Abstract
Chemistry drives many biological interactions between the microbiota and host animals, yet it is often challenging to identify the chemicals involved. This poses a problem, as such small molecules are excellent sources of potential pharmaceuticals, pretested by nature for animal compatibility. We discovered anti-HIV compounds from small, marine tunicates from the Eastern Fields of Papua New Guinea. Tunicates are a reservoir for novel bioactive chemicals, yet their small size often impedes identification or even detection of the chemicals within. We solved this problem by combining chemistry, metagenomics, and synthetic biology to directly identify and synthesize the natural products. We show that these anti-HIV compounds, the divamides, are a novel family of lanthipeptides produced by symbiotic bacteria living in the tunicate. Neighboring animal colonies contain structurally related divamides that differ starkly in their biological properties, suggesting a role for biosynthetic plasticity in a native context where biological interactions take place.
Introduction
Symbiotic bacteria interact with each other and their hosts in part through small molecules, the vast majority of which remain uncharacterized. These compounds represent a promising source of drugs, but discovering and delivering them is a serious problem due to their cryptic nature and technical hurdles in their discovery and characterization. While symbiosis-derived chemicals are found throughout nature, the oceans have traditionally been a particularly rich source of bioactive compounds, which may serve as chemical defenses for their host animals.1 For example, bryostatins and ecteinascidins are produced in low abundance within marine animals, yet are used in clinical application despite incredible supply and development hurdles.2,3
The finding that such compounds are often produced by symbiotic bacteria, none of which have yet been cultivated, has provided a potential way forward in the direct cloning and expression of whole biosynthetic gene clusters, with the dual goals of providing a source of rare, potentially therapeutic compounds, and understanding the pathways that produce them. However, only two examples employing this strategy exist, one involving marine animals,4 and the other the uncultivated human microbiota,5,6 leaving substantial hurdles to accessing chemical novelty. Collectively, such systems feature hundreds or thousands of biosynthetic pathways to bioactive compounds, only a few of which have been accessed. In the case of animals such as marine invertebrates, by far the majority of animal biodiversity is too small in sample size to be consistent with traditional drug discovery methods, yet seemingly every species within certain groups contains new and interesting chemistry.7 Thus, there is exceptional promise and challenge in accessing the chemistry from uncultivated microbes.
A good example can be found in tunicates living symbiotically with cyanobacteria of the genus Prochloron. These are true symbiotic, nutrient-sharing partnerships that have eluded cultivation despite >40 years of study, with no close free-living relative yet discovered.8 Prochloron spp. are prolific producers of natural products, and thus Prochloron-harboring tunicates have long been collected and extracted to provide bioactive materials.9 Many of the tunicates containing Prochloron are small and rare, preventing traditional chemical methods of extraction of large quantities of material for structural and biological characterization.
Here, we have developed a new approach for accessing rare and interesting new chemicals from the symbiotic microbiome that address the limitation of size and supply, leading to the discovery of the divamides in HIV assays. Our approach allowed us to conduct structure-activity relationship studies that are typically performed late in the drug discovery process. Additionally, we isolated divamides with different amino acid sequences that display remarkably different biological activities. Thus, our results have implications in both drug and enzyme biotechnology and in understanding the function of chemical diversity.
Results
Potent anti-HIV activity in small tunicate extracts
In the Eastern Fields of Papua New Guinea on a single coral reef, we collected two samples of the small marine tunicate, Didemnum molle (Fig. 1a). D. molle is ubiquitous throughout the tropical Western Pacific, and yet very few compounds have been reported because of the challenges of collecting and extracting these small, biologically and chemically diverse, mucus-laden animals. We performed organic extraction, identifying one tunicate (E11-036) that represented a promising hit in an anti-HIV assay (Fig. 1b). By contrast, the extract of a neighboring colony (E11-037) was inactive. By mass spectrometry, both animals clearly contained a family of related but non-identical compounds, indicating that subtle changes in structure were likely responsible for the anti-HIV activity.
Figure 1. Discovery of anti-HIV lanthipeptides, the divamides.
(a) The Didemnum molle tunicates E11-036 (left) and E11-037 (right), found living side-by-side in the Eastern Fields of Papua New Guinea, likely represent genetically distinct species (Supplementary Fig. 2).49 (b) Bioassay-guided fractionation of tunicate extracts revealed significant anti-HIV activity (p-value < 0.05) from E11-036 fractions compared with inactive E11-037 fractions (*; p-value = 0.0253; two-tailed, unequal variance t-test) or vehicle controls (**; p-value = 0.00632). Cell survival is reported relative to uninfected cells (U; 100 %) and infected cells (I; 0 %). AZT (n=11, at either 0.1 or 0.05 mg/ml) was used as a positive control (A; solid red line, average; dashed lines, ± s.d.). Distribution means and upper quartiles are represented by thick and thin horizontal lines, respectively. Fractions from all purification steps were screened at doses from 5–50 µg/ml. Points represent the average of 2–3 assay responses per fraction per dose. Multiple rounds of E11-036 fractionation were included. A single outlier from the E11-036 data was excluded (351%). (c) The chemical structure of the anti-HIV lanthipeptide divamide A (1), produced by E11-036, elucidated by NMR and chiral GC/MS (Supplementary Note). (d) Compounds 1, 2, and 4 incorporate lanthionine (yellow), lysinoalanine (red), and β-hydroxyaspartic acid (blue) within a common scaffold, while 1 and 2 also include N-terminal N-trimethylation (green). (e) The E11-036 div biosynthetic gene cluster (divA; top) exhibits nearly 100% identity with the partial E11-037 div gene cluster (divB; bottom), with the greatest disparity occurring within the core of divA (*).
To elucidate the nature of this activity, we performed assay-guided fractionation on E11-036 extract to obtain divamide A (1, ~150 nanomoles) as the major organic-soluble compound in the animal, which was solely responsible for the observed activity. Related compounds purified from sample E11-037, including divamides B (2) and C (3), were shown to be essentially inactive. Using spectroscopic methods, 1 was determined to be a novel peptide containing the modified amino acids lanthionine (Lan) and N-trimethylglutamate, and the partial peptide sequence “GTTR”. However, because of the limited amount of available material, the structure could not be completed using these methods.
The metagenome reveals novel lanthipeptide structures
We turned to metagenome sequencing as a novel method for structural confirmation. Lanthionine-containing peptides generally belong to a broad family of ribosomally and posttranslationally modified peptides (RiPPs),10,11 for which the precursor to the final natural product is genetically encoded. Enzymes act on this ribosomally produced precursor peptide, leading to maturation of a bioactive natural product. Therefore, we expected to find genes encoding both a precursor peptide containing the sequence “GTTR” and also enzymes that would introduce lanthionine and N-trimethylglutamate modifications.
DNA was extracted from macerated whole animal tissues, sequenced, and assembled. The putative tetramer “GTTR” was used to probe the assembled metagenome, yielding hundreds of hits. We searched for protein sequences with Cys, Thr, and Ser overrepresented at the C-terminal end and incorporating additional amino acids predicted from our structural analysis, producing a single hit with a short coding sequence (divA) for a precursor peptide from a RiPP biosynthetic pathway (Supplementary Table 1).10 The 20 C-terminal residues of DivA (ARD09202) encompassed the amino acids of 1 predicted by NMR as well as the sequence “GTTK”, suggesting the presence of lysine instead of the predicted arginine. The divA cluster was clearly related to that for the known compound, cinnamycin (4), a lanthipeptide produced by the soil-actinobacterium Streptomyces cinnamoneous.12 Cinnamycin contains a lysinoalanine bridge arising from a Michael adduct between lysine and dehydroalanine derived from serine.13 This modification explains the initial misassignment of GTTK in 1, since the lysine residue exhibited unusual chemical shifts by NMR in comparison to unmodified lysine.
Immediately adjacent to divA in the genome was a gene encoding DivM (ARD09203), bearing homology to the type II lanthionine synthetases.14 In addition, a series of other genes was predictive of the divamide A structure. The div cluster is comprised of genes putatively encoding five biosynthetic proteins, an export pump, and two genes of unknown function (Supplementary Table 1). In addition to DivA and DivM, these include an α-ketoglutarate-dependent Fe(II) monooxygenase/β-Asp hydroxylase (DivX; ARD09204), a homolog of DurN (WP_071962205.1) shown to be involved in lysinoalanine formation15–17 (DivN; ARD09209), a SAM-dependent methyltransferase (DivMT; ARD09206), and an ABC-type transporter (DivT; ARD09207).
Using these data, we predicted that 1 would incorporate three methyllanthionines (MeLan), a lysinoalanine (Lal), a β-hydroxy aspartic acid (Hya), and N-terminal trimethylation, a rare posttranslational modification in nature.18–20 The placement of these modified residues on the parent structure was performed by comparing the putative sequence of 1 to that of 4. Upon reexamination of our NMR data, every correlation from 2D-NMR matched the predicted structure, while we did not find a single correlation that violated the proposed structure (Fig. 1c). Thus, metagenomic sequencing data, in combination with NMR, represents a novel approach to elucidate the structure of a natural product. Finally, with the planar structure in hand, stereochemical analysis was performed using chemical degradation. We derivatized the resulting amino acids and compared their retentions by gas chromatography to those for authentic standards (Supplementary Note), enabling us to conclusively determine every configurational center except the N-terminal glutamate, which was unusually labile due to the N-trimethyl modifications.
With the structure of 1 from sample E11-036 complete, we then examined more challenging structures from an even smaller sample, E11-037. Interestingly, in contrast to the anti-HIV 1, the extract of this sample was biologically inactive in similar assays. E11-037 contained many different divamide-like peptides, which could readily be identified because of a characteristic Hoffman elimination in the mass spectrometer due to N-terminal trimethylation.21 In particular, we isolated and purified 2 and 3, which differ from 1 only in their amino acid sequences (Fig. 1d).
Only the structure of the major E11-037 compound, divamide B (2), could be conclusively determined (Supplementary Note). The interpretation of the NMR data for divamide B was much more challenging than those for 1 for two reasons: first, an overall lower yield of product was recovered; second, conformational flexibility around a proline residue not present in 1 made the NMR data less robust (Supplementary Note). We sequenced the metagenome of E11-037, discovering a gene for a peptide with the predicted 2 sequence. Metagenomic analysis revealed a nearly identical biosynthetic gene cluster to that of 1, although only a partial sequence was recovered due to difficulties in recovering quality DNA associated with the mucopolysaccharide-rich nature of the sample (Fig. 1e and Supplementary Table 2). As with 1, all available NMR data matched the predicted structure for 2 (Fig. 1d).
The divamides are encoded by the symbiotic microbiota
The div cluster was found not in the genomes of the host animal, but in those of symbiotic cyanobacteria, Prochloron didemni.8 P. didemni often produces bioactive, presumably defensive chemicals in the interior of tunicates,22 but lanthipeptides were not previously known as products of Prochloron. In fact, lanthipeptides in general are only rarely described from marine animals, and to the best of our knowledge no cinnamycin-like structure has been found encoded in any marine metagenome that is available in our lab or elsewhere except in the discovery described here. The variability of production of bioactive natural products within Prochloron spp. is well known, and this talented producer continues to surprise with the production of unprecedented compounds.
