Abstract
Lack of cell surface glycosylphosphatidylinositol (GPI)-anchored protein(s) has been used as a reporter of Pig-a gene mutation in several model systems. As an extension of this work, our laboratory initiated development of an in vitro mutation assay based on the flow cytometric assessment of CD90.2 expression on the cell surface of the mouse lymphoma cell line L5178Y/Tk+/−. Cells were exposed to mutagenic and non-mutagenic compounds for 24 hours followed by washout and incubation for an additional 7 days. Following this mutant manifestation time, cells were labeled with fluorescent antibodies against CD90.2 and CD45 antigens. These reagents indicated the presence of GPI-anchored proteins and general cell surface membrane receptor integrity, respectively. Instrument set-up was aided by parallel processing of a GPI anchor-deficient subclone. Results show that the mutagens reproducibly caused increased frequencies of mutant phenotype cells, while the non-mutagens did not. Further modifications to the method, including application of a viability dye and an isotype control for instrument set-up, were investigated. As a means to verify that the GPI-anchored protein-negative phenotype reflects bona fide Pig-a gene mutation, sequencing was performed on 38 CD90.2-negative L5178Y/Tk+/− clones derived from cultures treated with ethyl methanesulfonate. All clones were found to have mutation(s) within the Pig-a gene. The continued investigation of L5178Y/Tk+/− cells, CD90.2 labeling, and flow cytometric analysis as the basis of an in vitro mutation assay is clearly supported by this work. These data also provide evidence of the reliability of using GPI anchor-deficiency as a valid reporter of Pig-a gene mutation.
Keywords: gene mutation, Pig-a, L5178Y/Tk+/−, mutagenesis, phosphatidylinositol glycan complement class A gene, fluorescent antibody labeling, flow cytometry, phenotype, sequencing
Introduction
Current assays for mutagen detection comprise a range of test systems including bacteria [Ames assay; Ames et al., 1975; Gatehouse et al., 1994], cultured cells from various species [thymidine kinase gene assays; Moore et al., 1985, 2005] and whole animal models utilizing both nontransgenic [Walker et al., 2007] as well as specialized transgenic rodents [Big Blue®, Muta™Mouse; Morrison and Ashby, 1994; Nohmi et al., 2000]. One more recent method that has gained considerable attention within the last decade is based on the phosphatidylinositol glycan complement class A (Pig-a) gene. Methods based on this gene most commonly use flow cytometry to enumerate peripheral blood cells with a deficiency in glycosylphosphatidylinositol (GPI)-anchored proteins on the cell surface. The GPI anchors are formed by the concerted efforts of over 30 genes including many members of the Pig gene family, but Pig-a is the only one located on the X chromosome [Kinoshita, 2014]. Since this means there is only one functional Pig-a copy per cell, a single inactivating mutation in the gene is sufficient to eliminate Pig-a-mediated enzyme function and disrupt GPI anchor formation. This has formed the basis for the in vivo Pig-a gene mutation assays currently in existence.
In vivo methods for assessment of Pig-a gene mutation are well established [Dertinger and Heflich, 2011; Dertinger et al., 2011; Kimoto et al., 2016] and have demonstrated utility in numerous applications including genotoxicity hazard identification and risk assessment [Wills et al., 2016]. These methods have the advantage of being compatible with multiple species and thus can be readily applied to existing toxicology studies such as the 28 day repeat-dose study design [Dertinger et al., 2010, 2012]. The in vivo methods have also been the subject of expert working groups including the Health and Environmental Sciences Institute Genetic Toxicity Technical Committee (HESI GTTC) [Schuler et al., 2011] and the International Workshop on Genotoxicity Testing (IWGT) [Gollapudi et al., 2015]. Currently a submission process has been initiated to support the development of an Organization of Economic Co-operation and Development (OECD) guideline for these in vivo assays.
Another important aspect of methodologies that employ Pig-a gene-based assessment of mutagenesis is the ability to examine this marker using in vitro systems. This provides the means for studying the same biological endpoint across systems of varied complexity. Thus certain investigations can benefit from the ability to bridge the Pig-a gene mutation endpoint across model systems, especially in vitro to in vivo. Some of the earliest investigations that examined PIG-A mutation in vitro focused on using this indicator of specific gene function to examine the basic biology of mutation. Chen et al., [2001] examined the phenomenon of mutator phenotype (Mut) in human colon cancer cell lines. Mut cell lines were readily differentiated from non-Mut cell lines by comparing GPI anchor phenotype and demonstrating the lack of surface expression of GPI-anchored proteins as well as specific PIG-A gene mutation in Mut phenotype cells. As a means to study mutation rate in humans, Araten et al., [2013] selected PIG-A as a sentinel gene and examined immunofluorescent labeling of transformed human cord-blood myeloid cells analyzed by flow cytometry. This system provided a means to model mutation rate and make predictions regarding the contributions of this process to human carcinogenesis.
