Abstract
Background
Unruptured intracranial aneurysms (IAs) are typically asymptomatic and undetected except for incidental discovery on imaging. Blood-based diagnostic biomarkers could lead to improvements in IA management. This exploratory study examined circulating neutrophils to determine whether they carry RNA expression signatures of IAs.
Methods
Blood samples were collected from patients receiving cerebral angiography. Eleven samples were collected from patients with IAs and 11 from patients without IAs as controls. Samples from the two groups were paired based on demographics and comorbidities. RNA was extracted from isolated neutrophils and subjected to next-generation RNA sequencing to obtain differential expressions for identification of an IA-associated signature. Bioinformatics analyses, including gene set enrichment analysis and Ingenuity Pathway Analysis, were used to investigate the biological function of all differentially expressed transcripts.
Results
Transcriptome profiling identified 258 differentially expressed transcripts in patients with and without IAs. Expression differences were consistent with peripheral neutrophil activation. An IA-associated RNA expression signature was identified in 82 transcripts (p<0.05, fold-change ≥2). This signature was able to separate patients with and without IAs on hierarchical clustering. Furthermore, in an independent, unpaired, replication cohort of patients with IAs (n = 5) and controls (n = 5), the 82 transcripts separated 9 of 10 patients into their respective groups.
Conclusion
Preliminary findings show that RNA expression from circulating neutrophils carries an IA-associated signature. These findings highlight a potential to use predictive biomarkers from peripheral blood samples to identify patients with IAs.
Introduction
Intracranial aneurysm (IA) rupture can be fatal. An estimated 5% of Americans harbor unruptured IAs [1]. Because most unruptured aneurysms are asymptomatic, they remain dormant, often being found after rupture. Early IA detection would enable monitoring and treatment to prevent rupture and its devastating sequelae. Currently, most unruptured IAs are detected incidentally by cerebral imaging performed for other reasons. However, imaging is unsuitable for general IA screening due to potential risks and high costs [2]. Even for high-risk individuals (e.g., those with a family history of IA), it is debated whether imaging is cost-effective [2, 3]. Noninvasive and inexpensive strategies such as blood testing could offer a diagnostic alternative.
Recent studies have correlated RNA expression differences in the circulating blood with vascular diseases, such as thoracic aortic aneurysm [4]. We hypothesized that circulating neutrophils carry transcriptional signatures of IAs. Our rationale was that aneurysmal lesions are associated with persistent vascular wall inflammation, which can involve neutrophil responses [5]. Results from genome wide association studies suggest that IA-associated genes are involved in endothelial function, extracellular matrix maintenance, and inflammatory responses [6, 7]. Furthermore, gene expression profiling of IA tissue has demonstrated increased presence of inflammatory cytokines and chemoattractant proteins in the aneurysm wall [8, 9]. The presence of the neutrophil proteins, neutrophil gelatinase-associated lipocalin (NGAL), and myeloperoxidase (MPO) is consistent with a direct role of neutrophils in the degeneration of the vessel wall during IA natural history [10, 11]. As circulating neutrophils have been shown to display unique expression differences in other diseases characterized by inflammation [12], a continual interaction between neutrophils and an IA could leave imprints on the RNA expression of circulating neutrophils.
In this study, we investigated whether neutrophils have different RNA expression profiles in patients with IAs compared to patients without IAs. We recruited patients with and without aneurysms (confirmed on angiography) and paired them based on demographics and comorbidities. Next-generation RNA sequencing of circulating neutrophils was performed to identify an IA-associated expression signature in their transcriptomes. We further assessed if the IA-associated expression signature could distinguish patients with and without IA in a heterogeneous independent cohort of patients. Gene ontology analysis and physiological pathway modeling were used to determine the biological function of differentially expressed transcripts in IA. Results from this study could motivate future efforts towards developing blood-based biomarkers and shed light on the pathophysiology of IAs.
Materials and methods
Clinical study
This study was approved by our institutional review board (study no. 030–474433). Methods were carried out in accordance with the approved protocol. Written informed consent was obtained from all subjects. Between November 2013 and May 2014, 77 peripheral blood samples were collected from patients undergoing cerebral digital subtraction angiography (DSA) at our institute: 35 patients had a positive IA diagnosis and 42 had a negative IA diagnosis (controls). Positive or negative IA diagnosis was confirmed by imaging, and patient medical records were collected for pairing patients with IAs to controls. Additionally, each patient’s complete blood count, which was taken within 3 months of blood collection, was recorded.
Patients undergoing cerebral digital subtraction angiography (DSA) with positive and negative intracranial aneurysm (IA) diagnoses were enrolled in this study. Reasons for the patients to receive DSA included confirmation of findings from noninvasive imaging of the presence of IAs, vascular malformations, or carotid stenosis or follow-up noninvasive imaging of previously detected IAs. All consenting patients were older than 18 years, were English speaking, and had not received previous treatment for IA. To ensure that differences in the circulating neutrophils were not influenced by inherent inflammatory conditions, we excluded patients who potentially had altered leukocyte transcriptomes; this included patients who were pregnant, had recently undergone invasive surgery, were undergoing chemotherapy, had a body temperature above 37.78°C (100oF), had received solid organ transplants, had autoimmune diseases, and those who were taking prednisone or any other immunomodulating drugs. Furthermore, the included patients did not have any other known cerebrovascular malformations or extracranial aneurysms, including abdominal aortic aneurysms.
Sample preparation
Sixteen mL of blood was drawn from the access catheter in the femoral artery and transferred into two 8 mL, citrated, cell preparation tubes (BD, Franklin Lakes, NJ). Neutrophils were isolated within 1 hour of peripheral blood collection, as described elsewhere [12]. Cell preparation tubes were centrifuged at 1,700 × g for 25 minutes to separate erythrocytes and neutrophils from mononuclear cells and plasma in the peripheral blood samples. Erythrocytes and neutrophils were collected into a 3 mL syringe and placed into an erythrocyte lysis buffer that was made in-house. After all erythrocytes were lysed, the neutrophils were isolated by centrifugation at 400 × g for 10 min and disrupted and stored in TRIzol reagent (Life Technologies, Carlsbad, CA) at -80 oC until further processing. Neutrophils isolated in this fashion are more than 98% CD66b+by flow cytometry and contain no contaminating CD14+ monocytes [13].
Total neutrophil RNA was extracted using TRIzol, according to the manufacturer’s instructions. Trace DNA was removed by DNase I (Life Technologies, Carlsbad, CA) treatment. The RNA was purified using the RNeasy MinElute Cleanup Kit (Qiagen, Venlo, Limburg, Netherlands) and suspended in RNase-free water. After RNA isolation, the purity and concentration of RNA in each sample was measured by absorbance at 260 nm on a NanoDrop 2000 (Thermo Scientific, Waltham, MA), and 200–400 ng of RNA was sent to our university’s Next-Generation Sequencing and Expression Analysis Core facility for further quality control. Precise RNA concentration was measured at the core facility via the Quant-iT RiboGreen Assay (Invitrogen, Carlsbad, CA) with a TBS-380 Fluorometer (Promega, Madison, WI), whereas the quality of the RNA samples was measured with an Agilent 2100 BioAnalyzer RNA 6000 Pico Chip (Agilent, Las Vegas, NV). RNA samples with 260/280 ≥ 1.9 and an RNA integrity number (RIN) ≥ 6.0 were considered for RNA sequencing.
Cohort creation
Before sequencing, samples from IA patients and control subjects were paired by demographics and comorbidities to control for confounding variables. First, samples that did not have acceptable RNA quality for sequencing were excluded. Next, each patient in the IA group was paired with a control subject by factors that have been reported in the literature to correlate with IA. These included (in order of decreasing importance) age, sex, smoking status (yes or no), presence of hypertension, presence of hyperlipidemia, and presence of heart disease [14]. Matching criteria also included stroke history, presence of diabetes mellitus, and presence of osteoarthritis, when possible. With the exception of age, the factors used for matching were quantified as binary data points. The clinical factors were retrieved from the patients’ medical records via the latest Patient Medical History form (see S1 Fig) administered prior to imaging. Since this form in the medical record contained self-reported information, the presence of each comorbidity was corroborated with each patient’s reported list of medications (e.g., hypertension with lisinopril, hyperlipidemia with simvastatin, heart disease with metoprolol, stroke history with clopidogrel, diabetes mellitus with metformin, and osteoarthritis with nonsteroidal anti-inflammatory drugs/tramadol). We were able to corroborate 84% of the clinical data points for patients’ comorbidities through their medication history.
After performing the original experiments to identify an IA-associated neutrophil expression signature, we used the same clinical protocol to recruit an additional 5 patients with IAs and 5 IA-free controls into a small replication cohort (n = 10) to test whether the IA-associated signature could separate IA patients from controls in the second cohort. Blood samples and RNA were handled in the same manner as those in the original cohort, and the same RNA sequencing and data analysis protocols were followed. However, prior to sequencing, we did not control for demographics and comorbidities to obtain a more heterogeneous cohort.
