Abstract
Bevacizumab combined with cytotoxic chemotherapy is the backbone of metastatic colorectal cancer (mCRC) therapy; however, its treatment efficacy is hampered by therapeutic resistance. Therefore, understanding the mechanisms underlying bevacizumab resistance is crucial to increasing the therapeutic efficacy of bevacizumab. The Gene Expression Omnibus (GEO) database (dataset, GSE86525) was used to identify the key genes and pathways involved in bevacizumab-resistant mCRC. The GEO2R web tool was used to identify differentially expressed genes (DEGs). Functional and pathway enrichment analyses of the DEGs were performed using the Database for Annotation, Visualization, and Integrated Discovery(DAVID). Protein–protein interaction (PPI) networks were established using the Search Tool for the Retrieval of Interacting Genes/Proteins database(STRING) and visualized using Cytoscape software. A total of 124 DEGs were obtained, 57 of which upregulated and 67 were downregulated. PPI network analysis showed that seven upregulated genes and nine downregulated genes exhibited high PPI degrees. In the functional enrichment, the DEGs were mainly enriched in negative regulation of phosphate metabolic process and positive regulation of cell cycle process gene ontologies (GOs); the enriched pathways were the phosphoinositide 3-kinase-serine/threonine kinase signaling pathway, bladder cancer, and microRNAs in cancer. Cyclin-dependent kinase inhibitor 1A(CDKN1A), toll-like receptor 4 (TLR4), CD19 molecule (CD19), breast cancer 1, early onset (BRCA1), platelet-derived growth factor subunit A (PDGFA), and matrix metallopeptidase 1 (MMP1) were the DEGs involved in the pathways and the PPIs. The clinical validation of the DEGs in mCRC (TNM clinical stages 3 and 4) revealed that high PDGFA expression levels were associated with poor overall survival, whereas high BRCA1 and MMP1 expression levels were associated with favorable progress free survival(PFS). The identified genes and pathways can be potential targets and predictors of therapeutic resistance and prognosis in bevacizumab-treated patients with mCRC.
Introduction
Colorectal cancer (CRC) is the third most frequently diagnosed cancer and the second leading cause of cancer deaths worldwide, accounting for 10% of the worldwide cancer incidence and mortality [1]. Surgery is the treatment of choice for nonmetastatic CRC; however, approximately 20% of cases present with metastatic disease at the time of diagnosis and half of the patients experience recurrence and metastases even after complete resection of the primary tumor, leading to a poor prognosis and median overall survival (OS) of approximately 24 months [2, 3]. The inclusion of cytotoxic agents (irinotecan and oxaliplatin) in fluoropyrimidine (intravenous 5-fluorouracil or oral capecitabine)-based systemic chemotherapy has been reported to improve the associated response rates (RR) from 15%–20% to 30%–40%, time to progression from 5–6 to 8 months, and OS from 10–12 to 20–24 months [3–7]. Furthermore, therapeutic benefits have been demonstrated to increase through the use of targeted drugs, such as angiogenesis inhibitors (bevacizumab, ziv-aflibercept, and ramucirumab) and antiepidermal growth factor receptor antibodies (cetuximab and panitumumab), as the first and second lines of treatment in patients with with K-RAS-wild-type tumors tumors [8–12].
Bevacizumab is the first agent to influence OS in patients with metastatic CRC (mCRC); when combined with irinotecan-based chemotherapy, the median OS improved from 15.6 to 20.3 months, median PFS from 6.2 to 10.6 months and RR from 34.8% to 44.8%[10]. The addition of bevacizumab to oxaliplatin-based chemotherapy improved median PFS from 8.0 to 9.4 months though there was no significant difference in OS(19.9 to 21.3 months) [13], while in previously treated mCRC; oxaliplatin based therapy improved both OS and PFS (10.8 to 12.9 months and 4.7 to 7.3 respectively)[14]. When compared for effectiveness, the irinotecan based chemotherapy has shown to have an edge over oxaliplatin based chemotherapy with the addition of bevacizumab (OS = 31.4 vs 30.1 months, PFS = 12.1 vs 10.7 months)[15]. These results have also been echoed in the MAVERICC trial (OS = 27.5 vs 23.9 months, PFS = 12.6 vs 10.1 months)[16]. Bevacizumab is a humanized monoclonal antibody that binds to vascular endothelial growth factor A (VEGF-A) and thus prevents interaction with its receptors, VEGFR-1 (Flt-1) and VEGFR-2 (Flk-1/KDR), leading to the regression of existing tumor blood vessels, normalization of the remaining blood vessels, and consequently tumor inhibition [17]. However, the therapeutic effects of bevacizumab are strongly affected by the lack of biomarkers that can facilitate selecting a population that might benefit from this medication and can predict therapeutic resistance [18–20].
In this study, we investigated the predictive biomarkers and pathways of bevacizumab resistance in mCRC by using microarray data from the Genetic Expression Omnibus (GEO) database. The new biomarkers were assessed for their ability to predict OS and PFS. The identification of predictive and prognostic biomarkers can facilitate improving the therapeutic index of bevacizumab.
Materials and methods
Microarray data
The gene expression profile of GSE86525 was obtained from the GEO (http://www.ncbi.nlm.nih.gov/geo/) database [21], which was sequenced on the GPL16699 platform of Agilent-039494 SurePrint G3 Human GE v2 8 × 60K Microarray 039381 (Agilent Technologies, Santa Clara, CA, USA). The GSE86525 dataset includes microarray gene expression data derived from three bevacizumab-resistant HT29 xenograft tumors and three untreated HT29 xenograft tumors as controls. In brief, HT29 cells (1 × 107) suspended in phosphate-buffered saline were subcutaneously injected into the flanks of BALB/c nude mice, and the tumor-bearing mice were treated with bevacizumab (5 mg/kg, twice a week) for 3 weeks to obtain bevacizumab-resistant tumors. MTT colorimetric assays were used to determine the 50% inhibitory concentration for bevacizumab-resistant and untreated xenograft tumors; the tumor sizes were compared between the two groups. The sample tissues were immediately frozen under liquid nitrogen after isolation. Total RNAs were extracted from the samples, evaluated, labeled and hybridized, using a SurePrint G3 Human GE 8 × 60K microarray (Agilent Technologies). Array images were captured using a DNA microarray scanner (Agilent Technologies), and the data were analyzed using Feature Extraction Software (Agilent Technologies) to obtain background-corrected signal intensities. The expression data were further analyzed using GeneSpring GX software (version 11.0, Agilent Technologies), and the differentially expressed genes (DEGs) between the bevacizumab-resistant HT29 tumors vs untreated control were compared using the Fisher exact test, followed by multiple corrections using the Benjamini and Hochberg false discovery rate (FDR) method [22]. Gene sets with an FDR q-value of <0.05 were considered statistically significant, and all experiments were performed in triplicate.
Data preprocessing and DEGs screening
The data were recalculated using the GEO2R analytical tool to identify the DEGs associated with acquired bevacizumab-resistant CRC [23, 24]. The t test and Benjamini and Hochberg method were used to calculate the P values and FDR, respectively [22]. The genes were considered to be differentially expressed for an FDR value of <0.05 and fold change (FC) of >2 or <-2 (log2FC > 1 or < -1). The DEG expression data were extracted, and a bidirectional hierarchical clustering plot was constructed using MultiExperiment Viewer (MeV; version 4.8) software [25].
