). Densitometry and analysis were carried out with ImageQuant software (GE Healthcare).
In testing RNA inhibition of histone methyltranferase activity, the reaction was set up as described above except 2.4 µM mononucleosome was used and RNA was titrated into the reaction from 60 µM (2-fold dilutions).
RNA and nucleosome competition
The reaction was set up as above except 1000–2000 c.p.m radiolabeled nucleosome was mixed first with unlabeled nucleosome, and then the mixture was added into each reaction. After the initial 30 min incubation, unlabeled competitor RNA was titrated into each reaction and an additional 30 min incubation was carried out at 30°C before the reaction was loaded onto 0.7% agarose gel.
RNA and DNA competition assay
The reaction was set up as described for the RNA-nucleosome competition except DNA was radiolabeled.
Fluorescence polarization for binding reaction and competition assay
Each dsDNA containing 5’ Alexa-488 fluorescent dye was synthesized, annealed, and purified by IDT. Each 40 µl reaction contained 5 nM fluorescent DNA and PRC2 in binding buffer (50 mM Tris-HCl pH 7.5, 25 mM KCl, 2.5 mM MgCl2, 0.1 mM ZnCl2, 2 mM 2-mercaptoethanol, 0.1 mg/ml bovine serum albumin, 5% v/v glycerol). Reaction was incubated for 30 min at 30°C, followed by measuring fluorescence polarization in a 384-well plate with Synergy 2 multi-mode plate reader (BioTek). For competitor assays, initial signals were measured (defined as t = 0) before adding competitors. RNA or dsDNA cold competitors were added into the reactions and the measurements were taken every 20 s for 4 h.
MNase digestion analysis
To test the ability of PRC2 to protect linker regions of chromatin, an MNase-protection assay was set up. Micrococcal nuclease was diluted to concentrations of 3.2 Units/µL and 0.8 Units/µL in MNase reaction buffer (10 mM potassium HEPES pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA and 10% w/v glycerol). A PRC2-dodecanucleosome binding reaction was prepared in a final volume of 45 µL and incubated at 30°C for 30 min. Following incubation, 2.5 µl CaCl2 was added to a final concentration of 5 mM, and 2.5 µl of diluted MNase was added to the mixture. The MNase reaction was allowed to proceed for approximately 10 min before quenching with 12.5 µl of 500 mM EDTA. Then, 2.5 µl of a 10 mg/ml stock solution of proteinase K was added to digest PRC2 proteins and histones by incubating for 30–60 min at 50°C. The DNA was purified by phenol-chloroform extraction. Finally, the DNA was loaded onto a native 1.35% agarose gel (BP160) and run in 1× TBE buffer at 160 V for 3 h. DNA bands were visualized by ethidium bromide staining.
UV Crosslinking of RNA-PRC2 Complexes
RNA sample was transcribed with synthesized (GE-Dharmacon) with 4-thio-U incorporated into the oligo (24 mers). The 24mer-oligo was end- labeled with gamma-32P-ATP as described above. Trace amounts of hot RNA were refolded (as in8, except in some instances BSA and tRNA were omitted from the refold buffer) or used without refolding and incubated with 200 nM–1 µM PRC2 protein. PRC2 5-mer or 6-mer or PRC2 that contained an uncleavable MBP tag on SUZ12 was incubated with RNA for 30 min at 30 °C. Samples were moved to siliconized glass coverslips on ice, placed in a Stratalinker with 365 nm bulbs, and exposed to light for 10–30 min. Sample were diluted with SDS loading dye and loaded onto a Nupage 4–12% Bis-Tris gel (Life Technologies). Gel electrophoresis was carried out for 60 min at 180 volts. Gels were vacuum dried at 80 °C for 30 min on Whatman 3mm chromatography paper. Dried gels were exposed to phosphorimaging plates and signal acquisition was performed using a Typhoon Trio phosphorimager (GE Healthcare).
Supplementary Material
Acknowledgments
We thank Ci Ji Lim and other members of the Cech lab for useful conversations. We thank Chen Davidovich (Monash University, Clayton, Australia) for initial trials of expression and purification of JARID2 and for stimulating discussion. We thank Karolin Luger (U. Colorado Boulder) and her laboratory – especially Uma M. Muthurajan and Pamela Dyer – for providing plasmids and helpful discussion of reconstituting nucleosomes. We thank Juerg Mueller and his group (MPI Martiensreid, Germany) and Beat Fierz (EPFL, Lausanne, Switzerland) for sharing unpublished data and discussions. T.W.M is supported by National Institutes of Health grants R37-GM086868 and PO1-CA196539. T.R.C. is an investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
X.W., R.D.P., and T.R.C. designed the experiments. X.W. and RDP carried out experiments. A.R.G. carried out protein purification. Z.Z.B., E.J.G. and T.W.M. carried out the synthesis of modified histone H3. X.W., R.D.P., and T.R.C. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
T.R.C. is on the board of directors of Merck, Inc., which provides no funding for his research.
A Life Sciences Reporting Summary for this article is available online.
DATA AVAILABILITY
Source data for Figures 4d and 5d are available with the paper online. Other data are available from the authors upon request.
