Abstract
Purpose
Small cell lung cancer (SCLC) is an often-fatal neuroendocrine carcinoma usually presenting as extensive disease, carrying a 3% five-year survival. Despite notable advances in SCLC genomics, new therapies remain elusive, largely due to a lack of druggable targets.
Experimental Design
We used a high-throughput drug screen to identify a venetoclax sensitive SCLC sub-population, and validated the findings with multiple patient-derived xenografts of SCLC.
Results
Our drug screen consisting of a very large collection of cell lines demonstrated that venetoclax, an FDA-approved BCL-2 inhibitor, was found to be active in a substantial fraction of SCLC cell lines. Venetoclax induced BIM-dependent apoptosis in vitro and blocked tumor growth and induced tumor regressions in mice bearing high BCL-2-expressing SCLC tumors in vivo. BCL-2 expression was a predictive biomarker for sensitivity in SCLC cell lines and was highly expressed in a subset of SCLC cell lines and tumors, suggesting that a substantial fraction of SCLC patients could benefit from venetoclax. Mechanistically, we uncover a novel role for gene methylation that helped discriminate high BCL-2-expressing SCLCs.
Conclusions
Altogether, our findings identify venetoclax as a promising new therapy for high BCL-2 expressing SCLCs.
Keywords: small cell lung cancer, BCL-2, BH3 mimetic, apoptosis, drug screen, DNA methylation
Introduction
Despite initial sensitivity to platinum-based combination chemotherapies, SCLCs characteristically develop progressive chemotherapy resistance, resulting in a 5-year survival approximating 3%. In contrast to non-small cell lung cancer (NSCLC), which has significant druggable addictions to diverse receptor tyrosine kinases (RTKs) such as EGFR and ALK, genomic studies of SCLC tumors have not revealed widespread RTK-addiction (1-3).
Instead, these cancers demonstrate a predictable pattern of loss of tumor suppressors RB and p53 in almost all cases, and loss of PTEN or PIK3CA mutations in up to 20% of cases (3, 4). In addition, we have previously demonstrated that BCL-2, the founding pro-survival BCL-2 family member, is highly expressed in 65% of cases (5, 6), as is BIM (7), a pro-apoptotic BCL-2 family member which sensitizes SCLC to targeted therapies, in particular BH3 mimetics (8).
Since tumor suppressors and transcription factors are currently difficult drug targets, the development of targeted therapies for SCLC has been lacking. However, the recent addition of pro-survival BCL-2 family inhibitors to the repertoire of targeted therapies revealed marked preclinical activity in SCLC (7, 9). Unfortunately, in a phase II trial of the dual BCL-2 and BCL-xL inhibitor, navitoclax (ABT-263) in 39 SCLC patients, only 1 patient achieved a partial response and 8 patients had stable disease (10). While the response rate was disappointing, the disease control rate of 26% along with an objective response in a refractory population indicates detectable single-agent activity. Importantly, there was dose-limiting thrombocytopenia (10, 11) in the study, which is the result of BCL-xL inhibition (12). This strongly suggests the magnitude of BCL-2 inhibition was limited in this trial by the toxicities induced by BCL-xL inhibition.
The Genomics of Drug Sensitivity in Cancer (GDSC) platform is a comprehensive drug-screening initiative aimed at discovering new drug sensitivities (13). We used this platform to characterize the activity of venetoclax across cancer types and identify potential genomic predictors of response to this drug. We found that a large subset of SCLC cell lines is hypersensitive to the BCL-2 selective inhibitor venetoclax. Moreover, this activity correlated well with BCL-2 expression levels. These data demonstrate strong rationale for venetoclax therapy in high BCL-2-expressing SCLC tumors.
Materials and Methods
Cell lines
The SCLC cell lines, NCI-H510A, DMS-53, NCI-H187, NCI-H524, SW1271, COR-L303, IST-SL2, NCI-H1105, NCI-H211, NCI-H187, NCI-H146, NCI-H446, NCI-H196, NCI-H1688, NCI-H1048, and NCI-H82 were from the Center for Molecular Therapeutics at Massachusetts General Hospital Cancer Center, and are routinely checked by DNA STR for verification. The NCI-H2171 cell line was from the American Type Culture Collection (Manassas, VA, USA). Cell lines were routinely tested for mycoplasma by MycoAlert (Lonza), and if positive, treated with PlasmoCure from InvivoGen (San Diego, CA) until MycoAlert assays for the presence of mycoplasma were negative.
Drugs
Venetoclax for the cell culture and in vivo experiments was kindly provided by AbbVie. Cisplatin (cat# A8321) and Etoposide (cat# A1971) were purchased from ApexBio (Houston, TX).
High Throughput Drug Screen
Venetoclax was screened at the Center for Molecular Therapeutic at the Massachusetts General Hospital across authenticated cell lines from the GDSC collection essentially as previously described (13). Resazurin was used to measure viability for all the cell lines. Drug response estimates (IC50 and AUC: Area Under the dose response Curve) were obtained as previously described (13).
Analysis of mRNA expression
Expression of all genes in solid tumors, including venetoclax-sensitive and -resistant SCLC cell lines was obtained through the R2: Genomics Analysis and Visualization Platform (http://hgserver1.amc.nl) using the GDSC-based Celline Cancer Drug (Sanger) dataset (Array Express Accession: E-MTAB-3610). Expression data was also obtained from the R2 platform, using the CCLE expression dataset (GEO ID: GSE36133). For comparison between SCLC, NSCLC, and other solid tumors, blood-type tumors and all tumors in which histology was not determined were excluded from analyses.
Analysis of BCL2 promoter methylation
Illumina 450K array data from the GDSC (GEO ID: GSE68379) was used for the analysis of CpG methylation. Methylation status of the BCL2 promoter was performed on SCLCs with either 2- or 3- BCL2 copy numbers. Raw methylation data was imported into R and ChAMP (14) was used to normalize the array data. β-values were extracted for each cell line and individual CpG sites for the 1.5kb region immediately upstream of the BCL2 transcriptional start site were averaged for the top- and bottom-50% BCL2 expressers.
Lentivirus production
The lentiviral short-hairpin RNA (shRNA)-expressing constructs were purchased from Addgene (Cambridge, MA) and Open Biosystems (Huntsville, AL). The constructs were transfected into 293T packaging cells along with the packaging plasmids (Addgene) and the lentivirus-containing supernatants were used to transduce SCLC cells.