RNA sequencing
RNA libraries for these samples were constructed at our university’s Next-Generation Sequencing and Expression Analysis Core facility using the TruSeq RNA Library Preparation Kit (Illumina, San Diego, CA). All samples were subjected to 50-cycle, single-read sequencing in the HiSeq2500 (Illumina) and were demultiplexed using Bcl2Fastq v2.17.1.14 (Illumina). Gene expression analysis was completed using the Tuxedo Suite. Short RNA fragment data were compiled in FASTQ format and aligned to the human reference genome (human genome 19 [hg19]) using TopHat v2.1.13 [15]. Gene expression levels were calculated using fragments per kilobase of transcript per million mapped reads (FPKM) normalization in CuffLinks v2.2.1 [15]. RNA sequencing data files and processed transcript expression have been made available at NCBI’s GEO (accession no. GSE106520). To evaluate the quality of RNA sequencing, we performed quality control analysis using both FASTQC [16] before alignment and MultiQC [17] after alignment.
Differential expression analysis
Differential gene expression analysis was performed in CuffDiff v2.2.1 [18] and visualized in the CummeRbund v2.7.1 package in R [15, 19]. We used CuffDiff v2.2.1 (Trapnell Laboratory), which compared the log ratio of FPKM values in the IA and control groups against the log ratio of FPKM values of the IA group, and computed a test statistic [20]. The test statistic was calculated using the negative binomial distribution to model the variance of each sample and the square root of the Jensen-Shannon divergence to assess differences in relative abundance. The change in Jensen-Shannon divergence was then assigned a p-value, as described elsewhere [20].
Transcripts were considered significantly differentially expressed at p<0.05. We defined an IA-associated expression signature as those significant transcripts that also had an absolute fold-change ≥2. Post hoc power estimation was performed following Hart et al. [21] with α = 0.05, an average coefficient of variation of 0.404 (calculated from FPKMs), and counts per million mapped reads of 38. Multiple testing correction was performed by using the Benjamini-Hochberg method [22], and q-values were reported for each transcript.
Verification by RT-qPCR
To verify expression differences measured by RNA sequencing, quantitative reverse transcription polymerase chain reaction (RT-qPCR) was performed. We verified expression difference of 5 differentially expressed transcripts (CD177, SERPING1, GBP5, IL8, NAAA) in order to conserve RNA material. These 5 transcripts were chosen because they were among the most prominently differentially expressed transcripts, i.e., they were highly abundant (FPKM>10) and significantly differentially expressed (p<0.05) with an absolute fold-change >1.5. For each transcript, oligonucleotide primers were designed with a ~60°C melting temperature and a length of 15–25 nucleotides to produce PCR products with lengths of 50–200 base pairs using Primer3 software [23] and Primer BLAST (NCBI, Bethesda, MD). The replication efficiency of each primer set was tested by performing qPCR on serial dilutions of cDNA samples (primer sequences, annealing temperatures, efficiencies, and product lengths are shown in S1 Table).
For reverse transcription, first-strand cDNA was generated from total RNA using OmniScript Reverse Transcriptase kit (Qiagen, Venlo, Limburg, Netherlands) according to the manufacturer’s instructions. Quantitative PCR was run with 10 ng of cDNA in 25 μ reactions in triplicate in the Bio-Rad CFX Connect system (Bio-Rad, Hercules, CA) using ABI SYBR Green Master Mix (Applied Biosystems, Foster City, CA) and gene-specific primers at a concentration of 0.02 μM each. The temperature profile consisted of an initial step of 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute, and then a final melting curve analysis from 60°C to 95°C for 20 minutes. Gene-specific amplification was demonstrated by a single peak using the Bio-Rad dissociation melt curve. As previously described [12], GAPDH expression was used for normalization, and fold-changes between groups were calculated using the 2-ΔΔCt method.
Dimensionality reduction
We performed dimensionality reduction by unsupervised principal component analysis (PCA) and multidimensional scaling (MDS) using the transcriptomes of each sample in the CummeRbund and prcomp packages in R Bioconductor under the default settings [19]. For hierarchical clustering, we used the hclust package in R [24]. Dendrograms were created using Ward linkage from z-score normalized transcript levels.
Bioinformatics
We performed gene set enrichment analysis using the open-source software GO::TermFinder (Stanford University School of Medicine, Stanford, CA) [25]. This tool determined whether any gene ontology terms annotated two lists of genes (i.e., genes with higher expression in samples from patients with IAs than those without IA and genes with lower expression in samples from patients with IAs than those without IA) greater than what would be expected by chance. Significantly enriched ontologies were reported if the Q-Value was <0.05, based on significance testing using the hypergeometric distribution.
Networks of potential interactions were generated using Ingenuity Pathway Analysis (IPA) software (Qiagen Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) [26]. For IPA, each gene identifier was mapped to its corresponding gene object in the Ingenuity Knowledge Base and overlaid onto a molecular network derived from information accumulated in the Knowledge Base. Gene networks were algorithmically generated based on their “connectivity” derived from known interactions between the products of these genes. Networks were considered significant if their p-scores were >21.
Results
Study participants
During the 6-month study period, we collected 77 blood samples (35 from patients with IA, 42 from control subjects) as well as angiographic images and medical records data from individuals undergoing cerebral DSA. Of the blood samples collected, 37 (16 from IA patients, 21 from controls) had a sufficient quality of neutrophil RNA for sequencing. Pairing on the basis of demographics and comorbidities resulted in a final cohort of 22 individuals, including 11 IA patients and 11 IA-free controls (Table 1). These samples were of sufficient quality and had an average 260/280 of 2.02 and an average RIN of 7.04 (S2 Table). Patients with IAs had aneurysms ranging in size from 1.5–19 mm, and included 3 individuals with multiple IAs (S3 Table). There was no statistical difference in age (p>0.05, Student’s t-test), and other factors (p>0.05, χ2 test) (Table 1) as well as white blood cell populations between the two groups (p>0.05, Student’s t-test) (S2 Fig).
Table 1. Clinical characteristics*.
| Patients with IA (n = 11) |
Patients without IA (n = 11) |
P-Value | |
|---|---|---|---|
| Age (years) (Mean±SE) | 66.91±2.84 | 64.73±4.22 | 0.67 |
| Age (years) [Median (Q1/Q3)] | 67 (60.5/72) | 70 (60/71.5) | |
| Sex | |||
| Female | 63.64% | 54.55% | 0.66 |
| Current Smoker | |||
| Yes | 18.18% | 18.18% | 1.00 |
| Comorbidities | |||
| Hypertension | 63.64% | 81.82% | 0.34 |
| Hyperlipidemia | 45.45% | 54.55% | 0.67 |
| Heart disease | 18.18% | 27.27% | 0.61 |
| Stroke history | 0.00% | 9.09% | 0.31 |
| Diabetes mellitus | 18.18% | 36.36% | 0.34 |
| Osteoarthritis | 27.27% | 27.27% | 1.00 |
*We controlled for demographics and comorbidities so no factor was significantly higher in patients with IA or without IA (confirmed on imaging). There is no significant difference in age (p>0.05 by Student’s t-test) sex, smoking history, and comorbidities (χ2>0.05, chi-squared test) between the two groups. (IA = intracranial aneurysm, SE = standard error, Q = quartile)
Neutrophils have an IA-associated RNA expression signature
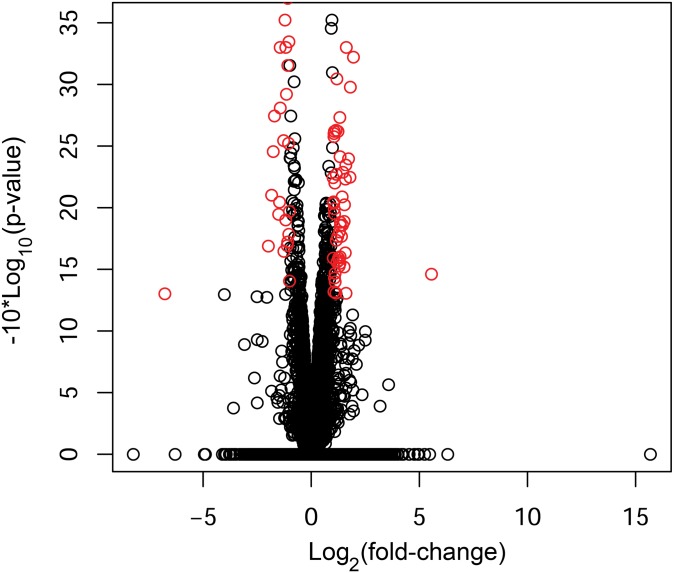
We performed RNA sequencing to identify differentially expressed neutrophil transcripts between 11 patients with IA and 11 paired controls. The sequencing had an average of 52.05 million sequences per sample and a 96.3% read mapping rate (% aligned) (S4 Table). The volcano plot in Fig 1 shows neutrophil expression differences between IA patients and controls in terms of average fold-change in expression and significance level. From 13,377 transcripts with testable expression differences, we identified 258 transcripts that were significantly differentially expressed (p<0.05) between the two groups. We defined an IA-associated RNA expression signature as significant transcripts that were increased or decreased by a factor of 2 or more. From the 258 transcripts, 82 met these criteria and are shown by the red circles in Fig 1 and detailed in Table 2. Post hoc power analysis [21] estimated that a power of 0.8 was achieved in detecting expression differences of at least 1.68 fold at α = 0.05. Therefore, our statistical criteria of p<0.05 and absolute fold-change≥2 had power >0.8 in detecting this signature.