Construction of PPI networks
Protein–protein interaction (PPI) networks were plotted using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; version 10.0; http://www.string-db.org/), an online database comprising comprehensive known and predicted interactions, to determine the interactive relationships among the DEG-encoded proteins. A combined score of >0.7 (high confidence) was used as the cutoff criterion [26]. PPI pairs were visualized using Cytoscape software (version 3.4.0; http://www.cytoscape.org/), and the CytoNCA tool was used to subcluster the plotted PPI networks [27–30]. Highly connected proteins with important biological functions were identified by calculating the degree (number of line connections between proteins) and the betweenness value (fraction of the number of shortest paths that pass through each node; A measure of how often nodes occur on the shortest paths between other nodes) of each node with a degree cutoff criterion of ≥2.
Enrichment analysis of DEGs
The Database for Annotation, Visualization, and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) was used to classify the DEGs involved in the PPI networks according to their biological processes, molecular functions, or cellular components by using the Gene Ontology (GO) Consortium Reference (http://www.geneontology.org/) [31, 32]. Gene sets with a P value of <0.05 and FDR value of <0.05 were considered statistically significant. In addition, the DAVID tool was used for pathway enrichment analysis, and the reference pathways were obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) database website to perform KEGG pathway enrichment analysis for the DEGs involved in the PPI networks, with a P value of <0.05 and FDR value of <0.05 being considered statistically significant [33, 34].
Clinical validation of the DEGs
The clinical assessment of DEGs associated with bevacizumab resistance was performed using the SurvExpress tool [35]. The colon metabase, which includes GSE12945[36], GSE14333[37], GSE17536[38], GSE17537[38], GSE31595, and GSE41258[39] with a total of 808 cases, was used in this study. Survival profiles were compared on the basis of a high or low mRNA expression level of a particular gene, and they were censored independently for OS and PFS in months and stratified further according to TNM clinical stages 3 and 4. A log-rank P value of <0.05 was considered statistically significant, and the data were analyzed using SPSS for Macintosh (version 21, IBM Corp Armonk, NY, USA; www-01.ibm.com) for plotting Kaplan–Meier survival curves.
Gene co-expression in colorectal cancer data
The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/) was used to obtain CRC data containing gene expression profiles. Level 3 RNASeq data containing gene expression profiles of 635 CRC cases (colon adenocarcinoma, N = 463; and rectal adenocarcinoma, N = 172) were obtained. The standard Pearson correlation coefficients (-1 to 1) and the coefficient of variation (the ratio of standard deviation to mean) of the desired gene pairs were calculated using SPSS for Macintosh (version 21, IBM Corp., Armonk, NY, USA; https://www-01.ibm.com). A P value of <0.05 was considered statistically significant and was used as the cutoff criterion.
Results
DEGs screening and heat map clustering analysis
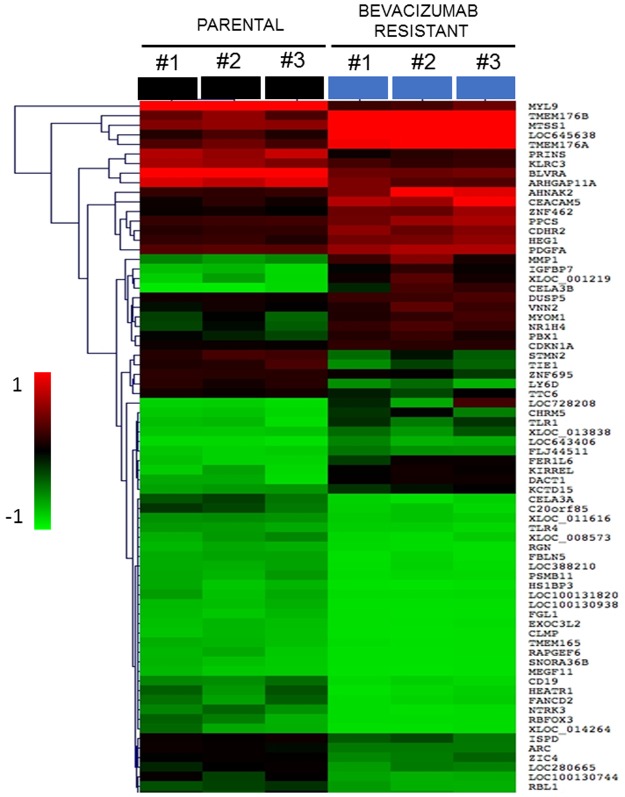
The GEO2R tool was used to identify DEGs from the data derived from the GPL16699 oligonucleotide microarray platform, comprising 62,976 probe sets. A total of 124 DEGs were determined to be associated with bevacizumab resistance, with 57 being upregulated and 67 being downregulated, as determined according to their log2FC and FDR values (S1 and S2 Tables). MeV software was used to construct a heat map to obtain the bidirectional hierarchical clustering of the DEGs and summarize the upregulated and downregulated DEGs (Fig 1).
Fig 1. Heat map showing up-regulated and down-regulated differentially expressed genes (DEGs) in bevacizumab-resistant colon cancer tumors.
A bidirectional hierarchical clustering heat map was constructed using MultiExperimental Viewer(MeV). The expression values are log2 fold changes (>1 or <−1, FDR <0.05)) between corresponding bevacizumab -resistant HT29 xenograft tumors and non-treated HT29 xenograft tumors. Black represents no change in expression, green represents down-regulation, and red represents up-regulation.
PPI network analysis
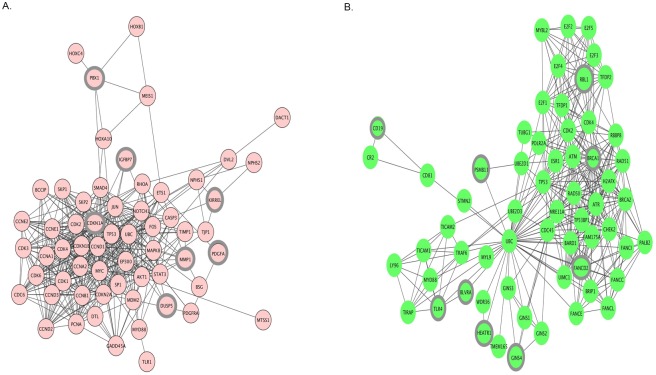
The PPI pairs obtained using the STRING database were visualized using Cytoscape software and analyzed using the CytoNCA plugin. The upregulated network had 88 nodes and 466 edges. Seven genes, namely cyclin-dependent kinase inhibitor 1A (CDKN1A; p21 and Cip1), matrix metallopeptidase 1 (MMP1; interstitial collagenase), pre-B cell leukemia transcription factor 1 (PBX1), platelet-derived growth factor alpha polypeptide (PDGFA), kin of IRRE-like (Drosophila) (KIRREL), insulin-like growth factor binding protein 7 (IGFBP7), and dual specificity phosphatase 5 (DUSP5), exhibited higher PPI degrees and betweenness values (Fig 2A and Table 1). In the downregulated network, containing 88 nodes and 350 edges, nine genes, namely breast cancer 1, early onset (BRCA1), retinoblastoma-like 1 (p107) (RBL1), toll-like receptor 4 (TLR4), CD19, HEAT repeat-containing 1 (CD19), Fanconi anemia complementation group D2 (FANCD2), proteasome subunit beta 11 (PSMB11), biliverdin reductase A (BLVRA), and GINS complex subunit 4 (GINS4), showed higher PPI degrees and betweenness values (Fig 2B, Table 2).
Fig 2. Protein–protein interaction (PPI) network of differentially expressed genes(A) up-regulated genes and (B) down-regulated genes.
The PPI pairs were imported into Cytoscape software as described in methods and materials. Pink nodes represent up-regulated genes while green nodes represent down-regulated genes. The lines represent interaction relationship between nodes. The highlighted DEGs represents degree = >2.