Immunoprecipitations and Western blot analyses
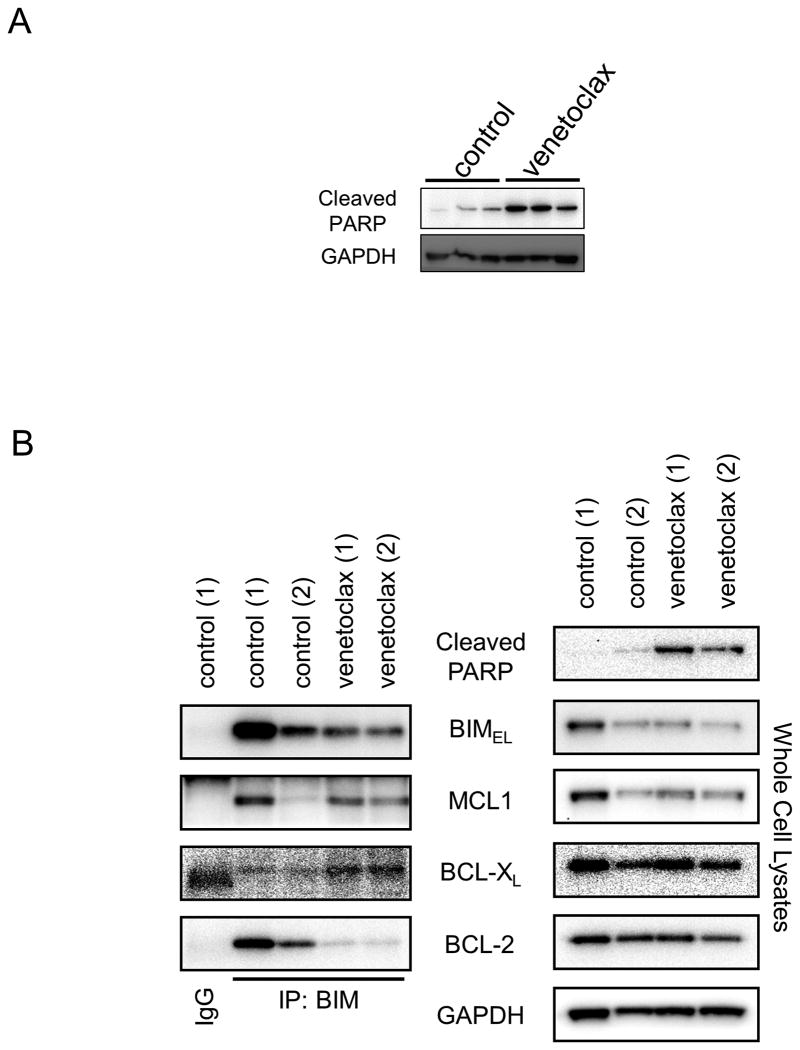
Antibodies were purchased as follows: BCL-2 (clone 100) from Sigma-Aldrich (St. Louis, MO) and Cell Signaling (GAPDH, BIM (C34C5), BCL-XL (54H6), and Cleaved PARP (D64E10) from Cell Signaling Technology (Danvers, MA); Noxa (114C307.1) from Thermo Fisher Scientific (Waltham, MA); MCL-1 from Enzo Life Sciences (Farmingdale, NY); BAX (N-20) from Santa Cruz Biotechnology (Santa Cruz, CA). Immunoprecipitation and Western blots were performed as described in (19). For Sup. Fig. 12, 500ug of each lysate were incubated with BIM antibody (500 ng; Cell Signaling Technology, cat# 2933S), or rabbit IgG (500 ng; Santa Cruz Biotechnology, cat# sc-2027). Following the addition of 25uL of 1:1 PBS: pre-washed Protein A beads to the antibody/lysate mix, samples were incubated with rotating motion overnight. Equal amounts of extracts (5% of immunoprecipitated protein) were prepared in parallel.
Cell viability assays
SCLC cells were seeded in triplicate (WST-1 assays) or quadruplicate (CTG-assays) in microtiter plates (96 wells) with a concentration of 1 × 104 cells per well in 100 μl growth medium (WST-1 assays) or a concentration of 2 × 103 cells per well containing 180 μl growth medium (CTG assays), and were treated with 0-10 μM venetoclax for 72 hr, 5-days, or 7-days. Cell survival was measured by the WST-1 assay (Fig. 4D) according to the manufacturer's instruction (Roche), and the CellTiter-Glo protocol (Promega) (Fig. 2, Sup. Fig. 2A, Sup. Fig. 6). For synergy assays in Sup. Fig. 2C, DMS-53, NCI-H2171, NCI-H82, and SW1271 were seeded at 1 × 104 cells /well in 96-well plates. A 3×4 dose matrix of Venetoclax and the combination of Cisplatin (C) and Etoposide (E) (ratio C:E 1/1.5) was performed and cell viability was measured at the end of 72 hrs using WST-1 as above. Drug synergy was determined using the Bliss independence model as previously described (15).
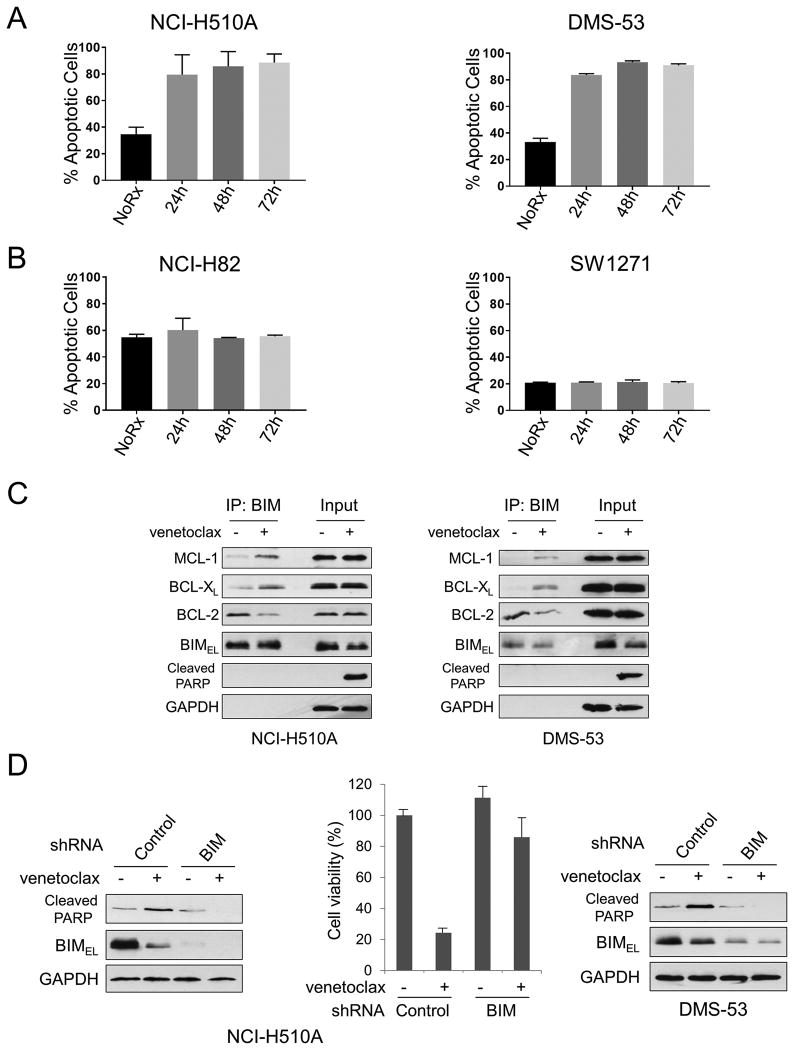
Figure 4. Venetoclax targets BCL-2 to disrupt BIM:BCL-2 complex and induces BIM-mediated apoptosis.
Apoptosis assays were performed in triplicate on (A) NCI-H510A and DMS-53 or (B) NCI-H82 and SW1271 cell lines treated with 1 μM venetoclax for the time indicated. (C) NCI-H510A or DMS-53 cells were treated with 1 μM venetoclax for 8 hours. Total cell lysates were subjected to immunoprecipitation with anti-BIM antibodies. Western blot analyses were carried out on precipitated samples with the indicated antibodies. (D) NCI-H510A (left panel) or DMS-53 (right panel) cells were infected with lentivirus encoding shRNA for non-targeting control or BCL2L11 (BIM). Cells were then treated with 1 μM venetoclax for 24 hours and equal amounts of total extracts were subjected to Western blot analyses using the indicated antibodies. Cell viability of NCI-H510A was determined by WST-1 assay at 24 hours after treatment. Average values from triplicate samples are shown as representative of two independent experiments. (D, center panel). All error bars were plotted using standard error of the mean.