Fig 1. Expression differences between patients with IAs and controls, and an IA-associated expression signature.
The volcano plot demonstrates differential RNA expression between the two groups. Red circles indicate an IA-associated signature of significantly differentially expressed transcripts (p<0.05) with an absolute fold-change≥2.
Table 2. The 82-transcript intracranial aneurysm-associated gene expression signature*.
| Transcript | Gene ID | Accession No. | Log2(F-C) | P-Value | Q-Value |
|---|---|---|---|---|---|
| MAOA | 4128 | M69226.1 | 5.56 | 0.03455 | 0.9999 |
| C21orf15 | 54055 | AY040090.1 | 2.38 | 0.00005 | 0.0836063* |
| CYP1B1 | 1545 | NM_000104.3 | 2.02 | 0.00005 | 0.0836063* |
| ARMC12 | 221481 | NM_145028.4 | 1.95 | 0.0006 | 0.4459 |
| CD177 | 57126 | NM_020406.3 | 1.81 | 0.001 | 0.585244 |
| OLAH | 55301 | NM_018324.2 | 1.79 | 0.0057 | 0.9999 |
| CYP1B1-AS1 | 285154 | NR_027252.1 | 1.73 | 0.004 | 0.9999 |
| FLT3 | 2322 | NM_004119.2 | 1.63 | 0.00005 | 0.0836063* |
| CD163 | 9332 | DQ058615.1 | 1.62 | 0.0005 | 0.393441 |
| KCNMA1 | 3778 | NM_001014797.2 | 1.61 | 0.050 | 0.9999 |
| DACT1 | 51339 | NM_016651.5 | 1.60 | 0.0045 | 0.9999 |
| FAM90A1 | 55138 | NM_018088.3 | 1.58 | 0.0059 | 0.9999 |
| SCT | 6343 | AF244355.1 | 1.58 | 0.0232 | 0.9999 |
| LOC100131289 | 100131289 | NR_038929.1 | 1.54 | 0.0095 | 0.9999 |
| NOG | 9241 | NM_005450.4 | 1.52 | 0.013 | 0.9999 |
| SCAMP5 | 192683 | NM_001178111.1 | 1.52 | 0.030 | 0.9999 |
| PTGDS | 5730 | NM_000954.5 | 1.47 | 0.0052 | 0.9999 |
| KIR2DS4 | 3809 | NM_012314.5 | 1.45 | 0.014 | 0.9999 |
| CYP4F35P | 284233 | NR_026756. | 1.43 | 0.0082 | 0.9999 |
| XKR3 | 150165 | NM_001318251.1 | 1.41 | 0.00005 | 0.0836063* |
| RPL39L | 116832 | NM_052969.2 | 1.40 | 0.017 | 0.9999 |
| CDHR2 | 54825 | NM_001171976.1 | 1.35 | 0.013 | 0.9999 |
| ENHO | 375704 | NM_198573.2 | 1.35 | 0.014 | 0.9999 |
| SLC12A7 | 10723 | NM_006598.2 | 1.35 | 0.00005 | 0.0836063* |
| FLJ27354 | 400761 | NR_033981.1 | 1.34 | 0.025 | 0.9999 |
| DGKH | 160851 | NM_152910.5 | 1.33 | 0.0039 | 0.9999 |
| SDC3 | 9672 | AF248634.1 | 1.33 | 0.028 | 0.9999 |
| THBS1 | 7057 | NM_003246.3 | 1.32 | 0.0019 | 0.85336 |
| RCVRN | 5957 | NM_002903.2 | 1.30 | 0.016 | 0.9999 |
| AKR1C1 | 1645 | NM_001353.5 | 1.30 | 0.030 | 0.9999 |
| SCRG1 | 11341 | NM_001329597.1 | 1.28 | 0.027 | 0.9999 |
| NRG1 | 3084 | NM_013959.3 | 1.26 | 0.030 | 0.9999 |
| AK5 | 26289 | NM_174858.2 | 1.24 | 0.0024 | 0.9999 |
| ITGA7 | 3679 | NM_001144996.1 | 1.23 | 0.017 | 0.9999 |
| PAM | 5066 | NM_000919.3 | 1.20 | 0.00005 | 0.0836063* |
| LYPD2 | 137797 | NM_205545.2 | 1.19 | 0.028 | 0.9999 |
| PRUNE2 | 158471 | NM_015225.2 | 1.19 | 0.0009 | 0.547241 |
| SLC22A17 | 51310 | NM_020372.3 | 1.16 | 0.018 | 0.9999 |
| ADTRP | 84830 | NM_001143948.1 | 1.16 | 0.0054 | 0.9999 |
| ADAMTS1 | 9510 | NM_006988.4 | 1.15 | 0.0024 | 0.9999 |
| ECRP | 643332 | NR_033909.1 | 1.15 | 0.049 | 0.9999 |
| LOC100507387 | 100507387 | NR_038402.1 | 1.11 | 0.040 | 0.9999 |
| KLRC2 | 3822 | NM_002260.3 | 1.11 | 0.034 | 0.9999 |
| AKR1C3 | 8644 | NM_003739.5 | 1.09 | 0.0063 | 0.9999 |
| SEPT10 | 151011 | NM_144710.4 | 1.08 | 0.011 | 0.9999 |
| CYYR1 | 116159 | NM_001320768.1 | 1.08 | 0.037 | 0.9999 |
| TCL1A | 8115 | NM_021966.2 | 1.07 | 0.0024 | 0.9999 |
| VWF | 7450 | NM_000552.4 | 1.06 | 0.010 | 0.9999 |
| GNLY | 10578 | NM_001302758.1 | 1.06 | 0.0025 | 0.9999 |
| C4BPA | 722 | NM_000715.3 | 1.05 | 0.0027 | 0.9999 |
| LINC00482 | 284185 | NR_038080.1 | 1.05 | 0.048 | 0.9999 |
| KIAA1598 | 57698 | BC022348.1 | 1.04 | 0.0092 | 0.9999 |
| PID1 | 55022 | NM_017933.4 | 1.03 | 0.0090 | 0.9999 |
| SERPINF2 | 5345 | NM_000934.3 | 1.02 | 0.027 | 0.9999 |
| VWA8 | 23078 | NM_015058.1 | 1.01 | 0.0057 | 0.9999 |
| CYP4F2 | 8529 | NM_001082.4 | -1.00 | 0.039 | 0.9999 |
| FADS2 | 9415 | NM_004265.3 | -1.01 | 0.011 | 0.9999 |
| VLDLR | 7436 | NM_003383.4 | -1.03 | 0.0005 | 0.393441 |
| CARD17 | 440068 | NM_001007232.1 | -1.04 | 0.016 | 0.9999 |
| IL8 | 576 | AF043337.1 | -1.04 | 0.0001 | 0.148633* |
| G0S2 | 50486 | NM_015714.3 | -1.05 | 0.003 | 0.9999 |
| FBXW8 | 26259 | NM_153348.2 | -1.08 | 0.0007 | 0.468195 |
| MFSD9 | 84804 | NM_032718.4 | -1.08 | 0.0002 | 0.26754 |
| CCL23 | 6368 | NM_005064.5 | -1.09 | 0.019 | 0.9999 |
| C1orf226 | 400793 | NM_001135240.1 | -1.11 | 0.020 | 0.9999 |
| GBP5 | 115362 | NM_052942.3 | -1.14 | 0.001 | 0.642096 |
| BATF2 | 116071 | NM_138456.3 | -1.17 | 0.0005 | 0.393441 |
| FCRL5 | 83416 | NM_031281.2 | -1.18 | 0.013 | 0.9999 |
| SERPING1 | 710 | NM_000062.2 | -1.21 | 0.0003 | 0.334425 |
| B4GALNT3 | 283358 | NM_173593.3 | -1.26 | 0.023 | 0.9999 |
| PDCD1LG2 | 80380 | NM_025239.3 | -1.28 | 0.0029 | 0.9999 |
| FBN1 | 2200 | NM_000138.4 | -1.33 | 0.00005 | 0.0836063* |
| PRSS21 | 10942 | NM_006799.3 | -1.43 | 0.0016 | 0.797475 |
| ETV7 | 51513 | NM_016135.3 | -1.43 | 0.0005 | 0.393441 |
| SEPT4 | 5414 | NM_004574.4 | -1.46 | 0.009 | 0.9999 |
| EGR2 | 1959 | J04076.1 | -1.50 | 0.011 | 0.9999 |
| GBP1P1 | 400759 | NR_003133.2 | -1.70 | 0.0018 | 0.85336 |
| PSORS1C3 | 100130889 | AB932952.1 | -1.75 | 0.0035 | 0.9999 |
| HRK | 8739 | NM_003806.3 | -1.83 | 0.0079 | 0.9999 |
| NEB | 4703 | NM_001164507.1 | -1.98 | 0.020 | 0.9999 |
| GPC4 | 2239 | NM_001448.2 | -2.32 | 0.00005 | 0.0836063* |
| LOC730441 | 207147 | BC039387.1 | -6.77 | 0.0498 | 0.9999 |
*Significantly differentially expressed transcripts with q-value<0.20 (20% FDR) are marked by “*”.