Table 1. Up-regulated genes which had interactions in the PPIs.
| Gene symbol | Gene name | Degree | Betweenness |
|---|---|---|---|
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 38.0 | 209.46272 |
| MMP1 | matrix metallopeptidase 1 (interstitial collagenase) | 10.0 | 20.48990 |
| PBX1 | pre-B-cell leukemia homeobox 1 | 5.0 | 192.13673 |
| KIRREL | kin of IRRE like (Drosophila) | 4.0 | 46.28517 |
| IGFBP7 | insulin-like growth factor binding protein 7 | 4.0 | 0.44311 |
| DUSP5 | dual specificity phosphatase 5 | 3.0 | 0.44311 |
| PDGFA | platelet-derived growth factor alpha polypeptide | 2.0 | 2.66667 |
Table 2. Down-regulated genes which had interactions in the PPIs.
| Gene symbol | Gene name | Degree | Betweenness |
|---|---|---|---|
| BRCA1 | breast cancer 1, early onset | 35.0 | 326.96054 |
| FANCD2 | fanconi anemia complementation group D2 | 19.0 | 33.54729 |
| RBL1 | retinoblastoma-like 1 (p107) | 13.0 | 12.75956 |
| TLR4 | toll-like receptor 4 | 8.0 | 31.75171 |
| GINS4 | GINS complex subunit 4 | 4.0 | 0.0 |
| CD19 | CD19 molecule | 2.0 | 0.0 |
| HEATR1 | HEAT repeat containing 1 | 2.0 | 0.0 |
| PSMB11 | proteasome subunit beta 11 | 2.0 | 0.0 |
| BLVRA | biliverdin reductase A | 2.0 | 0.0 |
Functional enrichment analysis
The DAVID tool was used to classify the DEGs involved in the PPI networks according to their common biological processes, molecular functions, or cellular components. Of the 1,454 GO gene sets included from the reference database, 111 were significantly enriched (P < 0.05; FDR < 0.05). Table 3 lists the top five gene sets, with those involved in the negative regulation of phosphate metabolic process and positive regulation of cell cycle process being the most significant and they include DUSP5, CDKN1A (p21 and Cip1), KIRREL, PDGFA, TLR4, PSMB11, BRCA1, and PBX1.
Table 3. Enriched Gene-Ontologies (GO’s).
| Gene-Ontology | Genes | p-value | FDRa |
|---|---|---|---|
| negative regulation of phosphate metabolic process (GO:0045936) | DUSP5, CDKN1A, KIRREL, PDGFA, TLR4 | 0.00002184 | 0.004597 |
| positive regulation of cell cycle process (GO:0090068) | PSMB11, CDKN1A, BRCA1, PBX1 | 0.00002337 | 0.004597 |
| regulation of lipid metabolic process (GO:0019216) | RBL1, PDGFA, IGFBP7, BRCA1 | 0.00003562 | 0.004597 |
| positive regulation of cell cycle arrest (GO:0071158) | PSMB11, CDKN1A, BRCA1 | 0.00003986 | 0.004597 |
| DNA damage response, signal transduction by p53 class mediator (GO:0030330) | PSMB11, CDKN1A, BRCA1 | 0.00004889 | 0.004597 |
As there were 111 enriched Gene-Ontologies, here we only present top 5 most significant terms according to P-value and
aFDR (False discovery rate).
GO: gene-ontology
KEGG pathway analysis
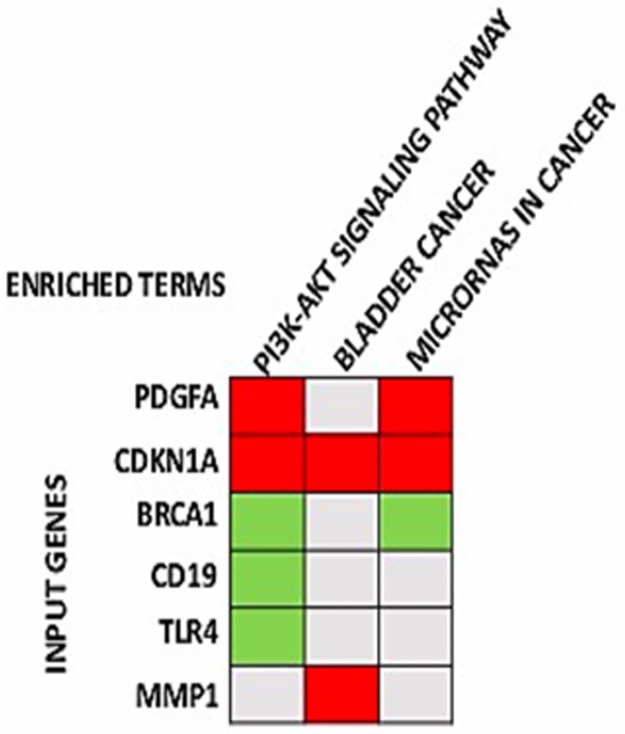
The DAVID tool was applied to classify the DEGs involved in the PPI networks by using the reference pathways from KEGG. KEGG pathway analysis revealed significant results (P < 0.05; FDR < 0.05) for three pathways: the phosphoinositide 3-kinase-serine/threonine kinase (PI3K-AKT) signaling pathway (involving CD19, BRCA1, PDGFA, CDKN1A, and TLR4), bladder cancer (involving CDKN1A and MMP1), and microRNAs in cancer (involving CDKN1A, PDGFA, and BRCA1; (Fig 3, Table 4)
Fig 3. Significant KEGG pathways and the genes involved.
Gene enrichment analysis showing KEGG pathways significantly enriched in bevacizumab resistant HT29 xenograft tumors and the genes involved in the pathways (the pathways are in order of their enrichment from left to right) (FDR <0.05, p-value <0.05).
Table 4. Enriched KEGG pathways.