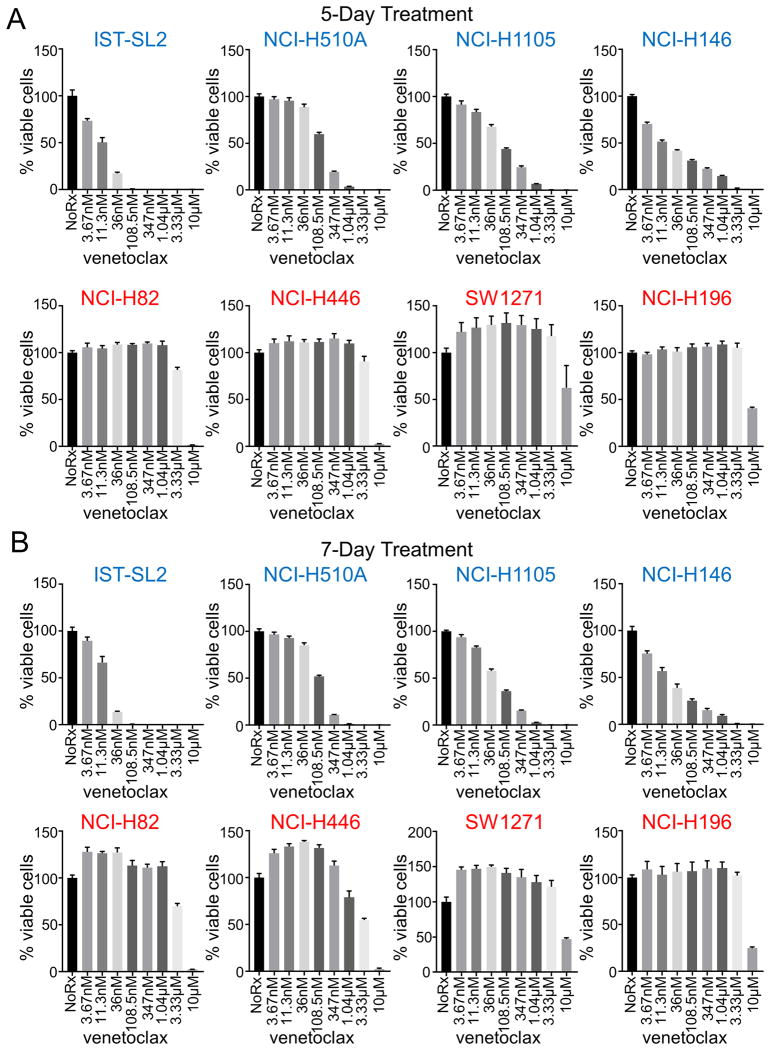
Figure 2. 5- and 7-day cell viability assays of venetoclax-sensitive and -resistant SCLC cell lines.
(A) 5-day CellTiter-Glo assays were performed on the indicated venetoclax-sensitive (top) and -resistant (bottom) cell lines. (B) 7-day CellTiter-Glo assays were performed on the cell lines in part A. Error bars are plotted as standard error. Venetoclax-sensitive cell lines are titled in blue, venetoclax-resistant cell lines are titled in red.
FACS apoptosis
DMS-53, NCI-H510A, NCI-H169, NCI-H82, and SW1271 cell lines were seeded at 1.0-1.5 × 106 cells in a 6-well tissue culture plate and either untreated or treated for 24, 48, or 72 hours with 1 μM venetoclax. Following treatment, cells were collected as per BD biosciences Annexin V/PI staining protocol. Cells were then assayed on a Millipore Guava flow cytometer. Cells were quantitated as either Annexin V-positive, PI-positive, double positive, or double negative using quadrants. Apoptotic cells were measured as 100% - double negative cell population.
SCLC cell line xenograft model
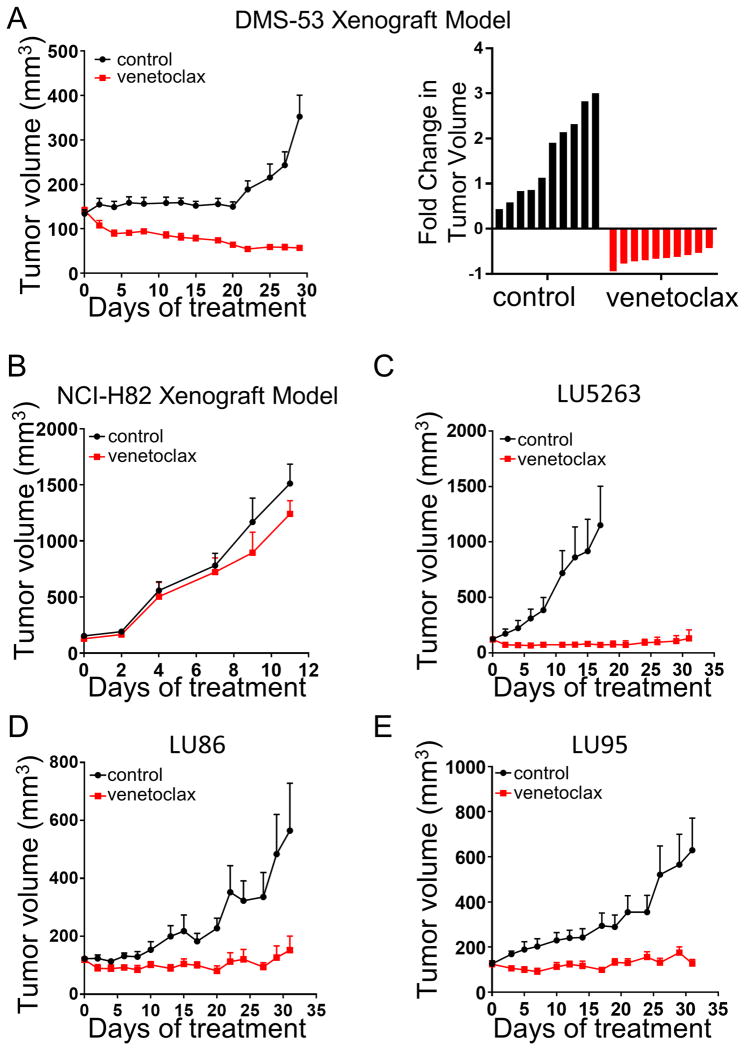
The DMS-53 cell line was injected into the right flank of 20 female NOD-SCID-gamma (NSG) mice at a concentration of 5 × 106 cells per mouse using a 1:4 ratio of cells to Matrigel (Corning, cat# 354248). The NCI-H82 cell line was injected into both flanks of 10 female NSG mice using the same protocol as above. Tumors were monitored until the average size of all tumors increased above the average injection volume of 100 mm3. Mice were then randomized into untreated and venetoclax-treated cohorts, with 10 mice per cohort. Venetoclax-treated mice were dosed with 100 mg/kg of venetoclax by oral gavage 6 days per week. Every 2-3 days, tumors were measured in two dimensions via calipers and tumor volume was calculated as: height × width × width × 0.52, where height was the larger of the two measurements. Mice were euthanized after 29 days of treatment. All animal experiments were approved by the Virginia Commonwealth University Institutional Animal Care and Use Committee (IACUC protocol# AD10001048).
Patient-derived xenograft models
The first patient derived xenograft (PDX) model (Crown Bioscience San Diego) is from a 71 year-old female patient with small cell carcinoma (LU5263). The second (LU86) and third (LU95) PDX models have been previously described (16) and were kindly provided by Stemcentrx (currently part of AbbVie). Briefly, a single-cell suspension was mixed with Matrigel at a 1:1 ratio and 0.5 × 106 cells were injected per mouse. Enrollment and treatment of this study was performed as above in the xenograft model, with 6 mice per cohort. Mice were euthanized after 31 days of treatment. A measure of apoptosis was obtained by treating three mice with venetoclax for three days. The mice were euthanized two hours after venetoclax treatment on the third day and the tumors were extracted, and immediately snap-frozen in liquid nitrogen for eventual lysate preparation.
Statistical Analyses
Mann-Whitney U tests were performed to determine significance of relationships in Fig. 1C, Sup. Fig. 4, and Sup. Fig. 5. A Spearman's correlation test was performed to determine significance and a correlation coefficient in Fig 3A, Sup. Fig. 1B and C, and Sup. Fig. 3A. ANOVA analysis was performed to determine the significance in Fig. 3C and D.