Multiple hypothesis correction identified 9 transcripts with FDR<0.20; C21orf15, CYP1B1, FLT3, XKR3, SLC12A7, PAM, IL8, FBN1, and GPC4. Although this correction effectively reduced the number of significant transcripts, it may be more beneficial to retain all 82 significant transcripts in the aneurysm-associated signature at this early stage of discovery. This prudence is warranted because the role of circulating neutrophils in IAs is unknown and may be highly complex. Genes tend to work together in intricate networks to carry out physiological and pathophysiological functions and influence complex traits. Therefore, individual genes in these systems that by themselves might not be significant (i.e., meet strict cutoffs of statistical tests that are not designed to find biologically relevant transcripts) could still play important roles in IA pathophysiology. To avoid missing potentially informative transcripts, we included all 82 transcripts in the IA-associated signature and in the clustering analysis.
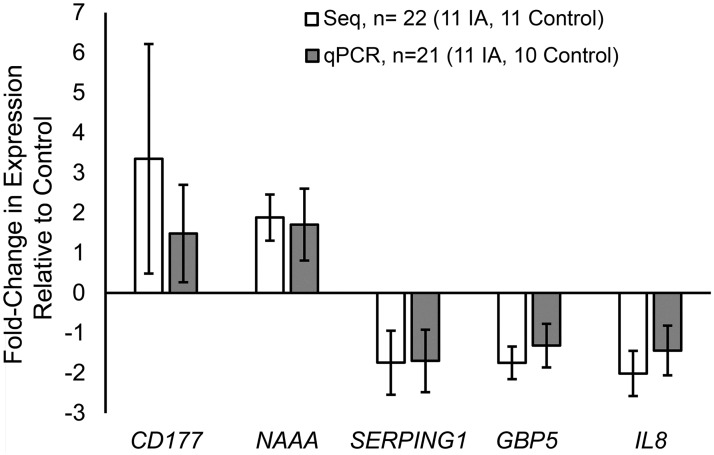
We confirmed differential expression of 5 prominent differentially expressed transcripts (CD177, NAAA, SERPING1, GBP5, and IL8) using RT-qPCR. Fig 2 demonstrates that the expression differences between patients with and without IA are of the same direction and of similar magnitudes whether calculated from RNA sequencing or RT-qPCR. There was no statistically significant difference in fold-change of expression measured by the two methods (all p-values>0.05, Student’s t-test).
Fig 2. Verification of RNA expression differences by RT-qPCR.
Quantitative PCR performed on 5 prominent differentially expressed transcripts demonstrates that both the magnitude and direction of the fold-change in expression measured by RNA sequencing are similar to that measured by qPCR. Only 21 samples were analyzed by RT-qPCR because sample C8 did not have enough RNA for the additional reactions. (Negative fold-change values calculated by negative inverse of fold-change, data points = average values, error bars = standard error.)
Neutrophil RNA expression discriminates IA from control groups
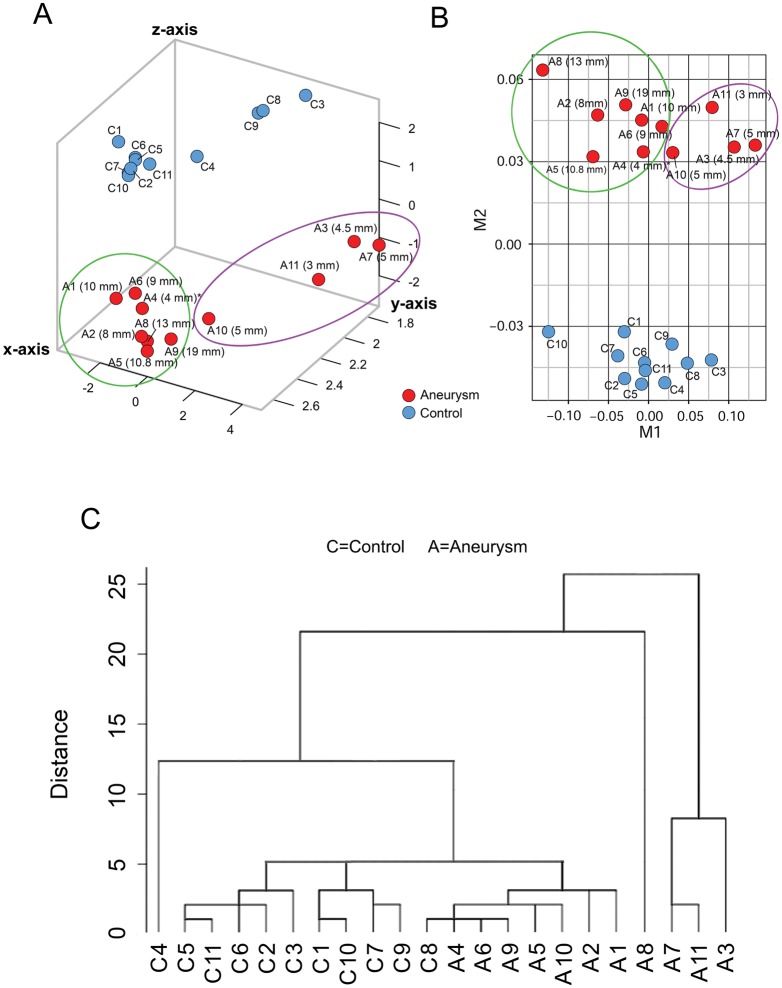
To visualize how well neutrophil RNA expression differentiated aneurysm samples from control samples, we performed dimensionality reduction analyses by PCA and MDS using all neutrophil transcriptome data (13,377 transcripts). Fig 3A shows that the IA and control groups separated in the principal component space. Similarly, MDS also showed separation of patients with IAs and control subjects (Fig 3B). We also found that the transcriptome data segregated the patients with IAs by the size of each patient’s largest IA, forming two groups on both the PCA and MDS plots: large (≥8 mm, with one exception) and small (≤5 mm) (Fig 3A and 3B).
Fig 3. Dimensionality reduction analyses separate blood samples from IA patients and controls.
(A) Principal component analysis (PCA) using all transcriptome data demonstrates aggregation of samples from IA patients (red) and controls (blue). Transcriptome data further separated samples from IA patients by aneurysm size, forming groups of large IAs (≥8 mm, with one exception) and small IAs (≤5 mm). (B) Multidimensional scaling (MDS) of transcriptome data further reduces dimensionality and mirrors the PCA results, also showing separation of IA (red) and control (blue) samples. (C) Hierarchical clustering using only the 82-transcript IA-associated signature also demonstrates separation of IA and control groups. Four aneurysms samples on the right were more distinct than others, while the rest of the samples segregated into three main clusters, two containing all control and one containing all IA (with one exception). Overall, 91% of the samples were grouped with their respective group.
Using the expression signature of 82 transcripts (p<0.05 and absolute fold-change ≥2), we performed supervised hierarchical clustering to determine whether it could also discriminate patients with IAs from controls. On the dendrogram in Fig 3C, samples from IA and control groups are separated. Four samples from the IA group on the right were more distinct than the others. In the rest of the samples, one control (to the left) was separate, and all other samples segregated into 3 groups. Two groups contained all control samples and 1 group contained all IA samples (with 1 exception). In general, hierarchical clustering congregated 91% of the samples with their respective groups.
Expression differences are consistent with leukocyte activation
To gain insight into the biology of neutrophil RNA expression differences we performed bioinformatics analyses using gene set enrichment analysis and physiological pathway modeling. Some tightly controlled pathways can be regulated by transcripts that show small but significant changes. To avoid missing potentially significant biological insights, we performed bioinformatics analysis using all 258 differentially expressed transcripts (p<0.05) regardless of fold-change. As detailed in Table 3, gene ontology analysis revealed that genes with higher neutrophil expression levels in the IA group were involved in defense response, leukocyte activation, stem cell maintenance, maintenance of cell number, cell activation, and stem cell development. Genes with lower expression levels in the IA group were involved in immune response and immune system process (Table 3).
Table 3. Gene ontology (GO) analysis*.
| GO ID | Term | P-value | Q-value | Annotated Genes |
|---|---|---|---|---|
| Genes Increased in IA | ||||
| GO:0006952 | Defense Response | 3.13E-06 | 2.36E-03 | KLRC2, VNN1, C4BPA, CD300E, SH2D1B, CD247, GNLY, INHBB, CD1D, KIR2DS4, PRF1, ORM2, STAB1, FCER1A, CD86 |
| GO:0045321 | Leukocyte Activation | 8.23E-06 | 6.21E-03 | VNN1, CD1D, CD7, PRF1, FCER1A, SH2D1B, SOX4, CD86, CD247 |
| GO:0019827 | Stem Cell Maintenance | 8.44E-06 | 6.37E-03 | NOG, TCL1A, KLF10, SCT, SOX4 |
| GO:0098727 | Maintenance of Cell Number | 8.95E-06 | 6.76E-03 | NOG, TCL1A, KLF10, SCT, SOX4 |
| GO:0001775 | Cell Activation | 1.18E-05 | 8.92E-03 | VNN1, CD7, VWF, SH2D1B, SOX4, CD247, CD1D, PRF1, FCER1A, CD86 |
| GO:0048864 | Stem Cell Development | 1.20E-05 | 9.10E-03 | NOG, DAB2, TCL1A, KLF10, SCT, SOX4 |
| Genes Decreased in IA | ||||
| GO:0006955 | Immune Response | 1.17E-07 | 6.85E-05 | AIM2, LILRA4, FCRL5, IL8, MOV10, CYSLTR2, IFI35, PDCD1LG2, RGS1, CD274, CCL23, DDX60, OLFM4, GBP1 |
| GO:0002376 | Immune System Process | 3.32E-07 | 1.94E-04 | MOV10, CEBPE, SMPD3, IFI35, PDCD1LG2, CCL23, OLFM4, GBP1, AIM2, FCRL5, LILRA4, IL8, CYSLTR2, RGS1, CD274, DDX60 |
* Gene set enrichment analysis was performed on significantly differentially expressed genes (p<0.05) in peripheral blood samples obtained from patients with intracranial aneurysms (IA). Significantly enriched ontologies present in genes with higher expression levels included defense response, leukocyte activation, stem cell maintenance, maintenance of cell number, cell activation, and stem cell development. Significantly enriched ontologies present in genes with lower expression levels included immune response and immune system process. Enriched ontologies from the GO database were considered significant at a false discovery rate-corrected p-value (q-value) <0.05.