Whole pathway expression in E. coli yields divamides
The lack of any apparent anti-HIV effects from the diverse peptides of E11-037, including 2 and 3, starkly highlights how even modest changes in structure govern pharmacology. We sought to determine the molecular basis for this disparity and to provide material to investigate the observed activity of 1. Given the scarcity of isolated material, we first aimed to produce divamides by recombinant expression. Such an in vivo total synthesis could also be used as a structural confirmation. Finally, it would represent the first example of a new strategy for drug discovery, marrying the best of traditional natural products chemistry with tools of metagenomics and synthetic biology.
Our expression strategy aimed to use these biosynthetic genes to reconstitute the whole div pathway in Escherichia coli. We constructed an expression vector, pDiv, that placed the whole div pathway, beginning with divA, under control of a single lac promoter. The pathway was constructed using synthetic DNA pieces (Supplementary Tables 3 and 4) such that the native arrangement of the div pathway was maintained, using native sequences for the intergenic regions and codon-optimized sequences only for the biosynthetic genes (Fig. 2a). The major difference from the native pathway organization in P. didemni was the rearrangement of divN. This gene and 120 bp upstream of its start codon, believed to contain its native promoter, were reoriented and appended to the pathway immediately following divMT. However, this initial vector yielded no detectable products. Through a series of trials, we found that replacement of codon-optimized divM with the native gene sequence in pDiv-2 (Fig. 2a), amplified from metagenomic DNA (Supplementary Figure 2), resulted in the production of a divamide intermediate with three methyllanthionine residues and dehydroalanine (Dha) but lacking lysinoalanine and methylation (5; Fig. 2b). Interestingly, this suggests that divN, the only gene in pDiv that differs from its native arrangement in the div pathway, is not functional in this context. In the expression system, we did not include a protease, since the native divamide protease was not part of the div cluster and was not apparent in the metagenome. Therefore, the cleavage products obtained were produced by an unidentified E. coli protease. Perhaps for this reason, we obtained not only the 20-mer product correctly cleaved before glutamate, but also a series of products that were incorrectly cleaved at the isoleucine and alanine residues (6–8). These products were purified by HPLC prior to further study.
Figure 2. The semi-in vivo synthesis of divamides A and B involves in vivo, chemical, and enzymatic steps.
(a) The expression vector pDiv-2 enabled in vivo production of intermediates of 1. The plasmid incorporates both E. coli codon-optimized (dark grey, biosynthetic; light grey, non-biosynthetic) and native codon sequence (black) divA pathway genes, and was constructed by assembling short synthetic DNAs (Supplementary Tables 3 and 4). A 120-bp region upstream of divN was placed immediately following divY and preceding divN in an attempt to include any native divN promoters (red). A single protease cleavage site within the precursor protein DivA is involved in native divamide production, while four cleavage sites have been recognized in E. coli divamide production. (b) MS spectra of the products of pDiv-2 (i) and pDiv-3 (ii) expression in E. coli, showing precursors of 1 and 2, respectively, with their extraproteolytic analogues, extended by up to three residues of the putative protease recognition site. (c) Synthetic scheme for production of 1. Expression of pDiv-2 in DH10β E. coli yields 5–8, incorporating three MeLan residues, Hya, and dehydroalanine (Dha) posttranslational modifications (i). Incubation of any of these products in base induces formation of Lal to yield 9–12 (ii). The desmethyl divamide A product 9 is the sole substrate of SAM-dependent N-trimethyltransferase DivMT (iii).
The challenge of producing the lysinoalanine residue was previously addressed in the synthesis of 4.17 In that event, a simple chemical method was developed wherein the lysinoalanine residue could be robustly produced by treatment with base. We followed this procedure with compounds 5–8, leading to efficient production of the lysinoalanine residue (Fig. 2c and Supplementary Fig. 2) with stereochemical fidelity (9–12, respectively; Supplementary Note). All that remained was to install the N-terminal trimethyl group by DivMT, which was expressed and purified from E. coli BL21(DE3) (Supplementary Fig. 2) and used in enzyme assays in vitro. Incubation of DivMT with S-adenosylmethionine and 9 efficiently yielded the natural product 1. By contrast, the use of precursors lacking lysinoalanine in the DivMT assay yielded no N-terminal trimethylation. This explains why we were unable to obtain divamide modified by methylation directly from the E. coli culture: in that event, lysinoalanine had not yet been formed, producing an intermediate that is not a substrate for methylation. In addition, we attempted methylation of 10–12, obtaining no product. Thus, it appears that the divamide methyltransferase exhibits relatively high specificity for the overall cinnamycin-like scaffold.
Because the enzymes for 2 appeared to be nearly identical to those of 1, we approached the synthesis of 2 by replacing just the coding sequence of the precursor peptide to generate plasmid pDiv-3. The genes for enzymes of 1 were left intact and unmodified. Expression of this construct in E. coli led to a 2 analog (13) along with several proteolytic fragments (14–16), each lacking methylation and the lysinoalanine residue. In short, we observed the same features as with expression of 1. Treatment of these compounds with base yielded 17–20, and methylation of 17 with DivMT led to the natural product. From our synthesis of 1 and 2, functional roles could be assigned to all biosynthetic genes in the pathway except divN.
Scale up of this initial process involved several improvements, the most notable of which was adding cysteine to the expression media,23 substantially increasing the in vivo production of divamides. Currently, the best method produces 13.7 µg of 5 per liter of E. coli culture broth, after purification to homogeneity (Supplementary Tables 5,6 and Supplementary Fig. 3). NMR analysis confirmed that the synthesized 1 was identical to the natural product, confirming the elucidated structure (Supplementary Note). Both 1 and 2 obtained by semi-in vivo synthesis exhibited identical properties to the natural products. This approach represents a novel strategy to discover and confirm the structures of natural products from the symbiotic microbiota.
SAR reveals potential for therapeutic optimization
With material in hand, we evaluated the biological activities and structure-activity relationships (SAR) of the divamides and their biosynthetic intermediates alongside cinnamycin. Initial experiments followed peptide effects on human CEM 1A2 tat/rev++ T-cells, which die upon infection with MC99IIIBΔTat-Rev HIV-1, enabling analysis of anti-HIV activity using an MTT-based viability assay (Fig. 3 and Supplementary Fig. 4).24 We found that recombinant and natural 1 displayed identical activities in this assay, reinforcing the accuracy of the structure determination. We also tested the analogs of 1 resulting from our recombinant methods in the same assay. These analogs included intermediates of 1 that were not methylated at the N-terminus, which consisted of i) the native peptide sequence (9); ii) a mixture consisting of divamide A derivatives with an extra alanine, alanine-alanine, or isoleucine-alanine-alanine residues on the N-terminus (10–12); iii) the native sequence lacking the lysinoalanine bridge (5). All derivatives containing lysinoalanine were active with similar potency. By contrast, 5, for which the lysinoalanine residue was not formed, was inactive in the cytoprotection assay, implying that the complete three-dimensional structure of divamide is necessary for activity.
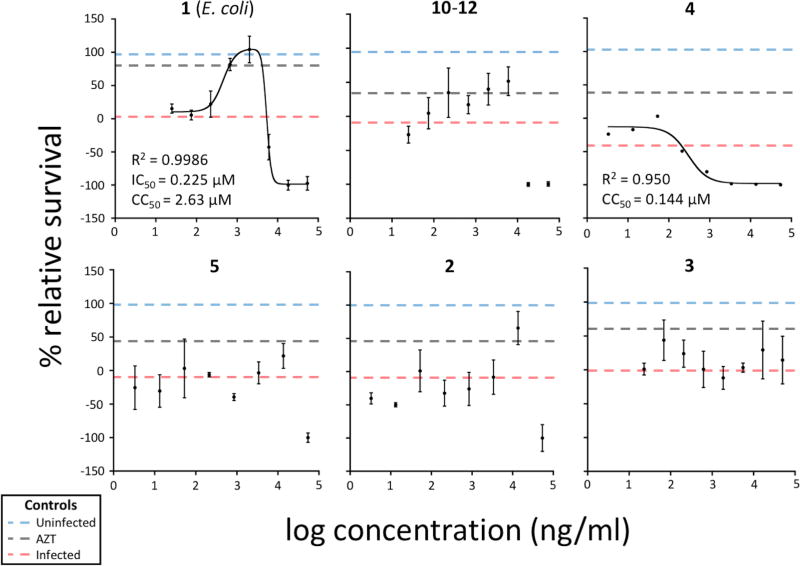
Figure 3. Cytoprotection assays reveal two biological activities of divamide A.
Cytoprotection assay dose-response curves for selected divamide species and 4. The dose-response curves of additional divamide analogues are presented in Supplementary Figure 4. Experimental controls include uninfected, infected, and AZT (at either 0.1 or 0.05 mg/ml) assay responses. Points represent an average of three assay responses per dose (error bars, s.d.). R2 values are included for compounds for which curves could be fit with equation 7 (see Online Methods) or a single four-parameter IC50 equation. Curves could not be fit for some HIV cytoprotective compounds, including the mixture of 10–12, due to abnormal curve shapes. HIV-inactive compounds are shown in the bottom row, two of which also display cytotoxicity (5 and 2), and for which the data did not fit any curve. 3 (bottom right) displayed neither cytotoxic nor anti-HIV activities.
Compounds 2 and 3, as well as 4, lacked any detectable cytoprotection in this assay. Compound 4 was potently cytotoxic to these T cells at ~ 200 nM. Compound 2 was also modestly cytotoxic in the low micromolar range. Compound 1 and derivatives were not notably cytotoxic below about 10 µM in the cytoprotection assay. These differences are noteworthy because all of the compounds have the same constrained shape, but differ only in the amino acid sequences. This implies that subtle changes in sequence allow the segregation of antiviral and cytotoxic properties.
In order to examine whether there is a direct relationship between the cytotoxicity and anti-HIV activities of the divamides, we adopted a flow cytometry-based approach to independently measure cell viability and viral infection of CEMTART T-cells, the parent strain of CEM 1A2 cells. Additionally, while the cytoprotection assay is amenable to high-throughput applications and parsing active from inactive samples, it is subject to quantitative inaccuracies caused by long post-treatment incubation times that introduce variation between replicate experiments, and we were interested in quantifying these properties. Cells were co-stained with a fluorescein-labeled antibody for the HIV-1 capsid protein p24 and a cell viability stain to track both anti-HIV and cytotoxic properties simultaneously. Under this assay, all divamide A analogues screened, as well as 4, displayed anti-HIV properties, with 4 being more potent than any divamide compound (Fig. 4a and Supplementary Fig. 4). However, the therapeutic index between HIV inhibition and cytotoxicity appeared wider for 1 and its derivatives than for 4 (Supplementary Table 8). The potency of both activities was greater in cytoprotection assays relative to flow-based experiments, likely reflecting a greater sensitivity of CEM 1A2 cells to HIV and/or drug than the parent strain. This may explain why anti-HIV activity was not previously observed for 4 in our cytoprotection assays: the effects of the two activities may have overlapped (Fig. 3). Additionally, the degree of cytotoxicity was similar between uninfected and HIV-infected cells, indicating that the mechanism of cytotoxicity is independent of infection (Fig. 4b). Overall, the results indicate that the prominent anti-HIV activity of 1 stems from its effective segregation of antiviral and cytotoxic properties.