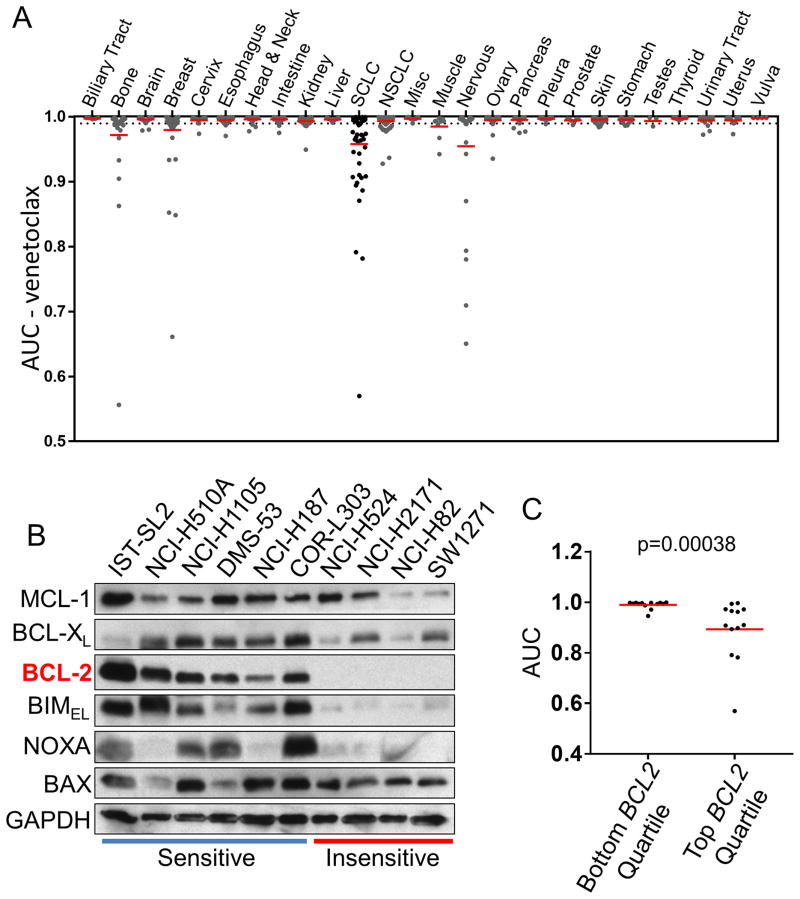
Figure 1. SCLC tumor cell lines are sensitive to venetoclax via increased BCL-2 expression.
(A) Venetoclax drug screen data against 26 different cancer types, with SCLC highlighted. (B) Expression of the BCL-2 family proteins in SCLC cell lines was determined by Western blot analyses using the indicated antibodies. The SCLC lines are indicated as venetoclax sensitive (AUC < 0.98, blue line) or insensitive (AUC > 0.98, red line). (C) SCLC expression data from the GDSC database was clustered into the bottom and top 25% BCL-2 RNA expressing cells and the AUC data were plotted. The red bar indicates the mean of each group. Statistical significance was determined using the Mann-Whitney U test.
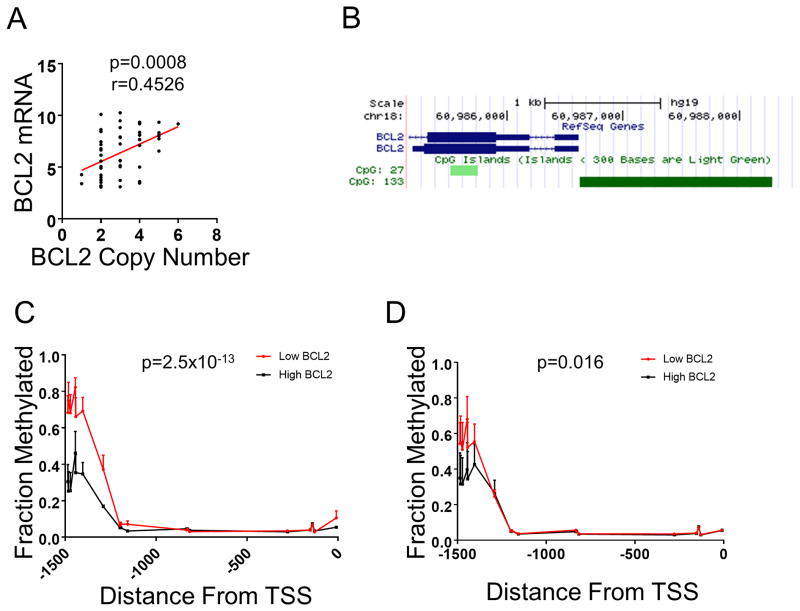
Figure 3. Methylation of the BCL2 promoter correlates with RNA expression.
(A) BCL2 expression correlates with DNA copy number. Correlation analysis was performed using Spearman's correlation. (B) A CpG island exists within a 1.5kb region upstream of the BCL2 transcriptional start site. (C) Methylation within the CpG island of BCL2. Top 50% of BCL2 expressing SCLC cell lines (black line; n=8) with two copies of BCL2 are compared to the bottom 50% BCL2 expressers (red line; n=8). (D) Methylation of within the CpG island of BCL2. Top 50% of BCL2 expressing SCLC cell lines (black line; n=6) with three copies of BCL2 are compared to the bottom 50% BCL2 expressers (red line; n=7). Statistical significance between methylation status was determined using ordinary ANOVA analysis.
Results
A subset of SCLC is hypersensitive to venetoclax
Venetoclax is a first in-class BCL-2 selective inhibitor, recently FDA-approved for chronic lymphocytic leukemia (CLL) patients with 17p deletion who have received at least one prior treatment. Based on its safety profile and potent anti-cancer activity, we performed a high-throughput screen of venetoclax across multiple cancer types with the goal of identifying other cancers that may respond to venetoclax. We screened over 950 cell lines including 791 lines derived from solid tumors. Among solid tumor cell lines, a subset of neuroblastomas (MYCN-amplified) (17) and SCLCs were particularly sensitive to venetoclax (Fig. 1A, Sup. Fig. 1A, and Sup. Table 1). Longer treatment times of 5- and 7-days did not change sensitivity profiles of the different SCLC cell lines to venetoclax (Fig. 2). Furthermore, a clear dose-response to venetoclax of the high BCL-2 NCI-H187 (AUC=0.965) cell line was observed (Sup. Fig. 2A). In contrast, the low BCL-2 NCI-H524 (AUC=0.987) was unaffected until the maximum dosage given (Sup. Fig. 2B). As SCLCs are sensitive to initial platinum-based chemotherapy (18) we tested venetoclax with cisplatin-etoposide at a 1:1.5 ratio (16). Here, we found some weak synergy at higher concentrations of chemotherapy in four of the six lines (Sup. Fig. 2C) across different venetoclax concentrations. These data suggest potential new indications for this clinical compound.
The sensitive SCLCs are distinguished by high BCL-2 expression
We next sought to investigate whether the expression level of BCL-2, the direct target of venetoclax, might predict response within SCLC. Through the GDSC database, RNA expression levels of BCL-2 were available in 52 out of the 56 SCLC cell lines screened. We found sensitivity to venetoclax positively correlated with BCL-2 expression (Fig. 1B, Fig. 1C, and Sup. Fig. 1B). In contrast, the mRNA and protein expression profiles of other prominent BCL-2 members, including BCL2L11 (BIM), did not further predict sensitivity (Fig. 1B, Sup. Fig. 3, Sup. Fig. 4). While large drug screen studies have shown that target expression is not consistently predictive of drug response (19), these data indicate that BCL-2 alone may constitute an efficient predictive biomarker for venetoclax in SCLC, with high BCL-2 expressing SCLCs yielding better responses.
Methylation of BCL-2 contributes to its expression
We next evaluated BCL-2 expression in SCLCs compared to other cancers, to confirm prior findings (20) that BCL-2 expression was generally higher in SCLC, suggestive of an underlying reliance on BCL-2. We used BCL2 mRNA expression data from both the GDSC and the Cancer Cell Line Encyclopedia (CCLE) (21, 22) resource and indeed found SCLC cell lines have higher BCL2 expression on average than other solid tumor cell lines including NSCLC (Sup. Fig. 5A). We next asked whether this relationship held true in patient tumors. Biomarker assessment in SCLC tumors is challenging due to the limitation of available tumor material. We identified two datasets (23, 24) with gene expression from SCLC tumors and found significantly (P<0.001) higher BCL-2 expression in SCLC tumors than in NSCLC tumors (Sup. Fig. 5B and 5C). These data confirm BCL2 expression is high in a substantial subset of SCLC cell lines and tumors.