| KEGGa pathway | Genes | p-value | FDRb |
|---|---|---|---|
| PI3K-Akt signaling pathway (hsa04151) |
CDKN1A, CD19, PDGFA, BRCA1, TLR4 | 0.000005237 | 0.0003037 |
| Bladder cancer (hsa05219) |
CDKN1A, MMP1 | 0.000483151 | 0.009340926 |
| MicroRNAs in cancer (hsa05206) |
CDKN1A, PDGFA, BRCA1 | 0.001572901 | 0.01303 |
aKEGG: Kyoto Encyclopedia of Genes and Genome
bFDR:False discovery rate
Survival analysis of the enriched DEGs
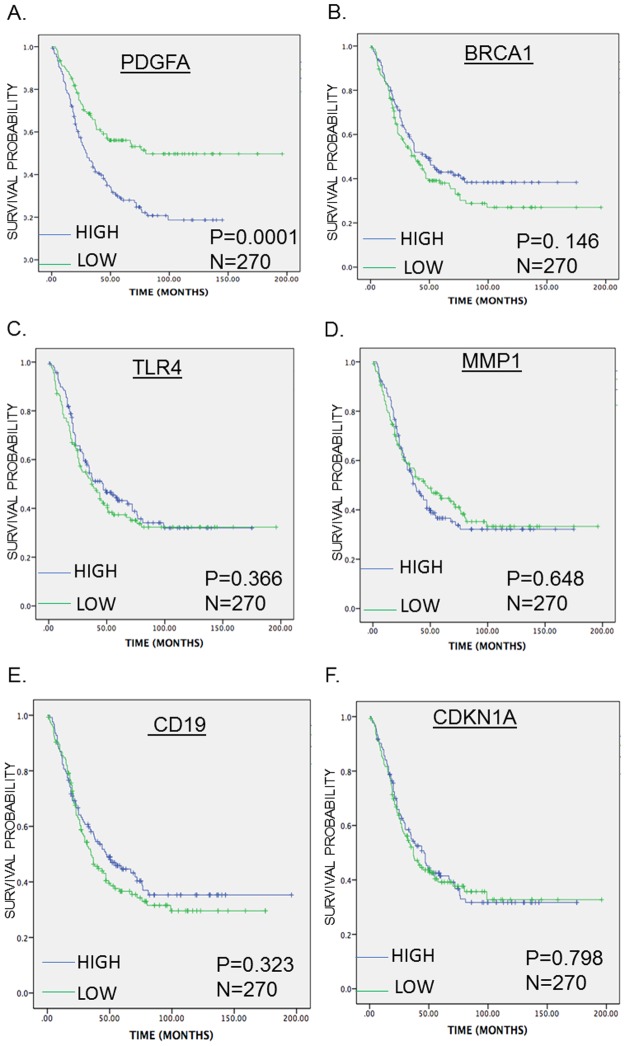
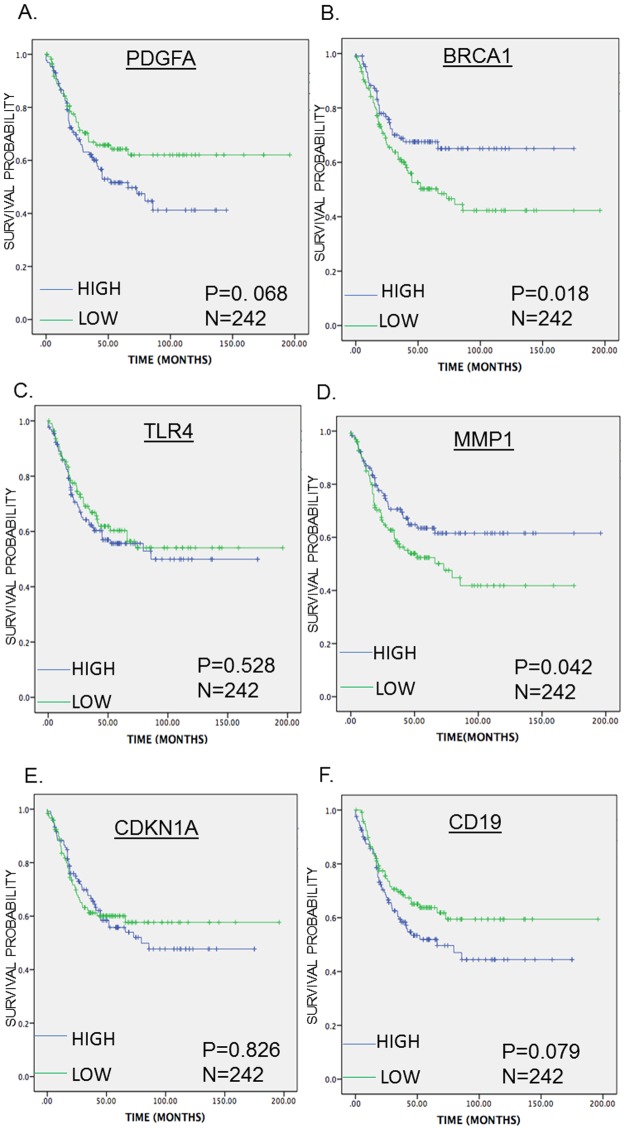
The SurvExpress tool was used to assess the enriched DEGs for their ability to predict OS and PFS in mCRC. High PDGFA expression levels were associated with poor OS, whereas high BRCA1 and MMP1 expression levels were associated with favorable PFS. However, the expression levels of CD19, CDKN1A, and TLR4 were neither associated with OS nor PFS (Figs 4 and 5).
Fig 4. Kaplan-Meier survival curves presenting the prognostic relationship between high and low expression of specific genes involved in bevacizumab resistance to overall survival (OS) in TNM clinical stage 3 and 4(A) PDGFA, (B) BRCA1, (C) TLR4, (D) MMP1 (E) CD19 and (F) CDKN1A expression.
The survival curves were plotted using the survExpress online tool. The specific DEGs expression levels were dichotomized by median value and stratified for TNM clinical stage. The results presented visually by Kaplan-Meier survival plots. p-values were calculated using log-rank statistics. Patient number(N) = 270, p = Logrank p-value.
Fig 5. Kaplan-Meier survival curves presenting the relationship between high and low expression of specific genes involved in bevacizumab resistance to Progress Free survival in TNM clinical stage 3 and 4 (A) PDGFA, (B) BRCA1, (C) TLR4, (D) MMP1 (E) CDKN1A and (F) CD19 expression.
The survival curves were plotted using the survExpress online tool. The specific DEGs expression levels were dichotomized by median value and stratified for TNM clinical stage. The results presented visually by Kaplan-Meier survival plots. P-values were calculated using log-rank statistics. Patient number(N) = 242, p = Logrank p-value.
Mechanism of gene correlation in tumor tissues
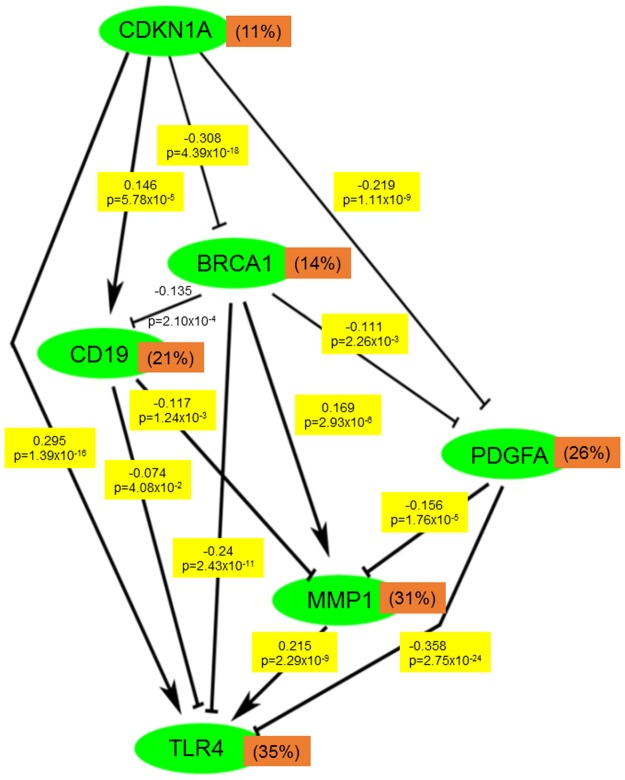
To elucidate the mechanism underlying the gene–gene correlation of the DEGs, TCGA RNASeq level 3 CRC data were used. BRCA1 was negatively correlated with PDGFA, CDKNA1, CD19, and TLR4 and positively correlated with MMP1. Moreover, PDGFA was negatively correlated with CDKNA1, BRCA1, MMP1, and TLR4. TLR4 was positively correlated with CDKNA1 and MMP1 and negatively correlated with CD19 and BRCA1. Furthermore, CD19 was positively correlated with CDKNA1 and negatively correlated with BRCA1, MMP1, and TLR4. However, PDGFA and CD19 were not significantly correlated (Fig 6).
Fig 6. Gene expression correlation of the DEGs involved in the pathways in the colorectal carcinoma tumor samples from TCGA data base.
TCGA RNASeq Level 3 data was used and Pearsons correlation coefficients (-1 to 1) were calculated. Number of patients = 653. The percentage presents the coefficient of variation of the genes, the lines presents negative correlation and the arrow presents positive correlation (p-value <0.05).