Figure 4. Segregation of anti-HIV and cytotoxic properties by flow cytometry.
Cytotoxicity and anti-HIV activities of divamides and 4 were monitored simultaneously via flow cytometry using a fluorescein-labeled antibody to HIV-1 capsid protein p24 (green; % live infected cells) and viability stain (purple; % live cells) with HIV-infected CEMTART T-cells. The biological marker and corresponding fluorescence channel are shown in parentheses along the vertical axes. Points represent an average of three independent experiments per dose (Supplementary Table 7; error bars, s.d.). Average uninfected and infected cell control responses are shown as horizontal lines for each biological marker. (a) Initial experiments employed only small doses (dose range 1), yielding full anti-HIV dose-response curves but only partial cytotoxicity curves. Sufficient material for assays at this dose range was available for only two additional divamide compounds (Supplementary Figure 4). (b) Later experiments were performed using more concentrated doses (dose range 2) to acquire full cytotoxic dose-response curves. Sufficient material for assays at these higher doses was available only for the compounds shown. Experiments performed on infected cells showed reduced cell viability at low compound doses comparable to untreated controls, preventing accurate curve fitting. Thus, experiments were performed on uninfected cells to enable quantification of cytotoxicity.
Compound 4 and its close relatives duramycin, duramycin B, and duramycin C are known to bind phosphatidylethanolamine (PE) in biological membranes.13,25–27 This property has been implicated in several biological activities reported for 4, including hemolytic, proapoptotic, and antiviral actions.28–33 With recombinant derivatives in hand, we fluorescently labeled the peptides and studied their interactions with membranes. Fluorescence microscopy images show that both cinnamycin and divamide interact with T-cell membranes (Fig. 5). Conjugation of cinnamycin and duramycin in this way has been shown to reduce the potency of these compounds but not their ability to bind PE.34 Thus, divamides, 4, and duramycin likely affect stages of infection where the virus interacts with the cell membrane, such as entry or exit.
Figure 5. Cinnamycin and divamide both interact with membranes.
Fluorescence microscopy shows the association of conjugated cinnamycin and divamide peptides with membranes. HIV-infected CEMTART T-cells were treated with FITC-streptavidin alone (a) or with biotin-LC-LC-4 (b) or biotin-LC-LC-9 (c), and were then stained with propidium iodide (PI). Uninfected cells display similar results (not shown). Scale bars, 20 µm.
The div pathway produces a diversity of peptides
Compounds 1–3 do not provide a complete picture of the chemical diversity within the tunicates described. While only two peptides could be isolated from E11-037, a number of other divamide-like ions were observed by LC/MS to display Hoffman elimination of trimethylamine,21 a characteristic of divamides possessing N-terminal trimethylation (Supplementary Fig. 5). Still other chemical species were observed resembling divamide but lacking N-trimethylation, likely representing partially or non-methylated intermediates (Supplementary Table 9). This is distinct from E11-036, from which a single divamide was observed and whose metagenome contains a single div pathway and precursor gene. The limited number of related lanthipeptides identified by searching sequenced genomes in GenBank encoded no more than two precursor genes per cluster (Supplementary Fig. 5). Thus, the div pathway appears to generate a substantially greater number of products than even closely related lanthipeptide pathways.
Discussion
This work represents the first example of a direct discovery-supply strategy of a symbiont-derived natural product. Natural products are targeted as potential pharmaceuticals because of their biological relevance, having evolved to mediate biological interactions. Historically, compounds have either been extracted directly from marine animals or synthesized to afford FDA-approved drugs and other bioactive materials. Extraction has proved fruitful but requires large amounts of animal material in order to elucidate a single structure, while syntheses of structurally complex natural products are often impractical, involving expensive, multi-step processes. Our integrated metagenome-synthetic biology approach circumvents these limitations for the first time. More recent work has focused on pathway mining from genomes or metagenomes.35 Chemical structures have been predicted from genomics, prompting synthesis of hypothetical analogs, but without reference to any initial activity.36 While novel chemistry can be discovered in this way, these methods suffer from the limitation that, in general, the potential biological activity is only determined after the extensive labor of producing compounds derived from biosynthetic pathways.
By integrating assay-based and genomic approaches, we were able to directly express a whole pathway from a very limited resource and obtain the natural product, allowing us to confirm our prediction of the product structure. This represents a new and efficient alternative to analyze the major active compounds in the world’s diverse symbiotic associations. We emphasize that biological activity within an ecologically relevant system is put first, with the resulting chemical and metagenomic approaches following promising biology, instead of the other way around. While our method focuses on RiPPs, which are particularly well suited to this approach, it is increasingly possible to predict aspects of chemical structures from non-ribosomal peptide synthase and polyketide synthase gene sequences,37–39 such that our NMR-metagenomics approach may become more broadly applicable as predictive methods improve further.
Our synthetic biology approach allowed us to determine which modifications are essential to activity, leading to a platform for further mechanistic and potentially preclinical evaluation. The biological activities of the divamides and their relatives reveal several key structure-activity relationships. For analogs of a single peptide sequence, derivatization does not notably alter the phenotype elicited by the peptide (Figures 3, 4 and Supplementary Table 8). Additionally, the tertiary structure conferred by the cyclic lysinoalanine residue is necessary to block infection. In contrast, biological responses vary greatly between different peptide sequences. Five substitutions between 1 and 2 lead to a lack of detectable HIV inhibition in 2, yet both display cytotoxicity, while compound 3 does not appear to evoke either activity within the dose range tested (Fig. 3). Compound 4 bears an identical scaffold and is more potently cytotoxic and anti-HIV than any of the divamides (Fig. 4 and Supplementary Table 8).
In future work, we could use these medicinal chemistry principles to create derivatives that may further expand the therapeutic index of cinnamycin-like compounds. Previous work on the impact of 4 and duramycin on viral infection will also inform these studies. T-cell immunoglobulin and mucin domain (TIM) proteins can act as receptors for viral entry by binding to phosphatidylserine (PS) within the phospholipid membrane of enveloped viruses like HIV, Ebola, and Dengue,40–42 and are also involved in the release of HIV from infected cells using the same PS-binding mechanism.40 Interestingly, duramycin inhibits entry of Ebola and Dengue viruses, indicating that TIM proteins also interact with PE.42 Thus, lipid binding appears a likely mechanism for the anti-HIV activity of the divamides and 4. However, the stark differences in potency and toxicity between individual divamides and 4 may reflect additional interactions with non-membrane components, potentially including TIM proteins, that are substantially affected by the peptide sequence. Moreover, our data indicate that the therapeutic index can be greatly modified by simple changes in peptide sequence. Collectively, the observations made here suggest that while posttranslational modifications establish the bioactive scaffold, it is the variation between amino acid sequences that ultimately determine the biological activities of the divamides.
For many years, evolutionary biologists have sought to explain the chemical diversity inherent in natural products. One attractive attempt to explain chemical diversity in the context of Darwinian evolution is a model suggesting selective mechanisms that maintain plastic biosynthetic pathways.43 In order to corroborate this or other ideas with experimental data, an appropriate system should: i) use wild organisms in their native contexts; ii) investigate thoroughly the chemical constituents of those organisms in nature; iii) define the biosynthetic gene sequences underlying a natural chemical variation; and iv) link a biological functional change to the observed chemical variation. The presence of several different divamide peptide sequences in E11-037 extracts suggests that the divA gene may have undergone gene duplication and divergence. Given the dramatic differences in biological activity between these gene products (albeit in an assays lacking direct ecological relevance), it is tempting to conclude that the diversification of substrates observed within the div pathway imparts a selective advantage to the host–microbiota system.44 Such variability, where identical enzymes use different substrates in different ecological contexts, is reminiscent of “diversity-generating” biosynthetic pathways.45,46 In diversity-generating biosynthesis, exceptionally plastic biosynthetic pathways remain relatively constant, while the substrates themselves are hypervariable.47, 48 While our data has several limitations, this study meets all four criteria, lending experimental support to the idea that selection pressure can maintain relaxed substrate specificity in biosynthetic pathways.
Online Methods
General
The majority of NMR spectra were recorded on a Varian INOVA 600 NMR spectrometer (1H 600 MHz, 13C 150 MHz) equipped with a 5 mm Nalorac IDTG cryogenic probe or a Varian INOVA 500 NMR (1H 500 MHz, 13C 125 MHz) with 5 mm Nalorac IDTG probe, using residual solvent signals for reference (δH = 4.80 ppm for D2O and δH = 4.87 ppm, δC = 49.15 ppm for D2O/CD3OD mixtures). For the initial structural characterization of divamide A, a Bruker Advance III HD 900 MHZ instrument (1H 900 MHz, 13C 225 MHz) with 5 mm 1H(13C/15N) cryogenic probe and a Bruker Advance III 500 MHz (1H 500 MHz, 13C 125 MHz) with a 5 mm 13C/15N(1H) cryogenic probe were also used. Liquid chromatography–MS spectra were recorded using a Micromass Q-ToF Micro mass spectrometer (Waters). High-resolution mass spectra were recorded on a LTQ-FTMS (ThermoElectron) or a maXis-II ETD Q-ToF (Bruker). Analytical HPLC was performed on a Beckman System Gold 126 solvent module with a 168 PDA detector or a Hitachi Primaide 1110 Pump with 1430 Diode Array Detector. An HP 6890 Series GC System equipped with an HP 7683 Series Injector and an HP 5973 Mass Selective Detector was used for all GC/MS experiments. A Fisher Multiscan FC plate reader was employed for biological assays. Amino acid standards included standard L and D amino acids (Cyclo Chemical or Sigma-Aldrich), DL-threo-β-hydroxyaspartic acid (Sigma-Aldrich), L-erythro-β-hydroxyaspartic acid (Wako Chemicals), and DL-lysinoalanine (Bachem). Methyllanthionine GC/MS standards were generously provided by Dr. John C. Vederas (University of Alberta). Cinnamycin was purified from spent culture broth of Streptomyces cinnamoneus ATCC 11874 grown in ISP-2 (1L) at 30°C in Fernbach flasks (2.8L) shaking at 150 rpm for three days.
Animal collection
Tunicates identified as Didemnum molle (Herdman, 1886) were collected by scuba from the Eastern Fields of Papua New Guinea (S 10° 16.217’; E 145° 38.679’) at a depth of approximately 5–10 m. Chemistry samples were immediately frozen, while samples for metagenome sequencing were first processed by slicing into or placing small samples in RNAlater, then leaving them at 4°C overnight with occasional inversion of tube, then freezing. Vouchers are maintained at the University of Utah under accession numbers E11-036 and E11-037, or PNG11-9-099 and PNG11-9-100, respectively.