As BCL-2 is amplified in SCLC (9), we explored whether DNA amplification alone was responsible for increased levels of BCL-2 expression. We found that BCL2 amplification did indeed correlate with BCL2 expression (Fig. 3A). However, we noticed that BCL2 RNA levels varied greatly within each copy-number category. The BCL2 gene has a CpG island immediately upstream of the transcriptional start site (TSS) (Fig. 3B). Given that CpG islands are often involved in the regulation of gene expression, we therefore asked whether DNA methylation may play a role in the expression of BCL-2. Using methylation data available from the GDSC, we found that both the 2- and 3-BCL2 copy number cell lines had differential methylation within this CpG island, with lower BCL-2 expressing cells having higher methylation (Fig. 3C and 3D). These results suggest that epigenetic regulation of BCL-2 expression plays an important role in SCLC, independent of BCL2 copy number.
Venetoclax kills SCLC cell lines by disrupting BIM:BCL-2 complexes
We next investigated the mode of venetoclax killing in SCLC cell lines. FACS analysis of PI/Annexin Cy5 stained SCLC cells demonstrated that 1 μM of venetoclax induced marked apoptosis over the treatment period of 72 hours in high BCL-2-expressing NCI-H510A and DMS-53 cells (Fig. 4A), corresponding to a sharp reduction in viable cells over the same treatment period (Sup. Fig. 6A and 6B). In contrast, venetoclax-resistant cell lines demonstrated no significant increase of apoptosis within the same 72-hour treatment (Fig. 4B). Importantly, the concentration used in these assays is clinically relevant based on steady state pharmacokinetics observed with venetoclax (25-27). Immunoprecipitation of BIM in NCI-H510A and DMS-53 whole cell lysates treated with 1 μM venetoclax revealed on-target activity of venetoclax as judged by the disassociation of BIM:BCL-2 complexes (Fig. 4C, left and right panels, respectively). Consistent with past reports, venetoclax led to an increase in BIM:MCL-1 complexes (17, 28) and we also noted increases in BIM:BCL-xL complexes (Fig. 4C). BIM knockdown mitigated venetoclax efficacy in NCI-H510A and DMS-53 cells (Fig. 4D), further demonstrating venetoclax kills in a BIM-mediated manner (17, 29). Altogether, these data demonstrate on-target killing by venetoclax in SCLC through disruption of BIM:BCL-2 complexes, leading to BIM-dependent apoptosis.
Venetoclax is effective in high BCL-2 expressing SCLC mouse models
We next assessed the ability of venetoclax to impede tumor growth in the high BCL-2 expressing DMS-53 mouse model. Treatment with 100 mg/kg of venetoclax delivered once daily (30) led to marked tumor shrinkage (Fig. 5A). Reflecting the established safety profile of venetoclax in humans (31), these mice exhibited little to no weight loss, nor other overt signs of toxicity (Sup. Fig. 7A). Consistent with our hypothesis, the low BCL-2 xenograft model, NCI-H82, was unresponsive to venetoclax treatment (Fig. 5B).
Figure 5. Venetoclax is effective in high BCL-2 expressing xenograft and PDX mouse models.
(A) Growth curves of the DMS-53 xenograft model. Mice were either untreated (black lines) or administered 100 mg/kg of venetoclax (red lines) by oral gavage six days per week. Right panel shows the total tumor change over the total course of treatment. (B) Growth curves of the venetoclax-resistant cell line, NCI-H82. Mice were treated identical as above, and are plotted as untreated (black lines) or venetoclax treated (red lines). (C-E) Growth curves of the PDX models LU5263 (C), LU86 (D), and LU95 (E), either untreated (black lines) or venetoclax treated (red lines). Dosing was identical to the xenograft model above. All error bars were plotted as standard error of the mean.
We expanded our analysis to three patient-derived xenografts (PDXs) that had high BCL-2 expression as measured by Western blot analysis (Sup. Fig. 8), including the chemorefractory LU86 model (16). Using the same treatment schedule, we found these tumors were also uniformly sensitive to venetoclax, with marked regressions in the LU5263 PDX model, and near total growth inhibition in the LU86 and LU95 models (Fig. 5C-E).
Analysis of LU5263 tumors treated for three days with venetoclax demonstrated marked apoptosis (Fig. 6A), consistent with the mode of activity of venetoclax [Figs. 4A, Sup. Fig. 6D, and ref. (30)]. Further, the in vitro mechanism of BIM:BCL-2 complex disruption was observed in the LU5263 tumors in vivo, including a noticeable increase in BIM:BCL-xL complexes (Fig. 6B).
Figure 6. Venetoclax disrupts BIM:BCL-2 complexes in vivo in the LU5263 PDX model.
(A) Induction of the apoptosis marker cleaved PARP (cPARP) in three control and three, 3-day venetoclax treated LU5263 tumors. GAPDH was used as a loading control. (B) Tumor lysates from 3-day venetoclax treated LU5263 tumors were immunoprecipitated using an anti-BIM antibody. Western blot analysis was carried out on precipitated and input samples using the antibodies indicated.
Discussion
Preclinical studies have demonstrated that venetoclax is an effective BCL-2 inhibitor that lacks the platelet killing capacity of dual BCL-2/BCL-xL inhibitors (30). This translates to more potent BCL-2 inhibition, and venetoclax has proven effective against BCL-2-dependent CLL without inducing marked thrombocytopenia (27, 31).
Despite promising preclinical activity, the BCL-2/BCL-xL inhibitor navitoclax did not translate to successful clinical activity in SCLC (10, 11). A leading hypothesis to explain this failure is that BCL-2 itself was not sufficiently inhibited due to the dose-limiting thrombocytopenia caused by BCL-xL inhibition. In this study, we have demonstrated that a significant population of SCLC cell lines – those bearing high BCL-2 levels – are susceptible to venetoclax therapy. Interestingly, while high BCL-2 (7, 32-35), high NOXA (36), high BIM (7) and low MCL-1 (7, 9) are key factors governing BH3 mimetic sensitivity in SCLC, high BCL2 mRNA alone predicted venetoclax sensitivity in SCLC (Figs. 1 and Sup. Fig. 1, 3, 4A) and neither BCL2L1, BCL2L11, nor MCL1 mRNA expression improved the ability of BCL-2 to predict response (Sup. Fig. 4B). Additionally, all of the sensitive cell lines we assayed for protein expression displayed varying levels of BCL-XL, MCL-1, and NOXA (Fig. 1B, Sup. Fig. 8), while displaying high levels of BCL-2 and BIM. However, not all high BCL2 mRNA expressers were sensitive to venetoclax (Fig. 1C, Sup. Fig. 1B). Further, another report using therapeutics that decrease MCL-1 expression found a more robust response with both navitoclax and venetoclax in a single SCLC xenograft and PDX model (34). Therefore, while our data indicate high BCL-2 expression may locate potential responders to venetoclax, more research is required to determine other potential co-factors in SCLC susceptibility to venetoclax.
Our in vivo data demonstrated that venetoclax effectively shrank or controlled tumors in high BCL-2 expressing xenografts and PDX models, while a low BCL-2 expressing xenograft was unresponsive (Fig. 5). Other studes using ABT-737 across a longer-scale treatment have found that tumors develop resistance (35, 37). Given the better tolerance for venetoclax due to a lack of thrombocytopenia development (27, 30, 31), it is also possible that this agent would be more efficacious in the treatment of SCLC. However, it remains unclear whether extended treatment times would lead to similar development of resistance using venetoclax.
Although the levels of BIM are variable even in sensitive cell lines (Fig. 1B, Sup. Figs. 3 and 4), down-regulation of BIM inhibited apoptosis following venetoclax treatment (Fig. 4D). In addition, the disruption of BIM:BCL-2 complexes is triggered by venetoclax (Fig. 4C), both suggesting BIM is necessary for venetoclax-induced apoptosis in SCLCs. Consistently, SW1271, in which the expression of BCL-2 and BIM are markedly low, was highly resistant to venetoclax (Fig. 1B).