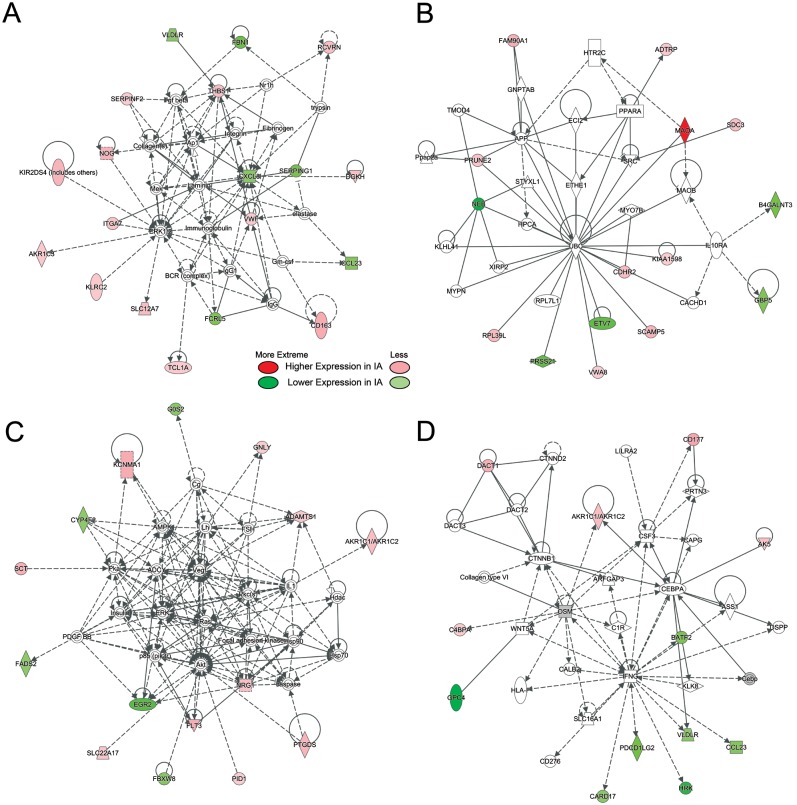
Physiological pathway modeling to identify networks of potential interactions revealed 4 networks with 7 signaling nodes forming hubs within the networks (Fig 4). These hubs included ERK1/2 and AP1; IL8 (CXCL8), AKT and VEGF; UBC; and IFNG. IPA indicated that these networks were consistent with activation of cellular movement and cardiovascular system function (network A), lipid metabolism (network B), cell-to-cell signaling and energy production (network C), and organismal injury, cell proliferation, and tissue morphology (network D). These functions are pertinent to neutrophil responses to intravascular perturbations.[27] See S5 Table for a list of names and biological functions of the transcripts in these 4 networks.
Fig 4. The 4 most regulated networks.
Networks were derived from IPA of differentially expressed transcripts (p<0.05) in neutrophils from IA patients and controls. Transcripts with increased expression levels in patients with IAs are red; transcripts with lower expression levels in patients with IAs are green; and fold-change is represented by intensity. Non-differentially expressed transcripts with known interactions are not colored. (A) This network (p-score = 41) shows regulation of transcripts with increased expression by an ERK1/2 and AP1. IL8, regulates transcripts with lower expression in samples from patients with IAs. (B) This network (p-score = 30) shows regulation of transcripts by UBC. (C) This network (p-score = 30) shows two nodes of regulation at AKT and VEGF. (D) This network (p-score = 23) shows regulation of transcripts with lower expression in IA samples by IFNG.
Replication study in a new, unpaired population
To determine whether expression of the IA-associated signature could separate patients with IAs from controls in an independent cohort, we performed a small replication study. We recruited 10 additional patients (5 with IA, 5 IA-free controls) but did not control for demographics and comorbidities in order to assess the signature’s potential for segregating patients in heterogeneous populations (see S6 Table for clinical characteristics). Patients with IAs had aneurysms ranging in size from 1.4–7 mm and included one individual with multiple aneurysms (S7 Table). From these patients’ peripheral blood samples, we isolated neutrophils and extracted neutrophil RNA and performed next-generation RNA sequencing to obtain FPKM levels of the 82 IA-associated transcripts. To visualize how these transcripts could distinguish the IA group from the control group, we performed PCA and hierarchical clustering. With the exception of one IA sample, PCA demonstrated separation of the two groups in the principal component space (Fig 5A). Hierarchical clustering mirrored this result, grouping the IA and control samples separately, with the exception of one IA sample (Fig 5B).
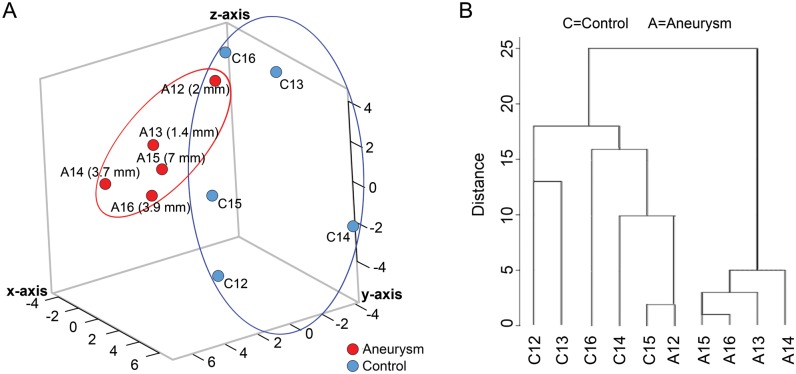
Fig 5. Replication study in a cohort of 10 new patients (5 with IAs).
(A) Principal component analysis performed using the 82 IA-associated transcripts shows separation of IA from control samples in this unmatched cohort. (B) Hierarchical cluster analysis demonstrates separation of the IA and control samples, with the exception of one IA sample that was grouped with controls.
Availability of RNA expression data
Raw RNA sequencing data files and processed transcript expression levels for the experiments described in this publication can be found at NCBI’s GEO (accession no. GSE106520). Deidentified patient metadata is presented in S8 Table and can also be found at NCBI’s GEO with the expression data.
Discussion
We performed transcriptome profiling on circulating neutrophils from paired patients with and without IAs and identified an aneurysm-associated signature of 82 transcripts. These transcripts discriminated patients with and without IA in hierarchical cluster analysis. In a replication study, this signature also distinguished patients with IAs from controls in an unpaired cohort. These findings present the exciting potential for developing predictive biomarkers that use this signature to identify patients with IAs.
Previous efforts in search of circulating aneurysm biomarkers
The search for circulating biomarkers for unruptured IAs has spanned more than two decades. A meta-analysis[28] of IA biomarker publications from 1994–2015 found 5 studies that linked IA presence to specific biomolecules in the blood. These studies found that serum elastase-to-A1AT ratios [29] and LPA [30], VEGF [31], MCP-1, IL-1β, TNF-α [32], and GM-CSF levels [33] were elevated in patients with unruptured aneurysms. However, in the present study, we did not observe significantly higher mRNA levels for these proteins in neutrophils from patients with IA. This may be because these proteins originate from sources other than neutrophils or may not be sufficiently unique to IA to be identified by our analysis.
One common trait of the previously-identified potential proteins markers, is that they are ubiquitous, being involved in a wide range of physiological and pathological functions. Thus, in addition to IA, they may also signify various vascular diseases. For example, serum VEGF is also increased during peripheral artery stenosis [34], plasma MCP-1 is also elevated in thromboembolic hypertension [35], and LPA is elevated in plasma of patients with vascular dementia [36]. Perhaps for this reason, significant follow-up efforts have not been made towards subsequent biomarker development and validation on the basis of these studies.
An alternative approach to identifying potential biomarkers is to profile the transcriptome of the circulating blood, which affords screening for multitudes of potential markers and can provide insight into novel disease mechanisms that may be specific to IA. Recently, circulating RNA expression signatures of unruptured IAs were found in microarray studies. In IA patients, Jin et al. [37] found 77 differentially expressed plasma microRNAs that were involved in proliferation, apoptosis, molecular activation, transport, and differentiation; Li et al. [38] discovered 119 differentially expressed plasma microRNAs related to inflammatory responses and connective tissue disorders; and Sabatino et al. [39] identified 53 differentially expressed mRNAs from peripheral blood mononuclear cells that were related to increased cell proliferation and apoptosis. These findings indicate that IA is associated with altered expression of a large number of transcripts from various circulating blood components. Each study benefited from the use of high-throughput technology (i.e. microarray). We believe that such technologies open the door to discovery of IA-specific signatures consisting of many transcripts.