Extraction and isolation
Frozen tunicate (200 g) was extracted three times in MeOH. Pooled extract was filtered by gravity and fractionated by Diaion HP20SS resin using isopropanol (IPA)/H2O mixtures of 25% increments of increasing IPA to generate five fractions with a cumulative weight (excluding 100% H2O fractions) of 581.8 mg for E11-036 and 685.2 mg for E11-037. E11-036 F1 fractions (eluted in 25% IPA; 211.3 mg) were subjected to C18 flash chromatography starting at 10% MeOH and increasing stepwise by 30% up to 100% MeOH, yielding eight fractions. Fraction E (70% MeOH; 19.1 mg) was further fractionated by HPLC. Using a Luna 5µ C18 column (100 Å, 250 × 4.60 mm) at 1 ml/min, the solvent 30% ACN/70% 0.2 M NaCl was held for 15 min then increased to 100% ACN over 5 min. Two peaks were collected and desalted using Waters SEPPAK C18 cartridges, resulting in 1 (tR = 8.87 min, 315.1 µg,) and oxidized 1 (tR= 8.09 min, not quantified). A degradation product, later identified as a methylated divamide A derivative (21), accumulated in the 1 sample over time. The resulting mixture was fractionated by C18 using a method similar to that described later for lysinoalanine formation, yielding 77.8 µg of pure 1 and 237.3 µg of a mixture of the two peptides.
C18 flash chromatography E11-037 F1 fractions (25% IPA; 172.9 mg) followed a similar procedure to generate 12 fractions. Fraction F (40% MeOH, 10.1 mg) was subjected to HPLC using the same Luna C18 column at 31% ACN/ 69% 0.2 M NaCl held for 11 min then increased to 60% ACN over 4 min and held at 60% for 8 min with a 1ml/min flow rate, yielding 2 (tR = 8.78 min). All elutions excluding the pure peptide were recombined (7.6 mg) and further purified by HPLC on a Luna 5µ Phenyl-Hexyl column (100 Å, 250×4.60 mm) at 1 ml/min starting at 20% ACN/80% 0.1% TFA for 4 min, then increasing to 24% ACN over 4 min and holding for 6 min, increasing to 30% ACN over 6 min and holding for 3 min, then finally to 60% ACN over 15 min. This yielded additional 2 (tR = 11.45 min, added to 2 described above for a combined weight of 100 µg) as well as 3 (tR = 20.12 min, 31.7 µg). Reported yields were determined by NMR and LC/MS quantification as described below.
Quantitative NMR
Quantitative NMR experiments followed previous protocols using external standards.50 Samples were suspended in deuterated solvents and dried multiple times to ensure full deuterium exchange. A 1:2 dilution series of an L-Trp stock solution was prepared in D2O encompassing a range of concentrations from 2.7 to 0.093 µg/ml to generate a standard curve. These concentrations were obtained by measuring UV absorbance using a Nanodrop 2000 (ε280, Trp = 5170.9 nm−1, ε276, Tyr = 1322.2 nm−1). Similarly, an L-Tyr standard was also prepared to assess the accuracy of the curve. L-Trp and L-Tyr samples were also fully exchanged in D2O prior to data acquisition. All samples were then suspended in D2O (120 µl) and transferred to new, dry Norell S-3-500-7 NMR tubes. The relaxation time t1 was determined for all proton signals intended for quantification and d1 was set to 10 times t1 of the highest observed d1. This value, as well as all other parameters, was maintained for all subsequent experiments. A 1D 1H spectrum was recorded for each sample and care was given to optimize shims between each experiment. Signals of interest were chosen based on isolation from other peaks and known proton assignment. Spectra were phased, baseline corrected, and integrated, and the absolute integral values were used to produce a standard concentration curve (Supplementary Figure 3). Concentrations of samples were calculated using the resulting linear equation. To reduce any signal-specific effects, three standard curves were generated from three separate L-Trp proton signals and the concentration of analyte was calculated from each of them. Similarly, at least two proton signals were chosen from each analyte spectrum and the concentrations determined from each L-Trp standard curve were averaged. Supplementary Table 5 summarizes the results of quantitative NMR. The concentration of L-Tyr was estimated at 0.351 ± 0.0095 mg/ml while the UV-determined concentration was 0.326 mg/ml. A 7.7% difference was calculated from the two L-Tyr concentrations, suggesting that analyte concentrations may be slightly overestimated.
Quantitative LC/MS
A 1:2 dilution series of 9, suspended at the same initial concentration as was determined by NMR (1.935 mM) and diluted 100-fold, was prepared to generate a standard curve by LC/MS (Supplementary Figure 3). Each sample (100 µl) was spiked with an internal cyanobactin standard (10 µl; m/z 883.5) to assess variability in injection volume between each run. Each concentration of 9 and each analyte were run in triplicate and in random order. The area under the curve (AUC) was determined for doubly charged ions in selected ion mode (SIM) for all peaks of interest within a sample. Concentrations were calculated from the average of the three AUC values. The internal standard AUC fluctuated between runs of like-samples within two standard deviations (SD = 35.7) of the average AUC (AUC = 132.3), or ± ~50%, indicating a moderate degree of run-to-run variability. This was deemed acceptable variation given the difficulty in obtaining an accurate mass of small amounts of material by weighing or by NMR. The internal standard AUC showed up to a 3-fold difference between runs of different samples, depending on the co-injected analyte. Biosynthetic 1, also quantified by NMR (0.664 mM), was used to assess the agreeability between NMR and LC/MS quantification curves. A concentration of 0.624 ± 0.0363 mM for E. coli-derived 1 was calculated – a difference of 5.92% less than the NMR concentration. A second curve was generated as described above for the quantification of a second stock solution of 4 (Supplementary Figure 3). This solution was prepared because the first solution was consumed in trial biological assays aimed at optimizing conditions prior to using divamide samples. The results of LC/MS quantification are summarized in Supplementary Table 6.
Chiral GC/MS
Amino acid standards included Gly and L and D configurations of all canonical amino acids as well as L- and D-allo-Ile and Thr. Noncanonical amino acid standards included: L-erythro-Hya (2S,3S), L-threo-Hya (2S,3R), D-threo-Hya (2R,3S), LL Lan (2R,6R), meso-DL Lan (2S,6R), LL MeLan (2R,3R,6R), DL MeLan (2S,3S,6R), L-allo-MeLan (2R,3S,6R), D-allo-MeLan (2S,3R,6R), LL Lal (2S, 9S), and DL Lal (2R, 9S). Peptides (less than 0.1 mg) and standards were hydrolyzed in 6 M HCl or 1 M NaOH at 100°C overnight then dried completely under a stream of nitrogen gas followed by high vacuum. Peptide hydrolysate and amino acid standards were then esterified with MeOH (200 µl) and acetyl chloride (50 µl) in a sealed reaction vial at 100°C for 1 hour, dried to completion, and finally treated with 100 µl each of DCM and PFP-anhydride at 100°C for 15 min. For the CP-Chirasil L-Valine column (25 m × 0.25 mm, 0.12 µm), an initial temperature of 80°C was held for 3 min before increasing linearly to 200°C over 30 min, then holding at 200°C for 12 min. For the Zebron ZB-5MSi Guardian column (30 m × 0.25 mm ID, 0.25 µm), a starting temperature of 60°C was held for 1.5 min before increasing linearly to 250°C over 31.7 min, then holding at 330°C for 13 min. All samples were prepared at 0.1 mg/ml in ethyl acetate and 5 µl injected per run using a split ratio of 2 and helium mobile phase gas.
DNA extraction
Extraction of DNA from D. molle samples E11-036 and E11-037 was complicated by the presence of copious amounts of polysaccharide-rich mucus. We utilized a protocol previously adapted from Sokolov for the precipitation of polysaccharides found in marine invertebrate slime.51,52 Briefly, tissue was sliced thinly and squashed between two aluminum foil sheets, then treated with proteinase K (Qiagen). Polysaccharides and proteins were then precipitated with saturated KCl. Additional KCl was required beyond what was called for in the original protocol. Samples were centrifuged and an equal volume of isopropanol added to the supernatant. White precipitate formed upon incubation at room temperature. The mixture was centrifuged again and the pellet washed with 70% ethanol. Excess solvent was removed by air drying and the pellet resuspended in QBT buffer (Qiagen). From this point forward, DNA extraction followed the standard protocol for the Qiagen Genomic-tip kit. An Illumina library was prepared using the E11-036 DNA with ~700 bp insert sizes and then sequenced on an Illumina MiSeq sequencer using a 250 bp paired end run. For D. molle sample E11-037, a 350 bp insert size library was prepared and sequenced (100 bp paired end) on an Illumina HiSeq 2500.
Assembly of the div cluster
To elucidate the full lanthipeptide cluster, E11-036 raw Illumina reads were filtered for quality and length (phred score >30 for greater than 40 bp) using the windowed adaptive trimming tool Sickle (Joshi, N. A. and Fass, J. N., Version 1.33 (2011)). The filtered reads were then assembled with the Velvet assembler v.1.2.09.53 The assembly (kmer size 61) was analyzed and produced a contig containing the precursor peptide gene and genes homologous to those of the gene cluster for 4 of S. cinnamoneus (Supplementary Table 1). In addition, analysis of this contig showed unique coding sequences, such as a putative methyltransferase. Based on the minimum genes required for in vitro 4 and duramycin biosynthesis,15,17 a second contig with a gene sharing 44% identity to durN, a protein shown to be involved in lysinoalanine formation, was included in the cluster for 1. Extensions of the contigs using matching reads were then optimized to produce a single contig 17 kbp in length containing the entire cluster (12 kbp from precursor gene to the durN homolog, divN). This sequence was deposited at GenBank under the accession number KY115608. E11-037 raw sequencing reads were assembled similarly. Two contigs were identified containing genes homologous to those observed in the div gene cluster. One contig 1152 bp in length contained the full divA gene and a partial divM sequence, consisting of 111 bp of the 3’ end. The second contig, 1655 bp in length, contained additional partial genes, consisting of 604 bp of the 5’ end of divX and 603 bp of the 3’ end of divMT, while also containing a complete but truncated version of divY, resembling only the 5’ end of E11-036 divY. By comparison with the gene cluster for 1, we estimate that a 3144 bp gap exists between these two contigs that would account for the remainder of the divM and divX genes. The E11-037 partial div cluster was deposited at GenBank under the accession number KY115609.