It should be noted that our previously published large cell line screen of navitoclax revealed that cell lines from many different solid tumors were susceptible this agent (7). This was not the case for venetoclax, however, where the only solid tumors with noticeable sensitivity were SCLCs and MYCN-amplified neuroblastomas (Fig. 1A, Sup. Fig. 1A, Sup. Table 1). We and others previously reported enhanced sensitivity of MYCN-amplified neuroblastoma to venetoclax (17, 38). Although MYCN is the predominant MYC family member amplified in neuroblastoma (39), in SCLC all three members of the MYC family are amplified independently of each other, with L-Myc (MYCL) being the most frequently amplified at 9% (3). Thus, future evaluation of how MYC family status could affect BCL-2 levels to predict venetoclax responses would be valuable.
We also report that while BCL-2 amplification plays a role in high BCL-2 expression in SCLC (9), methylation in the BCL2 promoter helps distinguish low from high BCL-2 expressers. Methylation of BCL2 has been linked to increased sensitivity to anti-mitotic agents in breast cancer patients (40), where no methylation was found in normal breast tissue. This is also consistent with studies demonstrating a reliance on another anti-apoptotic BCL-2 family member, MCL-1, for anti-mitotic efficacy (41, 42). However, in the case of BCL-2 inhibitors, lack of BCL2 methylation in SCLCs seem to have the opposite effect, sensitizing these cancers to venetoclax (Figs. 1C and 3), likely through the priming of BIM:BCL-2 complexes (Fig. 4).
Our data is consistent with a recent report identifying in vitro activity of venetoclax in SCLC cell lines, and a correlation between BCL-2 expression and venetoclax activity (32). In addition, Lam et al. very recently reported that venetoclax sensitizes SCLC cell lines to BET inhibition (33). These previous reports support the potential of venetoclax in SCLC. Here, we expand significantly on these findings by demonstrating that a subset of SCLC is uniquely sensitive to venetoclax among solid tumor cell types, with the exception of MYCN-amplified neuroblastoma (Fig. 1A and Sup. Table 1), and that single-agent venetoclax demonstrates anti-tumor efficacy in high BCL-2 expressing patient-derived xenografts (Fig. 5C-E), and a cell-line derived xenograft (Fig. 5A). These results provide a rationale for assessing the clinical efficacy of venetoclax in a biomarker selected SCLC population. Importantly, we also demonstrate that the expression of other BCL-2 family genes does not improve predictive power over BCL-2 expression alone (Sup. Fig. 3B). Lastly, we identify that methylation of the BCL2 promoter is increased in cell lines that express low levels of BCL2 mRNA despite evidence of gene amplification (Fig. 3), identifying a potential epigenetic component in the regulation of BCL2 in SCLC.
In conclusion, screening venetoclax against ∼800 solid tumor cancer cell lines uncovered activity across a large subset of SCLC lines. These sensitive cancers have high BCL-2 expression at the protein level and a high level of BCL2 mRNA expression. Together, this work suggests rationale for biomarker-directed clinical trials of the FDA-approved drug venetoclax in high BCL-2 expressing SCLCs.
Supplementary Material
Statement of Translational Relevance.
Through an unbiased high-throughput drug screen of 791 sold tumor cancer cell lines, we have identified the FDA-approved BCL-2 inhibitor to be active in high BCL-2 expressing SCLCs. This study should set the stage for clinical evaluation of venetoclax in SCLC patients with high BCL-2 expression.
Acknowledgments
The authors thank Ultan McDermott and Matthew J. Garnett, leaders of the GDSC studies at The Wellcome Trust Sanger Institute. This work is supported by a IH-NCI Cancer Center Support Grant (P30-CA016059; Director, Gordon Ginder) to H.H. and A.C.F. in the form of a Massey Pilot Project Massey Cancer Center Pilot grant (5P30CA016059-35), and a National Cancer Institute K22-CA175276 Career Development Award (A.C.F.). C.H.B and the GDSC efforts are supported by a grant from the Wellcome Trust (102696). A.C.F. is supported by the George and Lavinia Blick Research Fund and is a Harrison Endowed Scholar in Cancer Research. Services and products in support of the research project were generated by the VCU Massey Cancer Center Cancer Mouse Model Shared Resource, supported, in part, with funding from NIH-NCI Cancer Center Support Grant P30-CA016059.
Financial Support: This work is supported by a IH-NCI Cancer Center Support Grant (P30-CA016059; Director, Gordon Ginder) to H.H. and A.C.F. in the form of a Massey Pilot Project Massey Cancer Center Pilot grant (5P30CA016059-35), and a National Cancer Institute K22-CA175276 Career Development Award (A.C.F.). C.H.B and the GDSC efforts are supported by a grant from the Wellcome Trust (102696). Services and products in support of the research project were generated by the VCU Massey Cancer Center Cancer Mouse Model Shared Resource, supported, in part, with funding from NIH-NCI Cancer Center Support Grant P30-CA016059.
Footnotes
Conflict of Interest Disclosure: Y.K.M. is an employee of Crown Bioscience Inc. L.R.S. and S.J.D. are employees and shareholders of AbbVie Stemcentrx LLC. J.D.L. and A.J.S. are employees and shareholders of AbbVie. There are no other conflicts of interest.
References
- 1.McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, Bhutkar A, McKenna A, Dooley A, Vernon A, Sougnez C, Malstrom S, Heimann M, Park J, Chen F, Farago AF, Dayton T, Shefler E, Gabriel S, Getz G, Jacks T. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156(6):1298–311. doi: 10.1016/j.cell.2014.02.031. Epub 2014/03/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, Menon R, Koker M, Dahmen I, Muller C, Di Cerbo V, Schildhaus HU, Altmuller J, Baessmann I, Becker C, de Wilde B, Vandesompele J, Bohm D, Ansen S, Gabler F, Wilkening I, Heynck S, Heuckmann JM, Lu X, Carter SL, Cibulskis K, Banerji S, Getz G, Park KS, Rauh D, Grutter C, Fischer M, Pasqualucci L, Wright G, Wainer Z, Russell P, Petersen I, Chen Y, Stoelben E, Ludwig C, Schnabel P, Hoffmann H, Muley T, Brockmann M, Engel-Riedel W, Muscarella LA, Fazio VM, Groen H, Timens W, Sietsma H, Thunnissen E, Smit E, Heideman DA, Snijders PJ, Cappuzzo F, Ligorio C, Damiani S, Field J, Solberg S, Brustugun OT, Lund-Iversen M, Sanger J, Clement JH, Soltermann A, Moch H, Weder W, Solomon B, Soria JC, Validire P, Besse B, Brambilla E, Brambilla C, Lantuejoul S, Lorimier P, Schneider PM, Hallek M, Pao W, Meyerson M, Sage J, Shendure J, Schneider R, Buttner R, Wolf J, Nurnberg P, Perner S, Heukamp LC, Brindle PK, Haas S, Thomas RK. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nature genetics. 