Discussion
The overall mortality of CRC has remained unchanged over the past decades, despite advances in surgical and medical therapy [40, 41]. This is due to the difficulties associated with early detection of the disease and the development of acquired therapeutic resistance, leading to ineffective treatment in patients with metastatic diseases [42–44]. Therefore, the etiological factors and mechanisms of acquired therapeutic resistance must be explored to improve survival rates and prevent disease recurrence [43]. Microarray technology has been widely used in the identification of therapeutic targets for diagnosis and prognosis of cancers [45, 46]. Our study performed a systematic bioinformatic analysis of the microarray data of HT29 xenograft tumor models with acquired bevacizumab resistance and identified 124 DEGs, 57 of which were upregulated and 67 were downregulated. CD19, BRCA1, PDGFA, CDKNA1, MMP1, and TLR4 exhibited high PPI degrees and were enriched in the PI3K-AKT signaling pathway, bladder cancer, and microRNAs in cancer; however, only high PDGFA expression levels were associated with poor OS, whereas high BRCA1 and MMP1 expression levels were associated with favorable PFS. These discrepancies may be because the study cohort was not specifically on bevacizumab treatment, thus suggesting that biomarkers that predict OS do not specifically predict PFS. Therefore, to confidently interpret the study results, these biomarkers require further assessment in patients specifically treated with bevacizumab.
The results of this study reveal PDGFA overexpression to be associated with bevacizumab resistance and the prognosis of patients with mCRC. These results are consistent with those of a previous study, which identified PDGFA as a potential predictor of therapeutic resistance and an individual prognostic marker for bevacizumab treatment, because PDGFA expression was observed to be decreased after single-dose bevacizumab treatment in responders but remained unchanged in nonresponders [47]. PDGFA targeting with the PDGF receptor has been reported to increase chemotherapeutic sensitivity in different cancers [47–50]. Therefore, our study supports the current understanding that PDGFA acts not only as a predictor of treatment response but also as a prognostic factor, because PDGFA upregulation not only limited the response to bevacizumab but also affected the prognosis of patients with mCRC in this study. Notably, PDGF overexpression has been implicated in bevacizumab resistance and poor prognosis in bevacizumab-treated patients because the PDGF pathway is considered an alternative pathway in the development of bevacizumab resistance [51, 52].
The expression levels of MMP1 and BRCA1 were associated with PFS in patients with mCRC. Although this study is the first to demonstrate the aforementioned relationship in mCRC, MMPs have received attention in terms of their role in the mechanism underlying resistance to antiangiogenic therapy, because increased MMP2 and MMP9 expression levels have been associated with resistance to the anti-VEGF and antiplacental growth factor drug aflibercept and with poor OS [53, 54]. Furthermore, MMP1 expression has been strongly associated with tumor metastasis and adverse outcomes in mCRC and has been suggested as a potential prognostic and therapeutic target [55–59]. A previous study reported that BRCA1 is associated with early onset CRC and functions as a DNA repair gene to cytotoxic drugs [60]. BRCA1 has been considered as a predictor of treatment response and prognosis in breast, ovarian, and lung cancers [61–66]; however, its role in mCRC and bevacizumab resistance is yet to be explored. The present results suggest that BRCA1 may exert protective effects in mCRC; therefore, BRCA1 should be thoroughly studied because BRCA1 targeting might not only increase the prognostic and therapeutic effects of bevacizumab but also affect the expression levels of its associated genes, namely PDGFA, CDKN1A, TLR4, and MMP1.
CD19, CDKN1A, and TLR4 have also been reported to influence therapeutic resistance or overall prognosis in cancer. CD19 has been associated with chemotherapy and multidrug resistance in many hematological tumors, and plays a central role in targeted therapeutics against B-cell malignancies (because of its expression patterns throughout the B-cell lineage), and against most B-cell malignancies with successful preclinical experiments and first-generation clinical trials [67–72]. CDKN1A has been implicated in cell cycle regulation, cell death, DNA repair, and cell motility [73]. Studies have demonstrated CDKN1A overexpression to be associated with poor prognosis in gastric and esophageal carcinomas [74, 75]. Furthermore, studies have reported that TLR4 plays a role in CRC; polymorphisms increasing TLR4 signaling led to a highly aggressive CRC, whereas those reducing TLR4 signaling exerted protective effects [76, 77]. In addition, high TLR4 expression levels have been associated with highly advanced grades of colonic neoplasia and with lower OS, a high probability of CRC relapse, and the presence of liver metastases in humans [78–81]. Studies have also suggested TLR4 to promote angiogenesis in different cancers by activating the PI3K-AKT signaling pathway to induce VEGF expression. In addition, TLR4 inhibition is associated with VEGF inhibition [82–84]. This finding can explain TLR4 downregulation in the bevacizumab-resistant tumors in this study; however, in vitro validation of this finding is required.
Notably, five of the six genes that were commonly enriched as well as associated with bevacizumab resistance belonged to the PI3K-AKT signaling pathway. Therefore, we suggest that the PI3K-AKT signaling pathway is responsible for restraining the therapeutic efficacy of bevacizumab in mCRC. This observation is in accordance with the results of previous studies, which have suggested that modifications in the PI3K-AKT signaling pathway increase bevacizumab resistance as an alternative pathway to VEGF inhibition [85–87]. Moreover, the occurrence of mutations in the PI3K-AKT signaling pathway remains the main challenge for mCRC treatment with new biological agents [86, 88, 89].
The present findings provide novel data that could predict bevacizumab treatment response and the emergence of resistance. Furthermore, this approach can predict patient prognosis; however, additional studies are required to validate the study findings and determine their clinical applicability.
Supporting information
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254 . [DOI] [PubMed] [Google Scholar]
- 2.Nordlinger B, Van Cutsem E, Gruenberger T, Glimelius B, Poston G, Rougier P, et al. Combination of surgery and chemotherapy and the role of targeted agents in the treatment of patients with colorectal liver metastases: recommendations from an expert panel. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2009;20(6):985–92. Epub 2009/01/21. doi: 10.1093/annonc/mdn735 . [DOI] [PubMed] [Google Scholar]
- 3.Des Guetz G, Uzzan B, Morere JF, Perret G, Nicolas P. Duration of adjuvant chemotherapy for patients with non-metastatic colorectal cancer. The Cochrane database of systematic reviews. 2010;(1):Cd007046 Epub 2010/01/22. doi: 10.1002/14651858.CD007046.pub2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wadlow RC, Ryan DP. The role of targeted agents in preoperative chemoradiation for rectal cancer. Cancer. 2010;116(15):3537–48. Epub 2010/06/22. doi: 10.1002/cncr.25155 . [DOI] [PubMed] [Google Scholar]