Construction of pDiv and pRSFDuet-DivMT
Four DNA pieces containing portions of the div pathway with putative biosynthetic genes codon-optimized were synthesized by Integrated DNA Technologies (IDT) and pre-assembled into two larger pieces by overlapping PCR (Supplementary Table 3). Pieces 1a and 2 were joined via an initial reaction with 15 cycles of denaturing (98°C, 15 s), annealing (62°C, 30 s), and elongation (72°C, 1 min) using Phusion® DNA polymerase (New England BioLabs). This was followed by another 30 cycles of denaturing (98°C, 15 s), annealing (62°C, 15 s), and elongation (72°C, 2 min) with primers div1a-fwd and div2-rvs, using the first reaction directly as the template (Supplementary Table 4). Similarly, DNA pieces 3 and 1b were joined using primers div3-fwd and div1b-rev. The final products were gel purified using Ultrafree-DA Centrifugal Filters (EMD Millipore). The vector backbone was obtained by digestion of pPat with SacII and KpnI and gel purification of the resulting 4450 bp fragment.23
The backbone was transformed into the uracil auxotroph Saccharomyces cerevisiae BY4741 along with the PCR-joined 1a-2 and 3-1b DNA pieces for assembly by yeast recombination. The resulting colonies, grown on uracil-deficient SD agar, were combined and grown overnight in liquid uracil-deficient SD. Plasmid DNA was purified from the overnight culture using a QIAprep Spin Miniprep kit (Qiagen) with protocol modifications to enhance cell lysis: the cells were resuspended in buffer P1 with 0.5 mm glass beads and mixed by vortex on high for 10 min. Yeast plasmid was transformed into DH10β E. coli and the resulting colonies screened for the presence of div genes by sequencing. Additional plasmid was generated from one of the div-positive clones and fully sequenced using primers 5 through 15, confirming the desired sequence of pDiv.
Expression of pDiv in E. coli did not yield any 1-like products, so modifications were made to the vector to induce production. We had previously observed expression of soluble, recombinant DivM only with the native gene sequence (data not shown), and reasoned that the codon-optimization of divM might have resulted in poor expression or incorrect protein folding, rendering the initial biosynthetic step catalyzed by DivM nonfunctional in pDiv.54 The native sequence divM gene was cloned by PCR from E11-036 metagenomic DNA using primers intergenic-divM-native-fwd and intergenic-divM-native-rvs with Platinum® Taq DNA Polymerase High Fidelity (ThermoFisher Scientific) and 35 cycles of denaturation (94°C, 30 s), annealing (72°C, 30 s), and elongation (68°C, 3 min 30 s) to yield a faint band (Supplementary Figure 2). The product was diluted 100-fold and amplified by PCR with Phusion® with the same primers by 30 cycles of denaturation (98°C, 15 s), annealing (65/72°C gradient, 30 s), and elongation (72°C, 3 min 30 s) and purified using a QIAquick PCR Purification kit (Qiagen) (Supplementary Figure 2). pDiv was digested with MluI-HF and AflII to remove the majority of the codon-optimized divM gene and the resulting 8355 bp band was gel purified using a Qiaex II Gel Extraction kit (Qiagen). The native divM gene and linearized pDiv were assembled by yeast homologous recombination as previously described. Yeast lysis was further enhanced by incubation with buffer P2 at 55°C for 10 min after pulverization with glass beads by vortex. Plasmids were extracted from individual E. coli colonies transformed with yeast DNA and digested with SpeI and NdeI to confirm replacement of the codon-optimized divM with the native gene, then sequenced with primers 17–22 to verify the desired sequence of divM in pDiv-2.
The pDiv-3 expression vector for 2 was constructed in two separate phases, with replacement of divA in pDiv with a divA sequence encoding 2, followed by replacement of codon-optimized divM with the native divM gene. First, pDiv was digested with ClaI and NheI restriction enzymes. The product was then gel purified using a centrifugal filter as described previously. The linearized vector was recombined with synthetic DNA divA-divamideB (IDT; Supplementary Table 3) using yeast as described above. Plasmids obtained from pDiv-containing E. coli colonies transformed with extracted yeast DNA were sequenced with primer 1, confirming the altered sequence of divA with a single noncoding C to G mutation, the position of which is underlined in the sequence shown in Supplementary Table 3. The native divM gene was then introduced as described for the construction of pDiv-2, using the divamide B-encoding pDiv as a template for recombination. During the restriction digest of pDiv-2, XhoI was used in addition to AflII and MluI-HF to cut the codon-optimized divM gene and prevent reinsertion.
To construct the methyltransferase expression plasmid pRSFDuet-DivMT, the codon-optimized divMT gene was PCR amplified using pDiv as a template with primers divMT-MCS1-fwd and divMT-MCS1-rvs, Platinum® Taq Hi-Fi, and 2% DMSO by 30 cycles of denaturation (94°C, 30 s), annealing (45.5°C, 30 s), and elongation (68°C, 2 min). The PCR product was purified as described above. Both purified PCR product and the empty pRSFDuet vector were then digested with SacI and NotI restriction enzymes. Reaction mixtures were heat inactivated and the linearized pRSFDuet was dephosphorylated using Antarctic Phosphatase (New England BioLabs). The two pieces were then ligated together using T4 DNA ligase (New England BioLabs). The ligation reaction was transformed into DH10β E. coli and the transformed cells plated on LB kanamycin selection media. Individual colonies were picked and grown overnight in liquid culture. Cells were harvested and plasmids purified using the QIAprep Spin Miniprep kit. Plasmids were triple digested with ScaI, MluI, and PstI to verify the expected band sizes, and positive plasmids were sequenced for confirmation.
Expression and purification of the pDiv-2 E. coli product
pDiv-2 was transformed into DH10β E. coli and six colonies used to inoculate individual wells of a 24-well plate, each containing LB (6 ml) with ampicillin (50 µg/ml), which was shaken overnight at 150 rpm and 30°C. Wells were combined and 3 ml used to seed each of eight 2×YT cultures (1L), each containing ampicillin (50 µg/ml) and cysteine hydrochloride monohydrate (5 mM) from a freshly prepared, filter-sterilized stock solution (1.66 M), in Fernbach flasks (2.8L). These were grown at 30°C for 48 hours shaking at 150 rpm before harvesting cells by centrifugation at 4000 rpm and 4°C for 30 min and decanting the supernatant. Cell pellets were frozen and lyophilized before extracting with DCM and sonicating for 30 min. The DCM extract was removed by centrifugation at 4000 rpm, 4°C for 30 min. Residual DCM was removed by drying the pellets under N2 gas. A second extraction with 1:1 acetone:EtOAc followed the same procedure of sonication, centrifugation, and drying. The final extraction with 1:1 ACN:H2O was performed similarly and in duplicate, with both extractions combined. The organic solvent was removed by rotary evaporator and remaining aqueous extract frozen and lyophilized. Typically, about 1.2 g of dry extract was obtained per 24 L of culture. Extractions of more than 24 L of combined cultures resulted in poor compound recovery due to impaired chromatographic resolution.
Extract was either resuspended in water after weighing or dried directly onto 230–400 mesh grade 60 silica gel and added atop a silica flash column containing a 10-fold excess of silica gel relative to the estimated dry extract weight (i.e. 12 g of silica per extract from 24 L of culture), pre-equilibrated with CHCl3. The column was then eluted with three column volumes each of CHCl3, EtOAc, 1:1 EtOAc:MeOH, MeOH, and 1:5 H2O:MeOH. For fractions containing MeOH and H2O, a piece of filter paper was folded and held tightly in place at the mouth of the column to filter out any silica particulate co-eluting with the more polar solvents. The MeOH and 1:5 H2O:MeOH elutions were pooled and concentrated to dryness by rotary evaporator and/or lyophilization. Dried silica column elutions were resuspended in 10% MeOH in H2O and loaded onto a second column of 4 g Bakerbond 40 µm C18 resin per 24 L pooled culture, pre-equilibrated with MeOH, 50% MeOH:H2O, and 10% MeOH:H2O. The C18 column was then eluted with five column volumes each of 10%, 30%, 50%, and 100% MeOH.
The latter two C18 flash column elutions were pooled and dried, then resuspended in no more than 5 ml total volume. HPLC was performed using a Luna 5µ Phenyl-Hexyl 250 × 10 mm column (Phenomenex) running a 30% ACN/ 70% 0.1% TFA in H2O isochratic method at 4 ml/min, with peak A eluting at ~14.3 min and peak B eluting at ~16.1 min. Retention times tended to increase with repeated injections, and peak shapes broadened with injections of higher volume. Organic solvent was removed from HPLC fractions by rotary evaporator and the remaining acidic water removed by lyophilization. Typical yields by weight for 16 L of culture were ~0.6 mg of peak A and ~1.5 mg of peak B with some variability from batch to batch. Both peaks consisted of 5, with peak A representing the 20-mer peptide sequence and peak B consisting of a mixture of three different extra-proteolytic species (6–8). Currently, the best method produces 13.7 µg of 5 (peak A) per L of E. coli culture broth, after purification to homogeneity and quantification by NMR or LC/MS.
Expression and purification of the pDiv-3 E. coli product
Purification of intermediates of 2 from E. coli expressing pDiv-3, including 13 and 14–16, followed the same extraction protocol as described for pDiv-2 E. coli with some modifications. The 1:5 H2O:MeOH silica column fraction alone was subjected to further fractionation on C18. The 30% MeOH elution from the C18 flash column contained 13–16, owing to the increased polarity of divamide B derivatives relative to divamide A. This fraction was used as a substrate in subsequent lysinoalanine reactions to generate compounds 17–20, and 17 was methylated with DivMT to produce 2.
Chemical formation of lysinoalanine
HPLC-purified divamide intermediates were suspended in 0.1 M Tris buffer pH 10.8 and incubated at 37°C overnight. To remove Tris buffer and salt, the suspension was dried, resuspended in 10% MeOH in H2O, and loaded onto a small plug of Fluka C18 resin (90 Å pore size) in a Pasteur pipette column pre-equilibrated as described above for the large-scale C18 flash column. The plug was eluted with 10% MeOH (2–5 ml), 50% MeOH (1.2 ml), and 100% (1.2 ml) MeOH. The 50% and 100% MeOH elutions were pooled.
Lysinoalanine formation was confirmed by desulfurization of MeLan residues using in situ Ni2B, prepared from NiCl2 and NaBH4 as described for other lanthipeptides (Supplementary Figure 2).55,56 In our hands, the conditions that worked best were as follows: peptide (150 µg, relative to amount estimated by weight) was suspended in 7:1 MeOH:H2O (500 µl) with NiCl2 (1–2 mg) pre-dissolved in a 1.5 ml screw-cap reaction vial with stir bar. About a five-fold excess of dry NaBH4 was added directly to the vial, which was immediately capped and place in an oil bath at 50°C for 30 min. The liquid, containing black Ni2B precipitate, was transferred to a 1.7 ml microcentrifuge tube and centrifuged to remove the majority of precipitate. The supernatant was dried in vacuo and desalted on a C18 Pasteur pipette column as described above, washing with 10% MeOH in H2O and eluting in 50% and 100% MeOH. The latter two elutions were combined and suspended in 75% MeOH and subjected to LC/MS and LC/MS/MS analyses.