2012 doi: 10.1038/ng.2396. Epub 2012/09/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, Leenders F, Lu X, Fernandez-Cuesta L, Bosco G, Muller C, Dahmen I, Jahchan NS, Park KS, Yang D, Karnezis AN, Vaka D, Torres A, Wang MS, Korbel JO, Menon R, Chun SM, Kim D, Wilkerson M, Hayes N, Engelmann D, Putzer B, Bos M, Michels S, Vlasic I, Seidel D, Pinther B, Schaub P, Becker C, Altmuller J, Yokota J, Kohno T, Iwakawa R, Tsuta K, Noguchi M, Muley T, Hoffmann H, Schnabel PA, Petersen I, Chen Y, Soltermann A, Tischler V, Choi CM, Kim YH, Massion PP, Zou Y, Jovanovic D, Kontic M, Wright GM, Russell PA, Solomon B, Koch I, Lindner M, Muscarella LA, la Torre A, Field JK, Jakopovic M, Knezevic J, Castanos-Velez E, Roz L, Pastorino U, Brustugun OT, Lund-Iversen M, Thunnissen E, Kohler J, Schuler M, Botling J, Sandelin M, Sanchez-Cespedes M, Salvesen HB, Achter V, Lang U, Bogus M, Schneider PM, Zander T, Ansen S, Hallek M, Wolf J, Vingron M, Yatabe Y, Travis WD, Nurnberg P, Reinhardt C, Perner S, Heukamp L, Buttner R, Haas SA, Brambilla E, Peifer M, Sage J, Thomas RK. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls M, Baxi SM, Mehta PP, Liu KK, Zhu J, Estrella H, Li C, Zientek M, Zong Q, Smeal T, Yin MJ. Targeting small cell lung cancer harboring PIK3CA mutation with a selective oral PI3K inhibitor PF-4989216. Clin Cancer Res. 2014;20(3):631–43. doi: 10.1158/1078-0432.CCR-13-1663. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Ezra JM, Kornstein MJ, Grimes MM, Krystal G. Small cell carcinomas of the lung express the Bcl-2 protein. Am J Pathol. 1994;145(5):1036–40. [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang SX, Sato Y, Kuwao S, Kameya T. Expression of bcl-2 oncogene protein is prevalent in small cell lung carcinomas. J Pathol. 1995;177(2):135–8. doi: 10.1002/path.1711770206. Epub 1995/10/01. [DOI] [PubMed] [Google Scholar]
- 7.Faber AC, Farago AF, Costa C, Dastur A, Gomez-Caraballo M, Robbins R, Wagner BL, Rideout WM, 3rd, Jakubik CT, Ham J, Edelman EJ, Ebi H, Yeo AT, Hata AN, Song Y, Patel NU, March RJ, Tam AT, Milano RJ, Boisvert JL, Hicks MA, Elmiligy S, Malstrom SE, Rivera MN, Harada H, Windle BE, Ramaswamy S, Benes CH, Jacks T, Engelman JA. Assessment of ABT-263 activity across a cancer cell line collection leads to a potent combination therapy for small-cell lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1411848112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, Incio J, Digumarthy SR, Pollack SF, Song Y, Muzikansky A, Lifshits E, Roberge S, Coffman EJ, Benes CH, Gomez HL, Baselga J, Arteaga CL, Rivera MN, Dias-Santagata D, Jain RK, Engelman JA. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer discovery. 2011;1(4):352–65. doi: 10.1158/2159-8290.CD-11-0106. Epub 2011/12/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoemaker AR, Mitten MJ, Adickes J, Ackler S, Refici M, Ferguson D, Oleksijew A, O'Connor JM, Wang B, Frost DJ, Bauch J, Marsh K, Tahir SK, Yang X, Tse C, Fesik SW, Rosenberg SH, Elmore SW. Activity of the Bcl-2 family inhibitor ABT-263 in a panel of small cell lung cancer xenograft models. Clin Cancer Res. 2008;14(11):3268–77. doi: 10.1158/1078-0432.CCR-07-4622. Epub 2008/06/04. [DOI] [PubMed] [Google Scholar]
- 10.Rudin CM, Hann CL, Garon EB, Ribeiro de Oliveira M, Bonomi PD, Camidge DR, Chu Q, Giaccone G, Khaira D, Ramalingam SS, Ranson MR, Dive C, McKeegan EM, Chyla BJ, Dowell BL, Chakravartty A, Nolan CE, Rudersdorf N, Busman TA, Mabry MH, Krivoshik AP, Humerickhouse RA, Shapiro GI, Gandhi L. Phase II study of single-agent navitoclax (ABT-263) and biomarker correlates in patients with relapsed small cell lung cancer. Clin Cancer Res. 2012;18(11):3163–9. doi: 10.1158/1078-0432.CCR-11-3090. Epub 2012/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandhi L, Camidge DR, Ribeiro de Oliveira M, Bonomi P, Gandara D, Khaira D, Hann CL, McKeegan EM, Litvinovich E, Hemken PM, Dive C, Enschede SH, Nolan C, Chiu YL, Busman T, Xiong H, Krivoshik AP, Humerickhouse R, Shapiro GI, Rudin CM. Phase I study of Navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(7):909–16. doi: 10.1200/JCO.2010.31.6208. Epub 2011/02/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason KD, Carpinelli MR, Fletcher JI, Collinge JE, Hilton AA, Ellis S, Kelly PN, Ekert PG, Metcalf D, Roberts AW, Huang DC, Kile BT. Programmed anuclear cell death delimits platelet life span. Cell. 2007;128(6):1173–86. doi: 10.1016/j.cell.2007.01.037. Epub 2007/03/27. [DOI] [PubMed] [Google Scholar]
- 13.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O'Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–5. doi: 10.1038/nature11005. Epub 2012/03/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, Beck S. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics. 2014;30(3):428–30. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong M, Tan N, Zha J, Peale FV, Yue P, Fairbrother WJ, Belmont LD. Navitoclax (ABT-263) reduces Bcl-x(L)-mediated chemoresistance in ovarian cancer models. Mol Cancer Ther. 2012;11(4):1026–35. doi: 10.1158/1535-7163.MCT-11-0693. Epub 2012/02/04. [DOI] [PubMed] [Google Scholar]
- 16.Saunders LR, Bankovich AJ, Anderson WC, Aujay MA, Bheddah S, Black K, Desai R, Escarpe PA, Hampl J, Laysang A, Liu D, Lopez-Molina J, Milton M, Park A, Pysz MA, Shao H, Slingerland B, Torgov M, Williams SA, Foord O, Howard P, Jassem J, Badzio A, Czapiewski P, Harpole DH, Dowlati A, Massion PP, Travis WD, Pietanza MC, Poirier JT, Rudin CM, Stull RA, Dylla SJ. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med. 2015;7(302):302ra136. doi: 10.1126/scitranslmed.aac9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ham J, Costa C, Sano R, Lochmann TL, Sennott EM, Patel NU, Dastur A, Gomez-Caraballo M, Krytska K, Hata AN, Floros KV, Hughes MT, Jakubik CT, Heisey DA, Ferrell JT, Bristol ML, March RJ, Yates C, Hicks MA, Nakajima W, Gowda M, Windle BE, Dozmorov MG, Garnett MJ, McDermott U, Harada H, Taylor SM, Morgan IM, Benes CH, Engelman JA, Mosse YP, Faber AC. Exploitation of the Apoptosis-Primed State of MYCN-Amplified Neuroblastoma to Develop a Potent and Specific Targeted Therapy Combination. Cancer Cell. 2016;29(2):159–72. doi: 10.1016/j.ccell.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felip E, Pavlidis N, Stahel RA. ESMO Minimum Clinical Recommendations for diagnosis, treatment and follow-up of small-cell lung cancer (SCLC) Ann Oncol. 2005;16(Suppl 1):i30–1. doi: 10.1093/annonc/mdi829. Epub 2005/05/13. [DOI] [PubMed] [Google Scholar]
- 19.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Goncalves E, Barthorpe S, Lightfoot H, Cokelaer T, Greninger P, van Dyk E, Chang H, de Silva H, Heyn H, Deng X, Egan RK, Liu Q, Mironenko T, Mitropoulos X, Richardson L, Wang J, Zhang T, Moran S, Sayols S, Soleimani M, Tamborero D, Lopez-Bigas N, Ross-Macdonald P, Esteller M, Gray NS, Haber DA, Stratton MR, Benes CH, Wessels LF, Saez-Rodriguez J, McDermott U, Garnett MJ. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166(3):740–54. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikegaki N, Katsumata M, Minna J, Tsujimoto Y. Expression of bcl-2 in small cell lung carcinoma cells. Cancer Res. 1994;54(1):6–8. [PubMed] [Google Scholar]