References
- 1.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Müller J, Bienz M. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 1991;10:3147–3155. doi: 10.1002/j.1460-2075.1991.tb04876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mihaly J, Mishra RK, Karch F. A Conserved Sequence Motif in Polycomb-Response Elements. Mol. Cell. 1998;1:1065–1066. doi: 10.1016/s1097-2765(00)80107-0. [DOI] [PubMed] [Google Scholar]
- 4.Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014;28:1983–1988. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidovich C, et al. Toward a Consensus on the Binding Specificity and Promiscuity of PRC2 for RNA. Mol. Cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, et al. Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines. Mol. Cell. 2017;65:1056–1067.e5. doi: 10.1016/j.molcel.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory Interactions between RNA and Polycomb Repressive Complex 2. Mol. Cell. 2014;55:171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beltran M, et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 2016 doi: 10.1101/gr.197632.115. gr.197632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitges FW, et al. Histone Methylation by PRC2 Is Inhibited by Active Chromatin Marks. Mol. Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 13.Lewis PW, et al. Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science. 2013 doi: 10.1126/science.1232245. 1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu C, et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) Proc. Natl. Acad. Sci. 2010;107:19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350:aac4383. doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nekrasov M, Wild B, Müller J. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep. 2005;6:348–353. doi: 10.1038/sj.embor.7400376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendenhall EM, et al. GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells. PLOS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 19.Yuan W, et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- 20.Martin C, Cao R, Zhang Y. Substrate Preferences of the EZH2 Histone Methyltransferase Complex. J. Biol. Chem. 2006;281:8365–8370. doi: 10.1074/jbc.M513425200. [DOI] [PubMed] [Google Scholar]
- 21.Müller J, et al. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 22.Sanulli S, et al. Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Mol. Cell. 2015;57:769–783. doi: 10.1016/j.molcel.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikitina T, Wang D, Gomberg M, Grigoryev SA, Zhurkin VB. Combined micrococcal nuclease and exonuclease III digestion reveals precise positions of the nucleosome core/linker junctions: implications for high-resolution nucleosome mapping. J. Mol. Biol. 2013;425:1946–1960. doi: 10.1016/j.jmb.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Widlak P, Garrard WT. Unique features of the apoptotic endonuclease DFF40/CAD relative to micrococcal nuclease as a structural probe for chromatin. Biochem. Cell Biol. Biochim. Biol. Cell. 2006;84:405–410. doi: 10.1139/o06-063. [DOI] [PubMed] [Google Scholar]
- 25.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radman-Livaja M, Rando OJ. Nucleosome positioning: how is it established, and why does it matter? Dev. Biol. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ku M, et al. Genomewide Analysis of PRC1 and PRC2 Occupancy Identifies Two Classes of Bivalent Domains. PLOS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 29.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buck-Koehntop BA, et al. Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso. Proc. Natl. Acad. Sci. U. S. A. 2012;109:15229–15234. doi: 10.1073/pnas.1213726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Son J, Shen SS, Margueron R, Reinberg D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013;27:2663–2677. doi: 10.1101/gad.225888.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen X, et al. EZH1 Mediates Methylation on Histone H3 Lysine 27 and Complements EZH2 in Maintaining Stem Cell Identity and Executing Pluripotency. Mol. Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015;21:2007–2022. doi: 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holoch D, Margueron R. Mechanisms Regulating PRC2 Recruitment and Enzymatic Activity. Trends Biochem. Sci. 2017;42:531–542. doi: 10.1016/j.tibs.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Justin N, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 2016;7 doi: 10.1038/ncomms11316. ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riising EM, et al. Gene Silencing Triggers Polycomb Repressive Complex 2 Recruitment to CpG Islands Genome Wide. Mol. Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Piunti A, et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat. Med. 2017;23:493–500. doi: 10.1038/nm.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Portoso M, et al. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017:e201695335. doi: 10.15252/embj.201695335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fenouil R, et al. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res. 2012;22:2399–2408. doi: 10.1101/gr.138776.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lynch MD, et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012;31:317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartke T, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King AD, et al. Reversible Regulation of Promoter and Enhancer Histone Landscape by DNA Methylation in Mouse Embryonic Stem Cells. Cell Rep. 2016;17:289–302. doi: 10.1016/j.celrep.2016.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin B, et al. DNMT1 and DNMT3B modulate distinct polycomb-mediated histone modifications in colon cancer. Cancer Res. 2009;69:7412–7421. doi: 10.1158/0008-5472.CAN-09-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viré E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 49.Grijzenhout A, et al. Functional analysis of AEBP2, a PRC2 Polycomb protein, reveals a Trithorax phenotype in embryonic development and in ESCs. Dev. Camb. Engl. 2016;143:2716–2723. doi: 10.1242/dev.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sengupta AK, Kuhrs A, Müller J. General transcriptional silencing by a Polycomb response element in Drosophila. Development. 2004;131:1959–1965. doi: 10.1242/dev.01084. [DOI] [PubMed] [Google Scholar]
- 51.Klymenko T, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome Regulation by Polycomb and Trithorax Proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Mito Y, Henikoff JG, Henikoff S. Histone Replacement Marks the Boundaries of cis-Regulatory Domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 54.Shen X, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaneko S, et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen UTT, et al. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nat. Methods. 2014;11:834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.