On the basis of this vision, we conducted the current study, albeit using next-generation RNA sequencing. This latest high-throughput technology affords two key advantages over microarrays used in previous investigations: (1) it offers a larger dynamic range, facilitating detection of expression differences in low-abundance transcripts; and (2) it avoids predetermined probes, allowing examination of novel RNAs (i.e., splice variants, non-coding RNAs, gene isoforms) [40]. These capabilities led us to discover a signature of 82 transcripts, containing several uncharacterized and/or non-coding RNAs, which cannot be detected on conventional microarrays. They include C21orf15, LOC100131289, FLJ27354, LOC100507387, LINC00482, C1orf226, and LOC730441. To our knowledge, these novel transcripts have not been associated with any other diseases. Further research is required to determine their functions and how they contribute to IA pathophysiology.
We also designed our study to avoid common pitfalls of expression profiling studies [37–39]. First, to avoid misclassification, we used DSA to confirm that the control subjects did not have aneurysms. Previous studies did not perform such imaging. Second, to find RNA expression differences due to the presence of an IA and not confounding factors, we paired the subjects by demographics and comorbidities. Previous studies typically used healthy subjects or spouses as controls. Third, we performed a replication study in an independent, unpaired cohort to investigate whether the signature can distinguish patients with IA in a general population. These measures helped to increase the likelihood that the discovered signature is associated with IA presence.
Circulating neutrophils and intracranial aneurysms
Intracranial aneurysm natural history is characterized by mounting inflammatory responses and progressive degradation of the aneurysmal wall, starting from initial pro-inflammatory changes in smooth muscle cells that lead to overproduction of matrix metalloproteinases (MMPs) [5, 41]. Once the aneurysmal pouch is formed, it harbors a hemodynamic environment conducive to macrophage and neutrophil infiltration into the wall, which is aided by a local increase of plasma chemokines and cytokines (IL-1β, IL-17, TNF-α) in the lumen [11, 42]. These inflammatory infiltrates massively produce MMPs to further degrade the aneurysm wall and advance its growth and rupture [5, 43]. This is evidenced by gene expression studies of human aneurysmal tissues, which found increased matrix degradation processes, inflammatory processes, and production of inflammatory cytokines and chemoattractant proteins in the IA wall [8, 9]. Furthermore, Yu et al. found that differences in DNA methylation in aneurysmal tissue act to promote inflammatory signaling through the NF-KB, JNK-STAT, and ERK/JNK pathways [44], uncovering a potential epigenetic underpinning to dysregulated inflammation during IA.
The role of neutrophils in IA pathophysiology may be complex and is not well understood. Besides secreting MMP-9, activated neutrophils also release NGAL and MPO, which indirectly contribute to extracellular matrix degradation and cytotoxicity, respectively. Increased NGAL in aneurysm tissue modulates the activity of MMP-9, protecting it from degradation and thus aiding aneurysm progression [11]. Increased MPO, an inflammatory enzyme, elicits oxidative stress and pro-inflammatory cell signaling through production of reactive oxygen species [10]. It has been observed that plasma levels of NGAL and MPO are increased in the blood of patients with aneurysms [11, 45]. Furthermore, both of these proteins can have autocrine effects that promote neutrophil activation [46, 47], which could lead to expression changes observed in our study. Interestingly, we found significantly increased expression of SLC22A17, which is the NGAL receptor, in neutrophils from patients with IAs. This may reflect a possible interaction with circulating NGAL. However, we did not observe significantly higher levels of NGAL or MPO in circulating neutrophils, suggesting that these proteins may originate from the aneurysm sac itself, or other circulating cells.
Further analysis of our expression data supports an association between activated circulating neutrophils and IA presence. Gene set enrichment analysis reveals that neutrophils from IA patients have higher levels of gene expression associated with leukocyte activation. This is evidenced by increased expression levels of several CD antigens from the “leukocyte activation” ontology (CD1D, CD7, CD86, and CD247) as well as CD177, a marker of neutrophil activation. IPA also reveals functions indicative of activated neutrophils, showing networks consistent with activation of cellular movement, cell-to-cell signaling, and cell proliferation. The fact that neutrophil expression data segregated aneurysms by size in PCA and MDS (Fig 3A and 3B) may indicate a correlation between the degree of IA advancement and neutrophil activation. Overall, our findings suggest that peripheral neutrophil activation may play a role in IA development.
Limitations
This proof-of-concept study has three main limitations. First, our IA-associated expression signature was extracted from a small sample. However, by selecting transcripts with large effect size (fold-change ≥2), performing confirmatory qPCR, and demonstrating separation of IAs from controls in an independent cohort, we were able to increase confidence in the discovered signature. We hope this exploratory effort lays a foundation for future studies in larger cohorts with increased statistical power. Second, all of our study subjects were recruited from patients receiving cerebral imaging at our center; thus, there may be a potential for selection bias. Limiting subjects to those receiving imaging was necessary in our study to confirm the presence or absence of IA, but using broader, randomized patient populations from multiple centers would have been beneficial. Lastly, despite patient matching between the groups to remove confounding factors (potentially contributing to the IA signature), there is a possibility that the found differential neutrophil expression in the signature could be caused by other conditions. It is known that neutrophil activation can also occur in other vascular pathologies and inflammatory states. Further research is needed to investigate the specificity of this IA-associated signature to IA.
Conclusions
Early IA detection is important for preventing rupture; therefore, blood-based diagnostics could change the landscape of IA management. In this preliminary, exploratory effort, we identified an IA-associated RNA expression signature of 82 transcripts in circulating neutrophils. Despite our small sample size, this signature demonstrated a statistical power >0.80 and was able to distinguish patients with IAs from paired controls in several analyses. These transcripts also separated patients with IAs from unpaired controls in a small population. These findings need to be validated in larger, more diverse cohorts.
Supporting information
This form in the patient’s medical record was evaluated to retrieve the patient’s clinical information.
(TIF)
There was no significant difference in white blood cell count or leukocyte ratios between patients with IAs (n = 11) and controls (n = 7, no data were available for 4 of the controls). (A) Complete blood count data recorded within 3 months of blood collection showed no significant difference between-groups in the concentrations of leukocytes, erythrocytes, platelets, neutrophils, lymphocytes, or monocytes (p>0.05, Student’s t-test). (B) There was also no significant difference in the percentage (%) per 100 leukocytes for neutrophils, lymphocytes, monocytes, eosinophils, and basophils between patients with and without IA (p>0.05, Student’s t-test). (Data points = average values, error bars = standard error).
(TIF)
*Primers were selected using Primer3 and NCBI’s Primer Blast. All efficiencies were within the range of 0.90–1.10. (bp = base pair, Eff. = efficiency, Prod. = product, Temp. = temperature).
(DOCX)
*The quality of the RNA samples was assessed by the 260/280 ratio and the RIN. (RIN = RNA integrity number).
(DOCX)
*Aneurysm size ranged from 1.5mm to 19mm. Ten of 16 IAs (63%) were classified as small (greatest diameter <7mm) and 6 (37%) were classified as large (greatest diameter ≥7 mm). The aneurysms were situated at various locations in the Circle of Willis, with most being around the internal carotid artery (ICA) and its branches. Two patients with IAs had a family history of the disease. In general, digital subtraction angiography was performed for either confirmation of IA presence after an incidental finding of IA on noninvasive imaging, or for follow-up imaging of a previously detected IA. (ACA = anterior cerebral artery, AComA = anterior communicating artery, BT = basilar terminus, CT = computed tomography, DSA = digital subtraction angiography, IA = intracranial aneurysm, ICA = internal carotid artery, MCA = middle cerebral artery, MRA = magnetic resonance angiography, MRI = magnetic resonance imaging, PComA = posterior communicating artery, VB = vertebrobasilar).
(DOCX)
*The quality of the RNA sequencing experiments was measure pre-alignment via FASTQC and post-alignment via MultiQC. Overall, prior to alignment all samples had an average of 53.75 M sequences. MultiQC reported that the sequencing experiments had an average of 49.09 M mapped reads with a 96.13% read mapping rate, and detected an average of 17259 transcripts (transcripts with FPKM>0). (Align. = alignment, M. = million, Seqs. = sequences, Qual. = quality).
(DOCX)
*A table of the names of transcripts included in the top 4 networks derived from IPA, as well as the top diseases and functions of these transcripts. Neutrophil transcripts in bold were differentially expressed between patients with and without IA (p-value<0.05). Each network’s p-score was derived from its p-value [p-score = -Log10 (p-value)] calculated by the Fisher’s exact test. For a network with a p-score of 10, the odds of generating this network by chance alone is less than 1 out of 1010.
(DOCX)
*(IA = intracranial aneurysm, SE = standard error, Q = quartile).