- 5.Tebbutt NC, Cattell E, Midgley R, Cunningham D, Kerr D. Systemic treatment of colorectal cancer. European journal of cancer (Oxford, England: 1990). 2002;38(7):1000–15. Epub 2002/04/30. . [DOI] [PubMed] [Google Scholar]
- 6.Van Cutsem E, Tabernero J, Lakomy R, Prenen H, Prausova J, Macarulla T, et al. Addition of aflibercept to fluorouracil, leucovorin, and irinotecan improves survival in a phase III randomized trial in patients with metastatic colorectal cancer previously treated with an oxaliplatin-based regimen. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30(28):3499–506. Epub 2012/09/06. doi: 10.1200/jco.2012.42.8201 . [DOI] [PubMed] [Google Scholar]
- 7.Andre T, de Gramont A, Vernerey D, Chibaudel B, Bonnetain F, Tijeras-Raballand A, et al. Adjuvant Fluorouracil, Leucovorin, and Oxaliplatin in Stage II to III Colon Cancer: Updated 10-Year Survival and Outcomes According to BRAF Mutation and Mismatch Repair Status of the MOSAIC Study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33(35):4176–87. Epub 2015/11/04. doi: 10.1200/jco.2015.63.4238 . [DOI] [PubMed] [Google Scholar]
- 8.Vanneman M, Dranoff G. Combining immunotherapy and targeted therapies in cancer treatment. Nature reviews Cancer. 2012;12(4):237–51. Epub 2012/03/23. doi: 10.1038/nrc3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabbinavar FF, Schulz J, McCleod M, Patel T, Hamm JT, Hecht JR, et al. Addition of bevacizumab to bolus fluorouracil and leucovorin in first-line metastatic colorectal cancer: results of a randomized phase II trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23(16):3697–705. Epub 2005/03/02. doi: 10.1200/jco.2005.05.112 . [DOI] [PubMed] [Google Scholar]
- 10.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. The New England journal of medicine. 2004;350(23):2335–42. Epub 2004/06/04. doi: 10.1056/NEJMoa032691 . [DOI] [PubMed] [Google Scholar]
- 11.Loupakis F, Cremolini C, Masi G, Lonardi S, Zagonel V, Salvatore L, et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. The New England journal of medicine. 2014;371(17):1609–18. Epub 2014/10/23. doi: 10.1056/NEJMoa1403108 . [DOI] [PubMed] [Google Scholar]
- 12.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. The New England journal of medicine. 2008;359(17):1757–65. Epub 2008/10/24. doi: 10.1056/NEJMoa0804385 . [DOI] [PubMed] [Google Scholar]
- 13.Saltz LB, Clarke S, Diaz-Rubio E, Scheithauer W, Figer A, Wong R, et al. Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26(12):2013–9. Epub 2008/04/19. doi: 10.1200/jco.2007.14.9930 . [DOI] [PubMed] [Google Scholar]
- 14.Giantonio BJ, Catalano PJ, Meropol NJ, O'Dwyer PJ, Mitchell EP, Alberts SR, et al. Bevacizumab in combination with oxaliplatin, fluorouracil, and leucovorin (FOLFOX4) for previously treated metastatic colorectal cancer: results from the Eastern Cooperative Oncology Group Study E3200. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25(12):1539–44. Epub 2007/04/20. doi: 10.1200/jco.2006.09.6305 . [DOI] [PubMed] [Google Scholar]
- 15.Yamazaki K, Nagase M, Tamagawa H, Ueda S, Tamura T, Murata K, et al. Randomized phase III study of bevacizumab plus FOLFIRI and bevacizumab plus mFOLFOX6 as first-line treatment for patients with metastatic colorectal cancer (WJOG4407G). Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2016;27(8):1539–46. Epub 2016/05/15. doi: 10.1093/annonc/mdw206 . [DOI] [PubMed] [Google Scholar]
- 16.Lenz H-J, Lee F-C, Yau L, Koh HA, Knost JA, Mitchell EP, et al. MAVERICC, a phase 2 study of mFOLFOX6-bevacizumab (BV) vs FOLFIRI-BV with biomarker stratification as first-line (1L) chemotherapy (CT) in patients (pts) with metastatic colorectal cancer (mCRC). Journal of Clinical Oncology. 2016;34(4_suppl):493-. doi: 10.1200/jco.2016.34.4_suppl.493 [Google Scholar]
- 17.Kramer I, Lipp HP. Bevacizumab, a humanized anti-angiogenic monoclonal antibody for the treatment of colorectal cancer. Journal of clinical pharmacy and therapeutics. 2007;32(1):1–14. Epub 2007/02/09. doi: 10.1111/j.1365-2710.2007.00800.x . [DOI] [PubMed] [Google Scholar]
- 18.Maru D, Venook AP, Ellis LM. Predictive biomarkers for bevacizumab: are we there yet? Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19(11):2824–7. Epub 2013/04/04. doi: 10.1158/1078-0432.ccr-12-3409 . [DOI] [PubMed] [Google Scholar]
- 19.Heinemann V, Douillard JY, Ducreux M, Peeters M. Targeted therapy in metastatic colorectal cancer—an example of personalised medicine in action. Cancer treatment reviews. 2013;39(6):592–601. Epub 2013/02/05. doi: 10.1016/j.ctrv.2012.12.011 . [DOI] [PubMed] [Google Scholar]
- 20.Jain RK, Duda DG, Willett CG, Sahani DV, Zhu AX, Loeffler JS, et al. Biomarkers of response and resistance to antiangiogenic therapy. Nature reviews Clinical oncology. 2009;6(6):327–38. Epub 2009/06/02. doi: 10.1038/nrclinonc.2009.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic acids research. 2007;35(Database issue):D760–5. Epub 2006/11/14. doi: 10.1093/nar/gkl887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aubert J, Bar-Hen A, Daudin JJ, Robin S. Determination of the differentially expressed genes in microarray experiments using local FDR. BMC bioinformatics. 2004;5:125 Epub 2004/09/08. doi: 10.1186/1471-2105-5-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43(7):e47 Epub 2015/01/22. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics (Oxford, England). 2007;23(14):1846–7. Epub 2007/05/15. doi: 10.1093/bioinformatics/btm254 . [DOI] [PubMed] [Google Scholar]
- 25.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods in enzymology. 2006;411:134–93. Epub 2006/08/31. doi: 10.1016/S0076-6879(06)11009-5 . [DOI] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic acids research. 2015;43(Database issue):D447–52. Epub 2014/10/30. doi: 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics (Oxford, England). 2005;21(16):3448–9. Epub 2005/06/24. doi: 10.1093/bioinformatics/bti551 . [DOI] [PubMed] [Google Scholar]
- 28.Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Bio Systems. 2015;127:67–72. Epub 2014/12/03. doi: 10.1016/j.biosystems.2014.11.005 . [DOI] [PubMed] [Google Scholar]
- 29.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, et al. A travel guide to Cytoscape plugins. Nature methods. 2012;9(11):1069–76. Epub 2012/11/08. doi: 10.1038/nmeth.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13(11):2498–504. Epub 2003/11/05. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. Epub 2009/01/10. doi: 10.1038/nprot.2008.211 . [DOI] [PubMed] [Google Scholar]
- 32.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25(1):25–9. Epub 2000/05/10. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic acids research. 2016;44(D1):D457–62. Epub 2015/10/18. doi: 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28(1):27–30. Epub 1999/12/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E, Martinez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PloS one. 2013;8(9):e74250 Epub 2013/09/26. doi: 10.1371/journal.pone.0074250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staub E, Groene J, Heinze M, Mennerich D, Roepcke S, Klaman I, et al. An expression module of WIPF1-coexpressed genes identifies patients with favorable prognosis in three tumor types. Journal of molecular medicine (Berlin, Germany). 2009;87(6):633–44. Epub 2009/04/29. doi: 10.1007/s00109-009-0467-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15(24):7642–51. Epub 2009/12/10. doi: 10.1158/1078-0432.ccr-09-1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138(3):958–68. Epub 2009/11/17. doi: 10.1053/j.gastro.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):7131–6. Epub 2009/04/11. doi: 10.1073/pnas.0902232106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venook AP, Weiser MR, Tepper JE. Colorectal cancer: all hands on deck. American Society of Clinical Oncology educational book American Society of Clinical Oncology Meeting. 2014:83–9. Epub 2014/05/27. doi: 10.14694/EdBook_AM.2014.34.83 . [DOI] [PubMed] [Google Scholar]