Expression and purification of DivMT
The expression plasmid pRSFDuet-DivMT was transformed into BL21(DE3) E. coli, and individual colonies were picked to inoculate eight wells of a 24-well plate, each containing LB (6 ml) with kanamycin (50 µg/ml), which was shaken at 150 rpm and 30°C overnight. The wells were pooled and used to inoculate cultures of 2×YT (1 L) media with kanamycin (50 µg/ml) in Fernbach flasks (2.8 L), with 5 ml seed culture per flask. Cultures were shaken at 150 rpm and 30°C until an OD600 of 0.7 was reached, at which time cells were induced with IPTG (1 mM). The temperature was then reduced to 18°C and the cultures allowed to incubate overnight. Cells were harvested by centrifugation at 4000 rpm and 4°C and cell pellets of 2 L combined culture were frozen. Pellets were thawed on ice and suspended in lysis buffer (35 ml; 0.5 M NaCl, 25 mM imidazole, 10% glycerol) with lysozyme (0.4 mg/ml), PMSF (1 mM), MgCl2 (10 mM) and DNaseI (<1 mg) freshly added, using a glass stir rod. The mixture was sonicated on ice for three cycles of 2 min pulses at 40% amplitude (4 s pulse, 10 s delay), then centrifuged at 13 krpm and 4°C for 30 min. The supernatant was filtered using a 0.45 µm syringe filter onto Ni-NTA resin (2 ml; Qiagen) pre-equilibrated with lysis buffer and allowed to rock at 4°C for 30 min to equilibrate. The column was rinsed with lysis buffer (100 ml) followed by transition buffer (50 ml; 0.1 M NaCl, 25 mM MOPS, 25 mM imidazole, 10% glycerol, pH 8.0) and equilibrated another 10 min, then eluted with 10 ml each of the following imidazole concentrations, all prepared from transition buffer: 50 mM, 100 mM, 200 mM, and 500 mM imidazole. The resin was given a 10 min equilibration between each buffer change. SDS-PAGE was used to assess the elution time and purity of the 32.6 kDa protein (Figure 3c). The 200 mM elution was then dialyzed in dialysis buffer (1 L; 0.1 M NaCl, 25 mM MOPS, 10% glycerol, pH 8.0) at 4°C. The buffer was exchanged four times within 24 hours, adding BME at a final concentration of 5 mM to the first two buffer exchanges only. The concentration of the protein prep as determined by UV absorbance at 280 nm using a Nanodrop 2000 was 3.77 µM/0.123 mg/ml (MW = 32566.8 Da, ε280 (reduced) = 46215).
Enzymatic N-trimethylation
Compound 9 was suspended to an estimated concentration of 2 mg/ml in nanopure water. An analytical scale methylation assay was prepared by combining the solution of 9 (1 µl) with 1 µl of each of the following in 20 µl total volume: 0.5 M MOPS (pH 7.5), 50 mM DTT, 100 mM SAM, and 3.77 µM DivMT. Reactions were mixed and incubated at 37°C for 1–2 days using a thermocycler with a heated lid. MeOH was added to 20% of total volume and the entire sample subjected to LC/MS analysis. For the methylation of 17, a base-treated C18 fraction containing 17 from the extract of E. coli expressing pDiv-3 was used as a substrate. The methylation product was shown by HRESIMS to be identical to 2, with m/z 965.9194 [M+H]2+ by HRESIMS (calculated for C79H129N21O29S32+, 965.921325; Δ −1.99 ppm). For large-scale DivMT reactions, the entire desmethyl divamide solution was used (typically around 300 µl at 2 mg/ml was obtained per 24 L batch) and the remaining reagents scaled accordingly. The reaction solution was mixed and aliquoted into PCR tube strips, 80 µl per tube, and incubated at 37°C in a thermocycler for 2 days. All reactions were then pooled and dried in vacuo, then desalted using a C18 Pasteur pipette column, eluting with 10% MeOH (5–10 ml) followed by of 30%, 50%, and 100% MeOH (1.2 ml each). The latter two elutions were pooled and subjected to HPLC using a Gemini C6-Phenyl column (150 × 4.60 mm, 110Å, 3 µ) running an isochratic method of 27% ACN/ 68% 0.1 M NaCl/ 0.1% TFA aqueous buffer, yielding 1 at tR = 18.7 min. Residual extra-proteolytic divamide intermediates eluted earlier, at tR = 16.7 min. Organic solvent was removed from the collected peak prior to lyophilization. Dried material was suspended in 10% MeOH and de-salted using a C18 Pasteur pipette column as described before. As determined using quantitative NMR, 157 µg of pure 1 was obtained from 90 L of culture. This represents a 12.7% yield from the starting deslysinoalanine divamide A E. coli product.
Cytoprotection bioassays
T-cell lymphoid CEM 1A2 tat/rev++ cells are a clone of CEMTART tat/rev++ cells that die upon infection with HIV.57,58 These cells were employed for all HIV cytoprotection assays with divamide compounds. Cell lines were maintained in RPMI-1640 media supplemented with sodium bicarbonate (2 g/L), fetal bovine serum (20%), and antibiotic-antimycotic solution (100 X, GIBCO), and were cultivated in 5% CO2 at 37°C. Assays were run in 96-well round-bottom plates. Cells (200 µl) were added to wells at a concentration of 12,500 cells/well and sample compounds/controls applied in 1 µl DMSO. HIV-1 MC99IIIBΔTat-Rev was then applied at an appropriate titer.57 After a four-day incubation, yellow MTT (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide; 10 µl, 5 mg/ml) was added and the plates incubated for 1–2 hours. Plates were then centrifuged, the supernatant removed, and DMSO (100 µl) added to solubilize the MTT, now reduced and blue in color due to the metabolic activity of live cells. The plate was analyzed by plate reader at 590 nm and the results tabulated. Each dose of each compound was tested in triplicate, and each plate included the following controls, also in triplicate: untreated infected cells, uninfected cells, media only, and infected cells treated with azidothymidine (AZT), a positive control, at two different concentrations of 0.1 and 0.05 mg/ml in DMSO. The dose with the higher response is represented in Figure 3.
To best compare sample responses between multiple experiments, data was first normalized to the uninfected control, adjusted by the lowest normalized response, and then scaled for ease of comparison. In this way, data could be interpreted as the percent cell survival relative to cytotoxic concentrations. To obtain the normalized sample response, SN, equation 1 was used:
| (1) |
Variables represent average assay response values from three similarly treated wells of sample (S), blank (B), and uninfected control (U). Infected cells with and without AZT treatment were also normalized to obtain AN and IN, respectively. The highest AZT response of the two concentrations employed was used. Normalized data was then adjusted by a value, a, and scaled by a scaling factor, s, such that the uninfected control response (UN) was equal to 100 while the lowest cytotoxic response (DN), representative of the cytotoxic state, was set equal to −100:
| (2) |
| (3) |
| (4) |
Under these conditions, the scaled uninfected control response US and lowest cytotoxic response IS were equal to 100 % and −100% relative survival, respectively.
For samples lacking cytotoxic responses within the concentration range employed (Figure 3, curve 3, Supplementary Figure 4), data were scaled to fit a range of 0–100, with the 0 and 100 % thresholds representing the infected and uninfected states, respectively. The scaling factor, s, was calculated using equation 5, while sample data was scaled using equation 6:
| (5) |
| (6) |
Under these conditions, the scaled uninfected control response US and infected control response IS were equal to 100 % and 0% relative survival, respectively.
Dose-response curves were generated and fit using KaleidaGraph:
| (7) |
For a typical sigmoidal curve, A and D represent y-min and -max values, respectively, while x = log concentration, C is the IC50, and B the Hill coefficient. Because equation 7 combines two unrelated dose-dependent effects, A and D values do not accurately reflect y-min and -max values. B and C, however, are independent of y, the percent relative survival. C1 and C2 provide the IC50 and CC50 values, while B1 and B2 represent the steepness of their respective curve. Some sample curves showed a dip in the cytoprotection curve and could not be fit using the above model (Supplementary Figure 4). For these compounds, IC50 and CC50 values were estimated by eye (Supplementary Table 8).
Flow cytometry-based bioassays
CEMTART tat/rev++ cells were maintained as described for cytoprotection assays. Drugs suspended in DMSO were added to media (100 µl) in 96-well flat-bottom plates using an HP D300 Digital Dispenser. All wells were normalized with DMSO to 0.5% total volume. Divamide stock concentrations of either 5 or 2 µg/µl were utilized for assays employing a concentration range suitable for observing anti-HIV activity, while assays aimed at cytotoxicity used 20 µg/µl stock concentrations. AZT (5 µg/µl) was used as an anti-HIV control, while doxorubicin and etoposide (1 µg/µl) were used as cytotoxic controls. Cells were added (100 µl) and immediately infected with HIV-1 MC99IIIBΔTat-Rev, then incubated in 5% CO2 at 37°C for 65 hours.
To stain and fix cells, cells (200 µl) were transferred to polystyrene culture tubes. Phosphate buffered saline (PBS, 1 ml) was added, the tubes centrifuged (5 min, 2500 rpm), and supernatant decanted to remove media. Pellets were suspended in live/dead stain (100 µl; 0.25 µg/ml Fixable Viability Stain 450 (BD) in PBS) and mixed briefly by vortex, then incubated at 4°C for 30 min before diluting with PBS (1 ml) and centrifuging again. The cells were then suspended in fixing solution (200 µl; 1% paraformaldehyde, 0.1% Triton X-100 in PBS) and mixed briefly by vortex, then incubated at 4°C for 1 hr. Fixed cells were diluted with PBS (1 ml), centrifuged, and the supernatant removed. This was followed by a wash with PBS (1 ml) and second centrifugation. Cell pellets were suspended in HIV-1 p24 stain (100 µl; 0.5% Tween-20, 1 µg/ml BSA, and 1 µl/ml KC57-FITC (Beckman Coulter) in sterile PBS), mixed briefly by vortex, and incubated for 1 hr at 4°C. Stained cells were then diluted with flow cytometry buffer (1 ml; 0.05% Tween-20 and 1 µg/ml BSA in sterile PBS), centrifuged, and the supernatant removed. Pellets were transferred to a round-bottom 96-well plate in flow cytometry buffer (200 µl), mixing by both vortex and pipette suspension. Cells were subjected to flow cytometry using a BD FACSCanto RUO Special Order System (150 µl injection/well) with BD FACSDiva v.6.1.3 software. Details of each individual experiment are summarized in Supplementary Table 7.
Flow data was analyzed using Flowjo v.9. Live cells were gated by viewing scatter plots of DAPI fluorescence (408 nm laser, 450/50 filter; log-scale, 1–10,000) versus forward scatter (FSC; linear scale, 0–1000). Events with high DAPI responses were interpreted as dead cells. In general, three populations of cells were readily observed for both infected and uninfected controls. A gate was constructed encompassing the largest of the three populations, which displayed the lowest DAPI response. Live, infected cells were gated by viewing live cell scatter plots of an unused fluorophore channel on the y-axis (log-scale, 1–10,000), in this case PE (561 nm laser, 585/15 filter), versus FITC fluorescence (488 nm laser, 530/30 filter; log-scale, 1–10,000). For uninfected controls, a single population of cells was observed. A gate was constructed immediately to the right of this population to encompass all events in the upper range of FITC response within the full PE response range. A high FITC response generally ranged from 10 to 10,000, though the minimum FITC response of live cells tended to increase slightly with increasing PE response.