(DOCX)
*Aneurysm size ranged from 3.5 mm to 7 mm. Five of 6 IAs (83%) were classified as small (greatest diameter <7mm) and 1 (17%) was classified as large (greatest diameter ≥7 mm). The aneurysms were situated at various locations in the Circle of Willis, with most being in the anterior vasculature (ACA and MCA). (ACA = anterior cerebral artery, AComA = anterior communicating artery, BT = basilar terminus, CT = computed tomography, DSA = digital subtraction angiography, IA, intracranial aneurysm, MCA = middle cerebral artery, MRA = magnetic resonance angiography, MRI = magnetic resonance imaging).
(DOCX)
*(M = male, F = female, Y = yes, N = no, HT = hypertension, HL = hyperlipidemia, CAD = coronary artery disease, S Hx = stroke history, DM = diabetes mellitus, OA = osteoarthritis).
(DOCX)
Acknowledgments
We thank the patients who participated in this study; Maureen T. Donovan MSN and Mary L. Hartney RN CCRC for help obtaining consents; Ning Lin MD, Chandan Krishna MD, Marshall C. Cress MD, Leonardo Rangel-Castilla MD, and Maxim Mokin MD PhD for blood collection; Hussain Shallwani MD and Felix T. Chin for technical assistance; Jihnhee Yu PhD for assistance with statistical analyses; Jonathan Bard MA and Brandon Marzulo MS for sequencing assistance; Jennifer L. Gay CCRP for study protocol management; Paul H. Dressel BFA for illustration preparation; and Elaine C. Mosher MLS and Debra J. Zimmer for editorial assistance.
Data Availability
Raw RNA sequencing data files and processed transcript expression levels for the experiments described in this publication can be found at NCBI’s Gene Expression Omnibus (GEO) at accession no. GSE106520.
Funding Statement
This work was supported by the Brain Aneurysm Foundation Carol Harvey Chair of Research Grant (HM) and Team Cindy-Alcatraz Chair of Research Grant (HM), and NIH grant R01-AR-060604 (JNJ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vega C, Kwoon JV, Lavine SD. Intracranial aneurysms: current evidence and clinical practice. American family physician. 2002;66(4):601–8. Epub 2002/08/31. . [PubMed] [Google Scholar]
- 2.Bederson JB, Connolly ES Jr., Batjer HH, Dacey RG, Dion JE, Diringer MN, et al. Guidelines for the management of aneurysmal subarachnoid hemorrhage: a statement for healthcare professionals from a special writing group of the Stroke Council, American Heart Association. Stroke; a journal of cerebral circulation. 2009;40(3):994–1025. Epub 2009/01/24. doi: 10.1161/strokeaha.108.191395 . [DOI] [PubMed] [Google Scholar]
- 3.Group MRAiRoPwSHS. Risks and benefits of screening for intracranial aneurysms in first-degree relatives of patients with sporadic subarachnoid hemorrhage. The New England journal of medicine. 1999;341(18):1344–50. Epub 1999/10/28. doi: 10.1056/NEJM199910283411803 . [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Barbacioru CC, Shiffman D, Balasubramanian S, Iakoubova O, Tranquilli M, et al. Gene expression signature in peripheral blood detects thoracic aortic aneurysm. PloS one. 2007;2(10):e1050 Epub 2007/10/18. doi: 10.1371/journal.pone.0001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tulamo R, Frosen J, Hernesniemi J, Niemela M. Inflammatory changes in the aneurysm wall: a review. Journal of neurointerventional surgery. 2010;2(2):120–30. Epub 2010/06/01. doi: 10.1136/jnis.2009.002055 . [DOI] [PubMed] [Google Scholar]
- 6.Alg VS, Sofat R, Houlden H, Werring DJ. Genetic risk factors for intracranial aneurysms: a meta-analysis in more than 116,000 individuals. Neurology. 2013;80(23):2154–65. Epub 2013/06/05. doi: 10.1212/WNL.0b013e318295d751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussain I, Duffis EJ, Gandhi CD, Prestigiacomo CJ. Genome-wide association studies of intracranial aneurysms: an update. Stroke; a journal of cerebral circulation. 2013;44(9):2670–5. Epub 2013/08/03. doi: 10.1161/strokeaha.113.001753 . [DOI] [PubMed] [Google Scholar]
- 8.Pera J, Korostynski M, Krzyszkowski T, Czopek J, Slowik A, Dziedzic T, et al. Gene expression profiles in human ruptured and unruptured intracranial aneurysms: what is the role of inflammation? Stroke; a journal of cerebral circulation. 2010;41(2):224–31. Epub 2010/01/02. doi: 10.1161/strokeaha.109.562009 . [DOI] [PubMed] [Google Scholar]
- 9.Tromp G, Weinsheimer S, Ronkainen A, Kuivaniemi H. Molecular basis and genetic predisposition to intracranial aneurysm. Annals of medicine. 2014;46(8):597–606. Epub 2014/08/15. doi: 10.3109/07853890.2014.949299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gounis MJ, Vedantham S, Weaver JP, Puri AS, Brooks CS, Wakhloo AK, et al. Myeloperoxidase in human intracranial aneurysms: preliminary evidence. Stroke; a journal of cerebral circulation. 2014;45(5):1474–7. Epub 2014/04/10. doi: 10.1161/strokeaha.114.004956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serra R, Volpentesta G, Gallelli L, Grande R, Buffone G, Lavano A, et al. Metalloproteinase-9 and neutrophil gelatinase-associated lipocalin plasma and tissue levels evaluation in middle cerebral artery aneurysms. British journal of neurosurgery. 2014. Epub 2014/05/07. doi: 10.3109/02688697.2014.913777 . [DOI] [PubMed] [Google Scholar]
- 12.Jiang K, Sun X, Chen Y, Shen Y, Jarvis JN. RNA sequencing from human neutrophils reveals distinct transcriptional differences associated with chronic inflammatory states. BMC medical genomics. 2015;8:55 Epub 2015/08/28. doi: 10.1186/s12920-015-0128-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvis JN, Dozmorov I, Jiang K, Frank MB, Szodoray P, Alex P, et al. Novel approaches to gene expression analysis of active polyarticular juvenile rheumatoid arthritis. Arthritis research & therapy. 2004;6(1):R15–r32. Epub 2004/02/26. doi: 10.1186/ar1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wardlaw JM, White PM. The detection and management of unruptured intracranial aneurysms. Brain: a journal of neurology. 2000;123 (Pt 2):205–21. Epub 2000/01/29. . [DOI] [PubMed] [Google Scholar]
- 15.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7(3):562–78. Epub 2012/03/03. doi: 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews S. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc2010 [October 10, 2017].
- 17.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics (Oxford, England). 2016;32(19):3047–8. Epub 2016/06/18. doi: 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S, Chan CK. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods in molecular biology (Clifton, NJ). 2016;1374:339–61. Epub 2015/11/01. doi: 10.1007/978-1-4939-3167-5_18 . [DOI] [PubMed] [Google Scholar]
- 19.Goff L, Trapnell C, Kelley D. Analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. bioconductororg2016. p. 73.