- 41.Gallagher DJ, Kemeny N. Metastatic colorectal cancer: from improved survival to potential cure. Oncology. 2010;78(3–4):237–48. Epub 2010/06/05. doi: 10.1159/000315730 . [DOI] [PubMed] [Google Scholar]
- 42.Ross JS, Torres-Mora J, Wagle N, Jennings TA, Jones DM. Biomarker-based prediction of response to therapy for colorectal cancer: current perspective. American journal of clinical pathology. 2010;134(3):478–90. Epub 2010/08/19. doi: 10.1309/AJCP2Y8KTDPOAORH . [DOI] [PubMed] [Google Scholar]
- 43.El Khoury F, Corcos L, Durand S, Simon B, Le Jossic-Corcos C. Acquisition of anticancer drug resistance is partially associated with cancer stemness in human colon cancer cells. International journal of oncology. 2016. Epub 2016/10/18. doi: 10.3892/ijo.2016.3725 . [DOI] [PubMed] [Google Scholar]
- 44.Candeil L, Gourdier I, Peyron D, Vezzio N, Copois V, Bibeau F, et al. ABCG2 overexpression in colon cancer cells resistant to SN38 and in irinotecan-treated metastases. International journal of cancer. 2004;109(6):848–54. Epub 2004/03/18. doi: 10.1002/ijc.20032 . [DOI] [PubMed] [Google Scholar]
- 45.Levy P, Ripoche H, Laurendeau I, Lazar V, Ortonne N, Parfait B, et al. Microarray-based identification of tenascin C and tenascin XB, genes possibly involved in tumorigenesis associated with neurofibromatosis type 1. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13(2 Pt 1):398–407. Epub 2007/01/05. doi: 10.1158/1078-0432.ccr-06-0182 . [DOI] [PubMed] [Google Scholar]
- 46.Advani R, Burington B, Shi X, de Vos S, Ansell S, Forero-Torres A, et al. Evaluation of a gene signature to predict single agent dacetuzumab (SGN-40) activity in patients with DLBCL. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27(15_suppl):11063. Epub 2009/05/20. [Google Scholar]
- 47.Verstraete M, Debucquoy A, Dekervel J, van Pelt J, Verslype C, Devos E, et al. Combining bevacizumab and chemoradiation in rectal cancer. Translational results of the AXEBeam trial. British journal of cancer. 2015;112(8):1314–25. Epub 2015/04/14. doi: 10.1038/bjc.2015.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Cesne A, Blay JY, Reichardt P, Joensuu H. Optimizing tyrosine kinase inhibitor therapy in gastrointestinal stromal tumors: exploring the benefits of continuous kinase suppression. The oncologist. 2013;18(11):1192–9. Epub 2013/10/19. doi: 10.1634/theoncologist.2012-0361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luengo-Gil G, Gonzalez-Billalabeitia E, Chaves-Benito A, Garcia Martinez E, Garcia Garre E, Vicente V, et al. Effects of conventional neoadjuvant chemotherapy for breast cancer on tumor angiogenesis. Breast cancer research and treatment. 2015;151(3):577–87. Epub 2015/05/15. doi: 10.1007/s10549-015-3421-4 . [DOI] [PubMed] [Google Scholar]
- 50.Nagaraju GP, Park W, Wen J, Mahaseth H, Landry J, Farris AB, et al. Antiangiogenic effects of ganetespib in colorectal cancer mediated through inhibition of HIF-1alpha and STAT-3. Angiogenesis. 2013;16(4):903–17. Epub 2013/07/11. doi: 10.1007/s10456-013-9364-7 . [DOI] [PubMed] [Google Scholar]
- 51.Paulsson J, Ehnman M, Ostman A. PDGF receptors in tumor biology: prognostic and predictive potential. Future oncology (London, England). 2014;10(9):1695–708. Epub 2014/08/26. doi: 10.2217/fon.14.83 . [DOI] [PubMed] [Google Scholar]
- 52.Okamoto S, Nitta M, Maruyama T, Sawada T, Komori T, Okada Y, et al. Bevacizumab changes vascular structure and modulates the expression of angiogenic factors in recurrent malignant gliomas. Brain tumor pathology. 2016;33(2):129–36. Epub 2016/01/31. doi: 10.1007/s10014-016-0248-6 . [DOI] [PubMed] [Google Scholar]
- 53.de Groot JF, Piao Y, Tran H, Gilbert M, Wu HK, Liu J, et al. Myeloid biomarkers associated with glioblastoma response to anti-VEGF therapy with aflibercept. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17(14):4872–81. Epub 2011/06/03. doi: 10.1158/1078-0432.ccr-11-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu-Emerson C, Snuderl M, Kirkpatrick ND, Goveia J, Davidson C, Huang Y, et al. Increase in tumor-associated macrophages after antiangiogenic therapy is associated with poor survival among patients with recurrent glioblastoma. Neuro-oncology. 2013;15(8):1079–87. Epub 2013/07/06. doi: 10.1093/neuonc/not082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Said AH, Hu S, Abutaleb A, Watkins T, Cheng K, Chahdi A, et al. Interacting post-muscarinic receptor signaling pathways potentiate matrix metalloproteinase-1 expression and invasion of human colon cancer cells. The Biochemical journal. 2017;474(5):647–65. Epub 2016/12/23. doi: 10.1042/BCJ20160704 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davudian S, Shajari N, Kazemi T, Mansoori B, Salehi S, Mohammadi A, et al. BACH1 silencing by siRNA inhibits migration of HT-29 colon cancer cells through reduction of metastasis-related genes. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2016;84:191–8. Epub 2016/09/23. doi: 10.1016/j.biopha.2016.09.021 . [DOI] [PubMed] [Google Scholar]
- 57.Pietruszewska W, Bojanowska-Pozniak K, Kobos J. Matrix metalloproteinases MMP1, MMP2, MMP9 and their tissue inhibitors TIMP1, TIMP2, TIMP3 in head and neck cancer: an immunohistochemical study. Otolaryngologia polska = The Polish otolaryngology. 2016;70(3):32–43. Epub 2016/07/09. doi: 10.5604/00306657.1202546 . [DOI] [PubMed] [Google Scholar]
- 58.Slattery ML, Lundgreen A. The influence of the CHIEF pathway on colorectal cancer-specific mortality. PloS one. 2014;9(12):e116169 Epub 2014/12/30. doi: 10.1371/journal.pone.0116169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu ZH, Fang YJ, Wu XJ, Pan ZZ, Wan DS. [Expression of matrix metalloproteinase 1 in tissue of colon carcinoma and its clinical prognostic significance]. Zhonghua yi xue za zhi. 2011;91(41):2895–8. Epub 2012/02/16. . [PubMed] [Google Scholar]
- 60.Anders CK, Winer EP, Ford JM, Dent R, Silver DP, Sledge GW, et al. Poly(ADP-Ribose) polymerase inhibition: "targeted" therapy for triple-negative breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16(19):4702–10. Epub 2010/09/23. doi: 10.1158/1078-0432.ccr-10-0939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carser JE, Quinn JE, Michie CO, O'Brien EJ, McCluggage WG, Maxwell P, et al. BRCA1 is both a prognostic and predictive biomarker of response to chemotherapy in sporadic epithelial ovarian cancer. Gynecologic oncology. 2011;123(3):492–8. Epub 2011/09/17. doi: 10.1016/j.ygyno.2011.08.017 . [DOI] [PubMed] [Google Scholar]
- 62.Quinn JE, James CR, Stewart GE, Mulligan JM, White P, Chang GK, et al. BRCA1 mRNA expression levels predict for overall survival in ovarian cancer after chemotherapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13(24):7413–20. Epub 2007/12/21. doi: 10.1158/1078-0432.ccr-07-1083 . [DOI] [PubMed] [Google Scholar]