Fluorescence microscopy
Compound 4 (0.2 mg) was derivatized with NHS-LC-LC-biotin (10 µl of 100 mM solution in DMSO; Pierce) in 0.1 M ammonium bicarbonate (110 µl total volume) on ice and in darkness for 2 hrs. The reaction mixture was fractionated on Fluka C18 resin in a Pasteur pipette column, eluting with 10%, 30%, 50%, and 100% MeOH (2 ml). The 100% MeOH elution contained almost exclusively derivatized cinnamycin, which was purified by HPLC. LC/MS analysis indicated only singly biotinylated cinnamycin. Compound 9 (<1.0 mg) was derivatized with NHS-LC-LC-biotin (50 µl of 100 mM solution in DMSO) in 0.1 M ammonium bicarbonate (500 µl total volume) at room temperature and in darkness for 3 hrs. The reaction mixture was dried and resuspended in MeOH for HPLC. LC/MS analysis indicated that the collected peak contained exclusively the singly biotinylated IAA extraproteolytic derivative of divamide A. Biotinylated cinnamycin and biotinylated divamide A were quantified by LC/MS (Supplementary Table 6).
CEMTART tat/rev++ cells, either uninfected or HIV infected, were fixed with paraformaldehyde (200 µl; 1% paraformaldehyde, 0.1% Tritron X-100 in PBS), incubating for 30 min at 4°C. Paraformaldehyde was removed by diluting with wash buffer (1 ml; 0.5% Tween-20 in PBS), centrifugation at 2500 rpm for 5 min, and removal of the supernatant by decanting, followed by a second rinse with wash buffer (1 ml). Pellets were resuspended in wash buffer (100 µl) and incubated with biotinylated peptide (10 µg/ml, ~4 µM for both conjugated cinnamycin and divamide) for 1 hr at 4°C. At this concentration, none of the non-derivatized divamides displayed any cytotoxicity, while the CC50 of non-derivatized cinnamycin is 2 µM (Figure 5c). However, we observed no signs of cytotoxicity or cell lysis for biotin-cinnamycin-treated cells stained with streptavidin-FITC by microscopy. This is in agreement with previous observations that conjugation reduces cytotoxicity of 4 but not its ability to bind PE.34,59 As described for paraformaldehyde removal, cells were washed twice in wash buffer. Streptavidin-FITC (10 µg/ml; ThermoFisher) was incubated with resuspended pellets (100 µl total volume) for 30 min at 4°C, then washed twice to remove excess streptavidin-FITC. Cell pellets were then resuspended in PBS buffer with propidium iodide (PI) added (1 µg/ml) for visualization of cells at 25 °C. Cells were observed by fluorescence microscopy using a Nikon A1R confocal microscope with Plan Apo VC 20× objective and images recorded using NIS-Elements AR software at a resolution of 512×512 (Figure 5a, b) or 1024 × 1024 pixels (Figure 5c), corresponding to 0.6 and 0.36 µm/pixel, respectively, at a bit depth of 3 × 12 bits. Excitation and emission wavelengths used for the FITC channel were 488 and 525 nm, respectively, and 561.2 and 595 nm, respectively, for the Alexa Fluor 568 channel, used to detect PI fluorescence. For all images, display lookup tables (LUTs) were linear and covered the full data range. Acquired images were not processed further.
Statistics
The anti-HIV assay results of bioassay-guided fractionation are shown in Figure 1b as dot plots, with separate distribution means and upper quartiles shown for each plot. Each point represents the average of either two or three assay responses, recorded within a single experiment, per fraction per dose. All data presented are derived from three independent experiments. A two-tailed unequal variance t-test was used to compare data distributions between different conditions, and a p-value of less than 0.05 was considered significant. Comparisons made between E11-036 (n=139) and E11-037 (n=38) data or between E11-036 and vehicle control (n=12) data were significant (p-value = 0.0253 and 0.00632, respectively), whereas the comparison of E11-037 with vehicle control data was not (p-value = 0.112). A single outlier from the E11-036 data was excluded (351%). The sample size of E11-037 reflects all fractions generated within a single fractionation scheme, with each fraction screened at both 5 and 50 µg/ml. The larger sample size of E11-036 includes doses at 5, 10, 25, and 50 µg/ml for each fraction, as well as additional fractions generated in a second fractionation scheme using the remaining animal material. The mean response across all experiments of the higher of two AZT doses (at either 0.1 or 0.05 mg/ml) is shown with ± one standard deviation (n=11).
Cytoprotection assay dose-response curves are shown in Figure 3 and Supplementary Figure 4. Each point or line represents the mean of three assay responses, all recorded within a single experiment, per dose (error bars = ± standard deviation). Eight media-only assay responses, referred to as Bavg in equation 1, were recorded for each experiment (see Online Methods). The data presented are derived from five independent experiments, with most experiments testing two different compounds at once. R2 values are included for compounds for which curves could be fit with equation 7, or a single four-parameter IC50 equation for curve 4 (Figure 3).
Flow cytometry-based dose-response curves were generated from multiple, independent flow cytometry experiments. Data from three individual experiments (or two in some cases where the third experiment failed due to instrument clogging) per compound per dose were averaged to obtain each point shown in Figure 4 and Supplementary Figure 4 (error bars = ± standard deviation). Control responses, represented in Figure 4 and Supplementary Figure 4 as horizontal lines, were also averaged from multiple experiments per plate. Each plate included 57 independent flow cytometry experiments and tested either two different compounds when using dose range 1 with infected cells only, or a single compound using dose range 2 with both infected and uninfected cells. A total of four plates were required for 181 individual experiments. Each experiment varied in the total number of events counted, as well as the number of positive events counted, depending on whether administered doses were anti-HIV (many live cells, few infected cells), cytotoxic (few live cells, few infected cells), or neither (many live cells, few or many infected cells). Highly cytotoxic conditions yielded few events, such that at least 3400 events were recorded for all experiments. On average, however, at least 36,000 events were recorded per experiment. The details of each individual experiment, including exact n values, are described in Supplementary Table 7. Typically, >50% of all events were counted as live cells, and >50% of all live cells were counted as infected cells.
The fluorescence microscopy images shown in Figure 5 were representative of the entirety of each sample. Thus, a single image was recorded for each sample.
Data Availability
Annotated DNA sequences of E11-036 and E11-037 div gene clusters have been deposited to GenBank under the accession numbers KY115608 and KY115609, respectively.
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health (NIH) through Fogarty International Center Grant ICBG U01TW006671, R01 GM102602, and R01 GM107557, as well as the American Federation for Pharmaceutical Education (AFPE) Pre-Doctoral Fellowship and the American Chemical Society (ACS) Division of Medicinal Chemistry Pre-Doctoral Fellowship. Funding for the Varian INOVA 600 and 500 MHz NMR spectrometers was provided by NIH Grant RR06262. We thank the lab of J. C. Vederas for providing us with MeLan GC/MS standards and M. S. Donia for providing suggestions in the preparation of the manuscript.
Footnotes
Author Contributions: T. E. S., E. W. S., C. M. I., and L. R. B. designed the research. T. E. S. and E. W. S. wrote the paper. T. E. S. purified and characterized compounds from both tunicates and E. coli, constructed pDiv-2, pDiv-3, and pRSFDuet-DivMT expression vectors, performed all in vitro synthetic steps in divamide production, and designed Figures. C. D. P. and T. E. S. performed biological assays. E. P. constructed the pDiv expression vector. Z. P. H. aided in the purification of compounds from E. coli. J. K. sequenced and assembled the E11-036 cluster. M.M. Z. assembled the partial E11-037 cluster. M. K. H. collected animal material. T. P. W. and T. E. S. collected 900 MHz NMR spectra and the 500 MHz 13C NMR spectrum for compound 1. T. K. M. obtained permits for tunicate collection. T. S. B. provided access to the UW NMRFAM facility. L. R. B. and C. M. I. provided technical and conceptual assistance.
Competing Financial Interests Statement: The authors declare no competing financial interests.
The authors declare no conflicts of interest.