- 20.Eteleeb A, Rouchka E. Differential expression analysis methods for ribonucleic acid-sequencing data. OA Bioinformatics. 2013;1(1). [Google Scholar]
- 21.Hart SN, Therneau TM, Zhang Y, Poland GA, Kocher JP. Calculating sample size estimates for RNA sequencing data. J Comput Biol. 2013;20(12):970–8. Epub 2013/08/22. doi: 10.1089/cmb.2012.0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Pratical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 23.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in molecular biology (Clifton, NJ). 2000;132:365–86. Epub 1999/11/05. . [DOI] [PubMed] [Google Scholar]
- 24.Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. Journal of statistical software. 2012;46(11). Epub 2012/10/11. [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, et al. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics (Oxford, England). 2004;20(18):3710–5. Epub 2004/08/07. doi: 10.1093/bioinformatics/bth456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics (Oxford, England). 2014;30(4):523–30. Epub 2013/12/18. doi: 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillipson M, Kubes P. The neutrophil in vascular inflammation. Nature medicine. 2011;17(11):1381–90. Epub 2011/11/09. doi: 10.1038/nm.2514 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussain S, Barbarite E, Chaudhry NS, Gupta K, Dellarole A, Peterson EC, et al. Search for Biomarkers of Intracranial Aneurysms: A Systematic Review. World neurosurgery. 2015;84(5):1473–83. Epub 2015/06/29. doi: 10.1016/j.wneu.2015.06.034 . [DOI] [PubMed] [Google Scholar]
- 29.Baker CJ, Fiore A, Connolly ES Jr., Baker KZ, Solomon RA. Serum elastase and alpha-1-antitrypsin levels in patients with ruptured and unruptured cerebral aneurysms. Neurosurgery. 1995;37(1):56–61; discussion -2. Epub 1995/07/01. . [DOI] [PubMed] [Google Scholar]
- 30.Phillips J, Roberts G, Bolger C, el Baghdady A, Bouchier-Hayes D, Farrell M, et al. Lipoprotein (a): a potential biological marker for unruptured intracranial aneurysms. Neurosurgery. 1997;40(5):1112–5; discussion 5–7. Epub 1997/05/01. . [DOI] [PubMed] [Google Scholar]
- 31.Sandalcioglu IE, Wende D, Eggert A, Regel JP, Stolke D, Wiedemayer H. VEGF plasma levels in non-ruptured intracranial aneurysms. Neurosurgical review. 2006;29(1):26–9. Epub 2005/09/01. doi: 10.1007/s10143-005-0411-8 . [DOI] [PubMed] [Google Scholar]
- 32.Zhang HF, Zhao MG, Liang GB, Song ZQ, Li ZQ. Expression of pro-inflammatory cytokines and the risk of intracranial aneurysm. Inflammation. 2013;36(6):1195–200. Epub 2013/05/15. doi: 10.1007/s10753-013-9655-6 . [DOI] [PubMed] [Google Scholar]
- 33.Chalouhi N, Theofanis T, Starke RM, Zanaty M, Jabbour P, Dooley SA, et al. Potential role of granulocyte-monocyte colony-stimulating factor in the progression of intracranial aneurysms. DNA and cell biology. 2015;34(1):78–81. Epub 2014/11/13. doi: 10.1089/dna.2014.2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Han L, Xu X, Tang H, Wang H, Wei B. Serum biomarkers VEGF-C and IL-6 are associated with severe human Peripheral Artery Stenosis. Journal of inflammation (London, England). 2015;12:50 Epub 2015/08/19. doi: 10.1186/s12950-015-0095-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura H, Okada O, Tanabe N, Tanaka Y, Terai M, Takiguchi Y, et al. Plasma monocyte chemoattractant protein-1 and pulmonary vascular resistance in chronic thromboembolic pulmonary hypertension. American journal of respiratory and critical care medicine. 2001;164(2):319–24. Epub 2001/07/21. doi: 10.1164/ajrccm.164.2.2006154 . [DOI] [PubMed] [Google Scholar]
- 36.Ray L, Khemka VK, Behera P, Bandyopadhyay K, Pal S, Pal K, et al. Serum Homocysteine, Dehydroepiandrosterone Sulphate and Lipoprotein (a) in Alzheimer’s Disease and Vascular Dementia. Aging and disease. 2013;4(2):57–64. Epub 2013/05/23. [PMC free article] [PubMed] [Google Scholar]
- 37.Jin H, Li C, Ge H, Jiang Y, Li Y. Circulating microRNA: a novel potential biomarker for early diagnosis of intracranial aneurysm rupture a case control study. Journal of translational medicine. 2013;11:296 Epub 2013/11/28. doi: 10.1186/1479-5876-11-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li P, Zhang Q, Wu X, Yang X, Zhang Y, Li Y, et al. Circulating microRNAs serve as novel biological markers for intracranial aneurysms. Journal of the American Heart Association. 2014;3(5):e000972 Epub 2014/09/25. doi: 10.1161/JAHA.114.000972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabatino G, Rigante L, Minella D, Novelli G, Della Pepa GM, Esposito G, et al. Transcriptional profile characterization for the identification of peripheral blood biomarkers in patients with cerebral aneurysms. Journal of biological regulators and homeostatic agents. 2013;27(3):729–38. Epub 2013/10/25. . [PubMed] [Google Scholar]
- 40.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PloS one. 2014;9(1):e78644 Epub 2014/01/24. doi: 10.1371/journal.pone.0078644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng H, Tutino VM, Xiang J, Siddiqui A. High WSS or low WSS? Complex interactions of hemodynamics with intracranial aneurysm initiation, growth, and rupture: toward a unifying hypothesis. AJNR American journal of neuroradiology. 2014;35(7):1254–62. Epub 2013/04/20. doi: 10.3174/ajnr.A3558 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chalouhi N, Points L, Pierce GL, Ballas Z, Jabbour P, Hasan D. Localized increase of chemokines in the lumen of human cerebral aneurysms. Stroke; a journal of cerebral circulation. 2013;44(9):2594–7. Epub 2013/07/28. doi: 10.1161/strokeaha.113.002361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frosen J, Tulamo R, Paetau A, Laaksamo E, Korja M, Laakso A, et al. Saccular intracranial aneurysm: pathology and mechanisms. Acta neuropathologica. 2012;123(6):773–86. Epub 2012/01/18. doi: 10.1007/s00401-011-0939-3 . [DOI] [PubMed] [Google Scholar]
- 44.Yu L, Wang J, Wang S, Zhang D, Zhao Y, Wang R, et al. DNA Methylation Regulates Gene Expression in Intracranial Aneurysms. World neurosurgery. 2017;105:28–36. Epub 2017/04/24. doi: 10.1016/j.wneu.2017.04.064 . [DOI] [PubMed] [Google Scholar]
- 45.Chu Y, Wilson K, Gu H, Wegman-Points L, Dooley SA, Pierce GL, et al. Myeloperoxidase is increased in human cerebral aneurysms and increases formation and rupture of cerebral aneurysms in mice. Stroke; a journal of cerebral circulation. 2015;46(6):1651–6. Epub 2015/04/30. doi: 10.1161/strokeaha.114.008589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leopold JA. The Central Role of Neutrophil Gelatinase-Associated Lipocalin in Cardiovascular Fibrosis. Hypertension (Dallas, Tex: 1979). 2015;66(1):20–2. Epub 2015/05/20. doi: 10.1161/hypertensionaha.115.05479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stapleton PP, Redmond HP, Bouchier-Hayes DJ. Myeloperoxidase (MPO) may mediate neutrophil adherence to the endothelium through upregulation of CD11B expression—an effect downregulated by taurine. Advances in experimental medicine and biology. 1998;442:183–92. Epub 1998/06/23. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This form in the patient’s medical record was evaluated to retrieve the patient’s clinical information.
(TIF)
There was no significant difference in white blood cell count or leukocyte ratios between patients with IAs (n = 11) and controls (n = 7, no data were available for 4 of the controls). (A) Complete blood count data recorded within 3 months of blood collection showed no significant difference between-groups in the concentrations of leukocytes, erythrocytes, platelets, neutrophils, lymphocytes, or monocytes (p>0.05, Student’s t-test). (B) There was also no significant difference in the percentage (%) per 100 leukocytes for neutrophils, lymphocytes, monocytes, eosinophils, and basophils between patients with and without IA (p>0.05, Student’s t-test). (Data points = average values, error bars = standard error).
(TIF)
*Primers were selected using Primer3 and NCBI’s Primer Blast. All efficiencies were within the range of 0.90–1.10. (bp = base pair, Eff. = efficiency, Prod. = product, Temp. = temperature).
(DOCX)
*The quality of the RNA samples was assessed by the 260/280 ratio and the RIN. (RIN = RNA integrity number).
(DOCX)
*Aneurysm size ranged from 1.5mm to 19mm. Ten of 16 IAs (63%) were classified as small (greatest diameter <7mm) and 6 (37%) were classified as large (greatest diameter ≥7 mm). The aneurysms were situated at various locations in the Circle of Willis, with most being around the internal carotid artery (ICA) and its branches. Two patients with IAs had a family history of the disease. In general, digital subtraction angiography was performed for either confirmation of IA presence after an incidental finding of IA on noninvasive imaging, or for follow-up imaging of a previously detected IA. (ACA = anterior cerebral artery, AComA = anterior communicating artery, BT = basilar terminus, CT = computed tomography, DSA = digital subtraction angiography, IA = intracranial aneurysm, ICA = internal carotid artery, MCA = middle cerebral artery, MRA = magnetic resonance angiography, MRI = magnetic resonance imaging, PComA = posterior communicating artery, VB = vertebrobasilar).
(DOCX)
*The quality of the RNA sequencing experiments was measure pre-alignment via FASTQC and post-alignment via MultiQC. Overall, prior to alignment all samples had an average of 53.75 M sequences. MultiQC reported that the sequencing experiments had an average of 49.09 M mapped reads with a 96.13% read mapping rate, and detected an average of 17259 transcripts (transcripts with FPKM>0). (Align. = alignment, M. = million, Seqs. = sequences, Qual. = quality).
(DOCX)
*A table of the names of transcripts included in the top 4 networks derived from IPA, as well as the top diseases and functions of these transcripts. Neutrophil transcripts in bold were differentially expressed between patients with and without IA (p-value<0.05). Each network’s p-score was derived from its p-value [p-score = -Log10 (p-value)] calculated by the Fisher’s exact test. For a network with a p-score of 10, the odds of generating this network by chance alone is less than 1 out of 1010.
(DOCX)
*(IA = intracranial aneurysm, SE = standard error, Q = quartile).
(DOCX)
*Aneurysm size ranged from 3.5 mm to 7 mm. Five of 6 IAs (83%) were classified as small (greatest diameter <7mm) and 1 (17%) was classified as large (greatest diameter ≥7 mm). The aneurysms were situated at various locations in the Circle of Willis, with most being in the anterior vasculature (ACA and MCA). (ACA = anterior cerebral artery, AComA = anterior communicating artery, BT = basilar terminus, CT = computed tomography, DSA = digital subtraction angiography, IA, intracranial aneurysm, MCA = middle cerebral artery, MRA = magnetic resonance angiography, MRI = magnetic resonance imaging).
(DOCX)
*(M = male, F = female, Y = yes, N = no, HT = hypertension, HL = hyperlipidemia, CAD = coronary artery disease, S Hx = stroke history, DM = diabetes mellitus, OA = osteoarthritis).
(DOCX)
Data Availability Statement
Raw RNA sequencing data files and processed transcript expression levels for the experiments described in this publication can be found at NCBI’s Gene Expression Omnibus (GEO) at accession no. GSE106520.