- 63.Ignatov T, Eggemann H, Costa SD, Roessner A, Kalinski T, Ignatov A. BRCA1 promoter methylation is a marker of better response to platinum-taxane-based therapy in sporadic epithelial ovarian cancer. Journal of cancer research and clinical oncology. 2014;140(9):1457–63. Epub 2014/05/16. doi: 10.1007/s00432-014-1704-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lips EH, Michaut M, Hoogstraat M, Mulder L, Besselink NJ, Koudijs MJ, et al. Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast cancer research: BCR. 2015;17(1):134 Epub 2015/10/05. doi: 10.1186/s13058-015-0642-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andre F, Zielinski CC. Optimal strategies for the treatment of metastatic triple-negative breast cancer with currently approved agents. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2012;23 Suppl 6:vi46–51. Epub 2012/10/04. doi: 10.1093/annonc/mds195 . [DOI] [PubMed] [Google Scholar]
- 66.Villaruz LC, Socinski MA. Personalized therapy for non-small cell lung cancer: which drug for which patient? Seminars in thoracic and cardiovascular surgery. 2011;23(4):281–90. Epub 2011/01/01. doi: 10.1053/j.semtcvs.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaplan JB, Grischenko M, Giles FJ. Blinatumomab for the treatment of acute lymphoblastic leukemia. Investigational new drugs. 2015;33(6):1271–9. Epub 2015/09/19. doi: 10.1007/s10637-015-0289-4 . [DOI] [PubMed] [Google Scholar]
- 68.Wu J, Fu J, Zhang M, Liu D. Blinatumomab: a bispecific T cell engager (BiTE) antibody against CD19/CD3 for refractory acute lymphoid leukemia. Journal of hematology & oncology. 2015;8:104 Epub 2015/09/05. doi: 10.1186/s13045-015-0195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turtle CJ, Riddell SR, Maloney DG. CD19-Targeted chimeric antigen receptor-modified T-cell immunotherapy for B-cell malignancies. Clinical pharmacology and therapeutics. 2016;100(3):252–8. Epub 2016/05/14. doi: 10.1002/cpt.392 . [DOI] [PubMed] [Google Scholar]
- 70.Hay KA, Turtle CJ. Chimeric Antigen Receptor (CAR) T Cells: Lessons Learned from Targeting of CD19 in B-Cell Malignancies. Drugs. 2017;77(3):237–45. Epub 2017/01/23. doi: 10.1007/s40265-017-0690-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramos CA, Savoldo B, Torrano V, Ballard B, Zhang H, Dakhova O, et al. Clinical responses with T lymphocytes targeting malignancy-associated kappa light chains. The Journal of clinical investigation. 2016;126(7):2588–96. Epub 2016/06/09. doi: 10.1172/JCI86000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raufi A, Ebrahim AS, Al-Katib A. Targeting CD19 in B-cell lymphoma: emerging role of SAR3419. Cancer management and research. 2013;5:225–33. Epub 2013/09/12. doi: 10.2147/CMAR.S45957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cellular signalling. 2010;22(7):1003–12. Epub 2010/01/27. doi: 10.1016/j.cellsig.2010.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gomyo Y, Ikeda M, Osaki M, Tatebe S, Tsujitani S, Ikeguchi M, et al. Expression of p21 (waf1/cip1/sdi1), but not p53 protein, is a factor in the survival of patients with advanced gastric carcinoma. Cancer. 1997;79(11):2067–72. Epub 1997/06/01. . [DOI] [PubMed] [Google Scholar]
- 75.Taghavi N, Biramijamal F, Sotoudeh M, Moaven O, Khademi H, Abbaszadegan MR, et al. Association of p53/p21 expression with cigarette smoking and prognosis in esophageal squamous cell carcinoma patients. World journal of gastroenterology. 2010;16(39):4958–67. Epub 2010/10/19. doi: 10.3748/wjg.v16.i39.4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eyking A, Ey B, Runzi M, Roig AI, Reis H, Schmid KW, et al. Toll-like receptor 4 variant D299G induces features of neoplastic progression in Caco-2 intestinal cells and is associated with advanced human colon cancer. Gastroenterology. 2011;141(6):2154–65. Epub 2011/09/17. doi: 10.1053/j.gastro.2011.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Slattery ML, Herrick JS, Bondurant KL, Wolff RK. Toll-like receptor genes and their association with colon and rectal cancer development and prognosis. International journal of cancer. 2012;130(12):2974–80. Epub 2011/07/28. doi: 10.1002/ijc.26314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang CY, Shih CM, Tsao NW, Lin YW, Shih CC, Chiang KH, et al. The GroEL protein of Porphyromonas gingivalis regulates atherogenic phenomena in endothelial cells mediated by upregulating toll-like receptor 4 expression. American journal of translational research. 2016;8(2):384–404. Epub 2016/05/10. [PMC free article] [PubMed] [Google Scholar]
- 79.Wang EL, Qian ZR, Nakasono M, Tanahashi T, Yoshimoto K, Bando Y, et al. High expression of Toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. British journal of cancer. 2010;102(5):908–15. Epub 2010/02/11. doi: 10.1038/sj.bjc.6605558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Earl TM, Nicoud IB, Pierce JM, Wright JP, Majoras NE, Rubin JE, et al. Silencing of TLR4 decreases liver tumor burden in a murine model of colorectal metastasis and hepatic steatosis. Annals of surgical oncology. 2009;16(4):1043–50. Epub 2009/01/24. doi: 10.1245/s10434-009-0325-8 . [DOI] [PubMed] [Google Scholar]
- 81.Cammarota R, Bertolini V, Pennesi G, Bucci EO, Gottardi O, Garlanda C, et al. The tumor microenvironment of colorectal cancer: stromal TLR-4 expression as a potential prognostic marker. Journal of translational medicine. 2010;8:112 Epub 2010/11/10. doi: 10.1186/1479-5876-8-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Riddell JR, Bshara W, Moser MT, Spernyak JA, Foster BA, Gollnick SO. Peroxiredoxin 1 controls prostate cancer growth through Toll-like receptor 4-dependent regulation of tumor vasculature. Cancer research. 2011;71(5):1637–46. Epub 2011/02/24. doi: 10.1158/0008-5472.CAN-10-3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murad S. Toll-like receptor 4 in inflammation and angiogenesis: a double-edged sword. Frontiers in immunology. 2014;5:313 Epub 2014/07/30. doi: 10.3389/fimmu.2014.00313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun Y, Wu C, Ma J, Yang Y, Man X, Wu H, et al. Toll-like receptor 4 promotes angiogenesis in pancreatic cancer via PI3K/AKT signaling. Experimental cell research. 2016;347(2):274–82. Epub 2016/07/19. doi: 10.1016/j.yexcr.2016.07.009 . [DOI] [PubMed] [Google Scholar]
- 85.Piha-Paul SA, Wheler JJ, Fu S, Levenback C, Lu K, Falchook GS, et al. Advanced gynecologic malignancies treated with a combination of the VEGF inhibitor bevacizumab and the mTOR inhibitor temsirolimus. Oncotarget. 2014;5(7):1846–55. Epub 2014/04/20. doi: 10.18632/oncotarget.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu X, Kambrick S, Fu S, Naing A, Subbiah V, Blumenschein GR, et al. Advanced malignancies treated with a combination of the VEGF inhibitor bevacizumab, anti-EGFR antibody cetuximab, and the mTOR inhibitor temsirolimus. Oncotarget. 2016;7(17):23227–38. Epub 2016/03/05. doi: 10.18632/oncotarget.7594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiong YQ, Sun HC, Zhu XD, Zhang W, Zhuang PY, Zhang JB, et al. Bevacizumab enhances chemosensitivity of hepatocellular carcinoma to adriamycin related to inhibition of survivin expression. Journal of cancer research and clinical oncology. 2011;137(3):505–12. Epub 2010/05/22. doi: 10.1007/s00432-010-0914-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moench R, Grimmig T, Kannen V, Tripathi S, Faber M, Moll EM, et al. Exclusive inhibition of PI3K/Akt/mTOR signaling is not sufficient to prevent PDGF-mediated effects on glycolysis and proliferation in colorectal cancer. Oncotarget. 2016. Epub 2016/09/15. doi: 10.18632/oncotarget.11899 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maurel J, Postigo A. Prognostic and Predictive Biomarkers in Colorectal Cancer. From the Preclinical Setting to Clinical Practice. Current cancer drug targets. 2015;15(8):703–15. Epub 2015/10/11. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.