References
- 1.Blunt JW, Copp BR, Keyzers RA, Munro MH, Prinsep MR. Marine natural products. Nat. Prod. Rep. 2016;33:382–431. doi: 10.1039/c5np00156k. [DOI] [PubMed] [Google Scholar]
- 2.Miller IJ, Vanee N, Fong SS, Lim-Fong GE, Kwan JC. Lack of overt genome reduction in the bryostatin-producing bryozoan symbiont “Candidatus Endobugula sertula”. Appl. Environ. Microbiol. 2016;82:6573–6583. doi: 10.1128/AEM.01800-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schofield MM, Jain S, Porat D, Dick GJ, Sherman DH. Identification of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743. Environ. Microbiol. 2015;17:3964–3975. doi: 10.1111/1462-2920.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo CJ, Change FY, Wyche TP, Backus KM, Acker TM, Funabashi M, Taketani M, Donia MS, Nayfach S, Pollard KS, Craik CS, Cravatt BF, Clardy J, Voigt CA, Fischbach MA. Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell. 2017;168:517–526. doi: 10.1016/j.cell.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donia MS, Ruffner DE, Cao S, Scmidt EW. Accessing the hidden majority of marine natural products through metagenomics. Chembiochem. 2011;12:1230–1236. doi: 10.1002/cbic.201000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewin RA. Prochlorophyta as a proposed new division of algae. Nature. 1976;261:697–698. doi: 10.1038/261697b0. [DOI] [PubMed] [Google Scholar]
- 9.Donia MS, Fricke WF, Partensky F, Cox J, Elshahawi SI, White JR, Phillippy AM, Schatz MC, Piel J, Haygood MG, Ravel J, Schmidt EW. Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1423–1432. doi: 10.1073/pnas.1111712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willey JA, van der Donk WA. Lantibiotics: Peptides of diverse structure and function. Annu. Rev. Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 12.Widdick DA, Dodd HM, Barraille P, White J, Stein TH, Chater KF, Gasson MJ, Bibb MJ. Cloning and engineering of the cinnamycin biosynthetic gene cluster from Streptomyces cinnamoneus cinnamoneus DSM 40005. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4316–4321. doi: 10.1073/pnas.0230516100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredenhagen A, Fendrich G, Märki F, Märki W, Gruner J, Raschdorf F, Peter HH. Duramycins B and C, two new lanthionine containing antibiotics as inhibitors of phospholipase A2 – structural revision of duramycin and cinnamycin. J. Antibiot. 1990;43:1403–1412. doi: 10.7164/antibiotics.43.1403. [DOI] [PubMed] [Google Scholar]
- 14.Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 15.Huo L, Ökesli A, Zhao M, van der Donk WA. Insights into the biosynthesis of duramycin. Appl. Environ. Microbiol. 2017;83:e02698–16. doi: 10.1128/AEM.02698-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Rourke S, Widdick D, Bibb M. A novel mechanism of immunity controls the onset of cinnamycin biosynthesis in Streptomyces cinnamoneus DSM 40646. J. Ind. Microbiol. Biotechnol. 2016;44:563–572. doi: 10.1007/s10295-016-1869-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ökesli A, Cooper LE, Fogle EJ, van der Donk WA. Nine post-translational modifications during the biosynthesis of cinnamycin. J. Am. Chem. Soc. 2011;133:13753–13760. doi: 10.1021/ja205783f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cameron DM, Gregory ST, Thompson J, Suh MJ, Limbach PA, Dahlberg AE. Thermus thermophilus L11 methyltransferase, PrmA, is dispensable for growth and preferentially modifies free ribosomal protein L11 prior to ribosome assembly. J. Bacteriol. 2004;186:5819–5825. doi: 10.1128/JB.186.17.5819-5825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai X, Otake K, You C, Cai Q, Wang Z, Masumoto H, Wang Y. Identification of novel alpha-N-methylation of CENP-B that regulates its binding to the centromeric DNA. J. Proteome Res. 2013;12:4167–4175. doi: 10.1021/pr400498y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dognin MJ. The primary structure of L11, the most heavily methylated protein from Escherichia coli ribosomes. FEBS Lett. 1977;84:342–346. doi: 10.1016/0014-5793(77)80721-7. [DOI] [PubMed] [Google Scholar]
- 21.Zhang K, Yau PM, Chandrasekhar B, New R, Kondrat R, Imai BS, Bradbury ME. Differentiation between peptides containing acetylated or tri-methylated lysines by mass spectrometry: an application for determining lysine 9 acetylation and methylation of histone H3. Proteomics. 2004;4:1–10. doi: 10.1002/pmic.200300503. [DOI] [PubMed] [Google Scholar]
- 22.Donia MS, Fricke WF, Ravel J, Schmidt EW. Variation in tropical reef symbiont metagenomes defined by secondary metabolism. PLoS One. 2011;6:e17897. doi: 10.1371/journal.pone.0017897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tianero MD, Pierce E, Raghuraman S, Sardar D, McIntosh JA, Heemstra JR, Schonrock Z, Covington BC, Maschek JA, Cox JE, Bachmann BO, Olivera BM, Ruffner DE, Schmidt EW. Metabolic model for diversity-generating biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1772–1777. doi: 10.1073/pnas.1525438113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pannecouque C, Daelemans D, De Clercq E. Tetrazolium-based colorimetric assay for the detection of HIV replication inhibitors: revisited 20 years later. Nat. Protoc. 2008;3:427–434. doi: 10.1038/nprot.2007.517. [DOI] [PubMed] [Google Scholar]
- 25.Märki F, Hänni E, Fredenhagen A, van Oostrum J. Mode of action of the lanthionine-containing peptide antibiotics duramycin, duramycin B and C, and cinnamycin as indirect inhibitors of phospholipase A2. Biochem. Pharmacol. 1991;42:2027–2035. doi: 10.1016/0006-2952(91)90604-4. [DOI] [PubMed] [Google Scholar]
- 26.Iwamoto K, Hayakawa T, Murate M, Makino A, Ito K, Fujisawa T, Kobayashi T. Curvature-dependent recognition of ethanolamine phospholipids by duramycin and cinnamycin. Biophys. J. 2007;93:1608–1619. doi: 10.1529/biophysj.106.101584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machaidze G, Seeling J. Specific binding of cinnamycin (Ro 09-0198) to phosphatidylethanolamine. Comparison between micellar and membrane environments. Biochemistry. 2003;42:12570–12576. doi: 10.1021/bi035225b. [DOI] [PubMed] [Google Scholar]
- 28.Choung SY, Kobayashi T, Inoue J, Takemoto K, Ishitsuka H, Inoue K. Hemolytic activity of a cyclic peptide Ro09-0198 isolated from Streptoverticillium. Biochim. Biophys. Acta. 1988;940:171–179. doi: 10.1016/0005-2736(88)90192-7. [DOI] [PubMed] [Google Scholar]
- 29.Makino A, Baba T, Fujimoto K, Iwamoto K, Yano Y, Terada N, Ohno S, Sato SB, Ohta A, Umeda M, Matsuzaki K, Kobayashi T. Cinnamycin (Ro 09-0198) promotes cell binding and toxicity by inducing transbilayer lipid movement. J. Biol. Chem. 2003;278:3204–3209. doi: 10.1074/jbc.M210347200. [DOI] [PubMed] [Google Scholar]
- 30.Richard AS, Zhang A, Park SJ, Farzan M, Zong M, Choe H. Virion-associated phosphatidylethanolamine promotes TIM1-mediated infection by Ebola, dengue, and West Nile viruses. Proc. Natl. Acad. Sci. U. S. A. 2015;112:14682–14687. doi: 10.1073/pnas.1508095112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, Harris E, Pereira L. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe. 2016;20:155–166. doi: 10.1016/j.chom.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wakamiya T, Fukase K, Naruse N, Konishi M, Shiba T. Lanthiopeptin, a new peptide effective against Herpes simplex virus: structural determination and comparison with Ro 09-0198, an immunopotentiating peptide. Tetrahedron Lett. 1988;29:4771–4772. [Google Scholar]
- 33.Yates KR, Welsh J, Udegbunam NO, Greenman J, Maraveyas A, Madden LA. Duramycin exhibits antiproliferative properties and induces apoptosis in tumour cells. Blood Coagul. Fibrin. 2012;23:396–401. doi: 10.1097/MBC.0b013e3283538875. [DOI] [PubMed] [Google Scholar]
- 34.Zhao M. Lantibiotics as probes for phosphatidylethanolamine. Amino Acids. 2011;41:1071–1079. doi: 10.1007/s00726-009-0386-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medema MH, Fischbach MA. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chi J, Vila-Farres X, Inoyama D, Ternei M, Cohen LJ, Gordon EA, Reddy BV, Charlop-Powers Z, Zebroski HA, Gallardo-Macias R, Jaskowski M, Satish S, Park S, Perlin DS, Freundlich JS, Brady SF. Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Nat. Chem. Biol. 2016;12:1004–1006. doi: 10.1038/nchembio.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Challis GL, Ravel J. Coelichelin, a new peptide siderophore encoded by the Streptomyces coelicolor genome: structure prediction from the sequence of its non-ribosomal peptide synthetase. FEMS Microbiol. Lett. 2000;187:111–114. doi: 10.1111/j.1574-6968.2000.tb09145.x. [DOI] [PubMed] [Google Scholar]
- 38.Skinnider MA, Dejong CA, Rees PN, Johnston CW, Li H, Webster AL, Wyatt MA, Magarvey NA. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM) Nucleic Acids Res. 2015;43:9645–9662. doi: 10.1093/nar/gkv1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, de Los Santos ELC, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M, Alban SD, Miao C, Zheng Y-M, Fuller MS, Rennert PD, Maury W, Johnson MC, Freed EO, Liu SL. TIM-family proteins inhibit HIV-1 release. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3699–E3707. doi: 10.1073/pnas.1404851111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S, Meyerholz DK, Rennert P, Mullins RF, Brindley M, Sansdersfeld LM, Quinn K, Weller M, McCray PB, Jr, Chiorini J, Maury W. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. U.S.A. 2011;108:8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe. 2012;12:544–557. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Firn RD, Jones CG. A Darwinian view of metabolism: molecular properties determine fitness. J. Exp. Bot. 2009;60:719–726. doi: 10.1093/jxb/erp002. [DOI] [PubMed] [Google Scholar]
- 44.Kliebenstein DJ. A role for gene duplication and natural variation of gene expression in the evolution of metabolism. PLoS One. 2008;3:e1838. doi: 10.1371/journal.pone.0001838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Z, Torres JP, Tianero MD, Kwan JC, Schmidt EW. Origin of chemical diversity in Prochloron-tunicate symbiosis. Appl. Environ. Microbiol. 2016;82:3450–3460. doi: 10.1128/AEM.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr PJ, Joewono I, Rusch D, Chisholm SW, van der Donk WA. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sardar D, Pierce E, McIntosh JA, Schmidt EW. Recognition sequences and substrate evolution in cyanobactin biosynthesis. ACS Synth. Biol. 2015;4:167–176. doi: 10.1021/sb500019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cubillos-Ruiz A, Berta-Thompson JW, Becker JW, van der Donk WA, Chisholm SW. Evolutionary radiation of lanthipeptides in marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E5424–E5433. doi: 10.1073/pnas.1700990114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirose M, Nozawa Y, Hirose E. Genetic isolation among morphotypes in the photosymbiotic didemnid Didemnum molle (Ascidiacea, Tunicata) from the Ryukyus and Taiwan. Zoolog. Sci. 2010;27:959–964. doi: 10.2108/zsj.27.959. [DOI] [PubMed] [Google Scholar]
- 50.Burton IW, Quilliam MA, Walter JA. Quantitative 1H NMR with external standards: use in preparation of calibration solutions for algal toxins and other natural products. Anal. Chem. 2005;77:3123–3131. doi: 10.1021/ac048385h. [DOI] [PubMed] [Google Scholar]
- 51.Sokolov EP. An improved method for DNA isolation from mucopolysaccharide-rich molluscan tissues. J. Moll. Stud. 2000;66:573–575. [Google Scholar]
- 52.Tianero MD, Kwan JC, Wyche TP, Presson AP, Koch M, Barrows LR, Bugni TS, Schmidt EW. Species specificity of symbiosis and secondary metabolism in ascidians. ISME J. 2015;9:615–628. doi: 10.1038/ismej.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buhr F, Jha S, Thommen M, Mittelstaet J, Kutz F, Schwalbe H, Rodnina MV, Komar AA. Synonymous codons direct cotranslational folding toward different protein conformations. Mol. Cell. 2016;61:341–351. doi: 10.1016/j.molcel.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He Z, Kisla D, Zhang L, Yuan C, Green-Church KB, Yousef AE. Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl. Environ. Microbiol. 2007;73:168–178. doi: 10.1128/AEM.02023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawulka KE, Sprules T, Diaper CM, Whittal RM, McKay RT, Mercier P, Zuber P, Vederas JC. Structure of subtilosin A, a cyclic antimicrobial peptide from Bacillus subtilis with unusual sulfur to α-carbon cross-links: formation and reduction of α-thio-α-amino acid derivatives. Biochemistry. 2004;43:3385–3395. doi: 10.1021/bi0359527. [DOI] [PubMed] [Google Scholar]
- 57.Chen H, Boyle TJ, Malim MH, Cullen BR, Lyerly HK. Derivation of a biologically contained replication system for human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U.S.A. 1992;89:7678–7682. doi: 10.1073/pnas.89.16.7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiser R, Makovsky S, Terpening SJ, Laing N, Clanton DJ. Assessment of a cytoprotection assay for the discovery and evaluation of anti-human immunodeficiency virus compounds utilizing a genetically-impaired virus. J. Virol. Methods. 1996;58:99–109. doi: 10.1016/0166-0934(95)01998-7. [DOI] [PubMed] [Google Scholar]
- 59.Hou S, Johnson SE, Zhao M. A One-step staining probe for phosphatidylethanolamine. Chembiochem. 2015;16:1955–1960. doi: 10.1002/cbic.201500127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Annotated DNA sequences of E11-036 and E11-037 div gene clusters have been deposited to GenBank under the accession numbers KY115608 and KY115609, respectively.