- 21.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. Epub 2012/03/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Cell Line Encyclopedia C, Genomics of Drug Sensitivity in Cancer C. Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015;528(7580):84–7. doi: 10.1038/nature15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rohrbeck A, Neukirchen J, Rosskopf M, Pardillos GG, Geddert H, Schwalen A, Gabbert HE, von Haeseler A, Pitschke G, Schott M, Kronenwett R, Haas R, Rohr UP. Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. J Transl Med. 2008;6:69. doi: 10.1186/1479-5876-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF, Kipps TJ, Anderson MA, Brown JR, Gressick L, Wong S, Dunbar M, Zhu M, Desai MB, Cerri E, Heitner Enschede S, Humerickhouse RA, Wierda WG, Seymour JF. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N Engl J Med. 2016;374(4):311–22. doi: 10.1056/NEJMoa1513257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salem AH, Agarwal SK, Dunbar M, Enschede SL, Humerickhouse RA, Wong SL. Pharmacokinetics of Venetoclax, a Novel BCL-2 Inhibitor, in Patients With Relapsed or Refractory Chronic Lymphocytic Leukemia or Non-Hodgkin Lymphoma. J Clin Pharmacol. 2017;57(4):484–92. doi: 10.1002/jcph.821. [DOI] [PubMed] [Google Scholar]
- 27.Roberts AW, Stilgenbauer S, Seymour JF, Huang DC. Venetoclax in patients with previously treated chronic lymphocytic leukemia. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-16-0955. [DOI] [PubMed] [Google Scholar]
- 28.Phillips DC, Xiao Y, Lam LT, Litvinovich E, Roberts-Rapp L, Souers AJ, Leverson JD. Loss in MCL-1 function sensitizes non-Hodgkin's lymphoma cell lines to the BCL-2-selective inhibitor venetoclax (ABT-199) Blood Cancer J. 2016;6:e403. doi: 10.1038/bcj.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khaw SL, Merino D, Anderson MA, Glaser SP, Bouillet P, Roberts AW, Huang DC. Both leukaemic and normal peripheral B lymphoid cells are highly sensitive to the selective pharmacological inhibition of prosurvival Bcl-2 with ABT-199. Leukemia. 2014 doi: 10.1038/leu.2014.1. Epub 2014/01/10. [DOI] [PubMed] [Google Scholar]
- 30.Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, Huang DC, Hymowitz SG, Jin S, Khaw SL, Kovar PJ, Lam LT, Lee J, Maecker HL, Marsh KC, Mason KD, Mitten MJ, Nimmer PM, Oleksijew A, Park CH, Park CM, Phillips DC, Roberts AW, Sampath D, Seymour JF, Smith ML, Sullivan GM, Tahir SK, Tse C, Wendt MD, Xiao Y, Xue JC, Zhang H, Humerickhouse RA, Rosenberg SH, Elmore SW. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nature medicine. 2013;19(2):202–8. doi: 10.1038/nm.3048. Epub 2013/01/08. [DOI] [PubMed] [Google Scholar]
- 31.Stilgenbauer S, Eichhorst B, Schetelig J, Coutre S, Seymour JF, Munir T, Puvvada SD, Wendtner CM, Roberts AW, Jurczak W, Mulligan SP, Bottcher S, Mobasher M, Zhu M, Desai M, Chyla B, Verdugo M, Enschede SH, Cerri E, Humerickhouse R, Gordon G, Hallek M, Wierda WG. Venetoclax in relapsed or refractory chronic lymphocytic leukaemia with 17p deletion: a multicentre, open-label, phase 2 study. Lancet Oncol. 2016;17(6):768–78. doi: 10.1016/S1470-2045(16)30019-5. [DOI] [PubMed] [Google Scholar]
- 32.Polley E, Kunkel M, Evans D, Silvers T, Delosh R, Laudeman J, Ogle C, Reinhart R, Selby M, Connelly J, Harris E, Fer N, Sonkin D, Kaur G, Monks A, Malik S, Morris J, Teicher BA. Small Cell Lung Cancer Screen of Oncology Drugs, Investigational Agents, and Gene and microRNA Expression. J Natl Cancer Inst. 2016;108(10) doi: 10.1093/jnci/djw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam LT, Lin X, Faivre EJ, Yang Z, Huang X, Wilcox DM, Bellin RJ, Jin S, Tahir SK, Mitten M, Magoc T, Bhathena A, Kati WM, Albert DH, Shen Y, Uziel T. Vulnerability of small cell lung cancer to apoptosis induced by the combination of BET bromodomain proteins and BCL2 inhibitors. Mol Cancer Ther. 2017 doi: 10.1158/1535-7163.MCT-16-0459. [DOI] [PubMed] [Google Scholar]
- 34.Inoue-Yamauchi A, Jeng PS, Kim K, Chen HC, Han S, Ganesan YT, Ishizawa K, Jebiwott S, Dong Y, Pietanza MC, Hellmann MD, Kris MG, Hsieh JJ, Cheng EH. Targeting the differential addiction to anti-apoptotic BCL-2 family for cancer therapy. Nat Commun. 2017;8:16078. doi: 10.1038/ncomms16078. Epub 2017/07/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hann CL, Daniel VC, Sugar EA, Dobromilskaya I, Murphy SC, Cope L, Lin X, Hierman JS, Wilburn DL, Watkins DN, Rudin CM. Therapeutic efficacy of ABT-737, a selective inhibitor of BCL-2, in small cell lung cancer. Cancer Res. 2008;68(7):2321–8. doi: 10.1158/0008-5472.CAN-07-5031. Epub 2008/04/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakajima W, Hicks MA, Tanaka N, Krystal GW, Harada H. Noxa determines localization and stability of MCL-1 and consequently ABT-737 sensitivity in small cell lung cancer. Cell Death Dis. 2014;5:e1052. doi: 10.1038/cddis.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardner EE, Connis N, Poirier JT, Cope L, Dobromilskaya I, Gallia GL, Rudin CM, Hann CL. Rapamycin rescues ABT-737 efficacy in small cell lung cancer. Cancer Res. 2014;74(10):2846–56. doi: 10.1158/0008-5472.CAN-13-3460. Epub 2014/03/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bate-Eya LT, den Hartog IJ, van der Ploeg I, Schild L, Koster J, Santo EE, Westerhout EM, Versteeg R, Caron HN, Molenaar JJ, Dolman ME. High efficacy of the BCL-2 inhibitor ABT199 (venetoclax) in BCL-2 high-expressing neuroblastoma cell lines and xenografts and rational for combination with MCL-1 inhibition. Oncotarget. 2016;7(19):27946–58. doi: 10.18632/oncotarget.8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, Kim J, Lawrence MS, Lichenstein L, McKenna A, Pedamallu CS, Ramos AH, Shefler E, Sivachenko A, Sougnez C, Stewart C, Ally A, Birol I, Chiu R, Corbett RD, Hirst M, Jackman SD, Kamoh B, Khodabakshi AH, Krzywinski M, Lo A, Moore RA, Mungall KL, Qian J, Tam A, Thiessen N, Zhao Y, Cole KA, Diamond M, Diskin SJ, Mosse YP, Wood AC, Ji L, Sposto R, Badgett T, London WB, Moyer Y, Gastier-Foster JM, Smith MA, Guidry Auvil JM, Gerhard DS, Hogarty MD, Jones SJ, Lander ES, Gabriel SB, Getz G, Seeger RC, Khan J, Marra MA, Meyerson M, Maris JM. The genetic landscape of high-risk neuroblastoma. Nature genetics. 2013;45(3):279–84. doi: 10.1038/ng.2529. Epub 2013/01/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stone A, Cowley MJ, Valdes-Mora F, McCloy RA, Sergio CM, Gallego-Ortega D, Caldon CE, Ormandy CJ, Biankin AV, Gee JM, Nicholson RI, Print CG, Clark SJ, Musgrove EA. BCL-2 hypermethylation is a potential biomarker of sensitivity to antimitotic chemotherapy in endocrine-resistant breast cancer. Mol Cancer Ther. 2013;12(9):1874–85. doi: 10.1158/1535-7163.MCT-13-0012. [DOI] [PubMed] [Google Scholar]
- 41.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, Xiao Y, Christie AL, Aster J, Settleman J, Gygi SP, Kung AL, Look T, Nakayama KI, DePinho RA, Wei W. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471(7336):104–9. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, Belmont LD, Kaminker JS, O'Rourke KM, Pujara K, Kohli PB, Johnson AR, Chiu ML, Lill JR, Jackson PK, Fairbrother WJ, Seshagiri S, Ludlam MJ, Leong KG, Dueber EC, Maecker H, Huang DC, Dixit VM. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471(7336):110–4. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.