Since these initial studies, several other investigators have explored the utility of in vitro Pig-a based methods, primarily for applications associated with safety assessment. Kruger et al. [2015, 2016] examined human TK6 cells as a model for differentiation of mutagenic agents from non-mutagens. They also used gene sequencing to identify mutations in both PIG-A and PIG-L genes as contributing to GPI anchor deficiency. The same group recently reported positive correlations between DNA adducts and GPI anchor deficiency in benzo[a]pyrene diol-epoxide-treated TK6 cells [Piberger et al., 2017]. The role of other Pig genes, including PIG-L, in GPI anchor deficiency of TK6 cells was also reported by Nicklas et al. [2015]. Investigators at Swansea University [Rees et al., 2017] initially studied TK6 cells, but ultimately developed an optimized flow cytometric method using human MCL-5 cells. Finally, Nakamura et al. [2012] used chicken DT40 cells to demonstrate the role of the Pig-o gene (present on the Z-chromosome in avian species) as a target for mutation detection in an analogous in vitro assay.
For the experiments reported here, we employed mouse lymphoma L5178Y/Tk+/− −3.7.2C cells to develop a flow cytometric method for the phenotypic identification of Pig-a mutant cells. The mouse species has been shown to be compatible with mutagenicity assays based on the Pig-a gene (Olsen et al., 2017). These particular cells were chosen due to their utilization in other genotoxicity assays such as the Mouse Lymphoma Assay and in vitro micronucleus studies. Also these cells do not suffer from the exceptionally high baseline mutant frequency observed in the TK6 cell line used for in vitro Pig-a studies by several other investigators. This relieves a user of the need to purify or otherwise isolate wildtype cells prior to using them for experiments.
The initial, method-development studies described here examine exposure to mutagenic/non-mutagenic cytotoxic compounds, as well as specific assay modifications devised to optimize assessment of the mutant population. More work will be necessary to fully identify the optimal experimental design and data analysis strategies for this methodology. Importantly, we also investigate sequencing of mutant phenotype clones to directly establish the link between genomic alterations in the Pig-a gene and the GPI anchored-protein deficient phenotype.
Materials and Methods
Cell line and chemicals
The L5178Y/Tk+/− −3.7.2C mouse lymphoma cell line was obtained from ATCC (Manassas, VA) and grown in DMEM supplemented with fetal bovine serum, glutamine, and Pen/Strep. Cells were maintained at ≤ 1×106 cells/ml in a 5% CO2, 37°C humidified incubator using 125 mL tissue culture flasks for routine passage and exposure to test articles. For some experiments, a GPI anchor-deficient L5178Y/Tk+/− clone was employed. This mutant cell line was generated by treatment of L5178Y/Tk+/− cells with ethyl methanesulfonate (EMS) and subsequent isolation and cultivation of clones with an aerolysin-resistant phenotype.
All chemicals were obtained from Sigma-Aldrich (St. Louis, MO) unless stated otherwise. Antibodies (Ab) against CD90.2-PE (Clone 53-2.1), CD45-APC (Clone 30-F11), CD45-FITC (Clone 30-F11) and Isotype Control (Rat IgG2a, Clone RTK2758) were from BioLegend (San Diego, CA), DRAQ7 was from Cell Signaling (Danvers, MA). The CD90.2 target is a protein associated with GPI anchors on the cell surface, thus it serves as the indicator of wildtype vs mutant cells. The CD45 reagent is included as both a process control and an indicator of cell health. Thus only cells with sufficient levels of CD45 staining are considered for determination of GPI anchor deficiency and cells lacking appropriate CD45 conceivably caused by overt damage - which could also affect CD90.2 surface expression - can be avoided. The compounds selected for study were dissolved in dimethylsulfoxide (DMSO) and included the mutagens ethyl nitrosourea (ENU), EMS, methyl methanesulfonate (MMS), 1,3-propane sultone (1,3-PS), 4-nitroquinoline oxide (4-NQO), cisplatin (CSP), vinblastine (VIN), etoposide (ETO), camptothecin (CAM) and the non-mutagens D-mannitol (MAN), dexamethasone (DEX), diethanolamine (DEA), phenformin HCl (PHE), cycloheximide (CYC) and carbonyl cyanide m-chlorophenyl hydrazone (CCCP).
Cell exposure and processing
Treatment of cells with a range of concentrations of the mutagenic or non-mutagenic agents was performed for 24 hours in T125 flasks. All exposures for the definitive studies were performed in triplicate flasks and all experiments were conducted as three independent repeats. After 24 hours of exposure, cells were collected from all treatment vessels and counts were performed using the volumetric counting function on a MACS Quant flow cytometer (Miltenyi Biotec, Bergisch Gladbach, Germany). These data enabled the determination of cytotoxicity based on the comparison of cell counts obtained from treated cultures relative to vehicle control counts. The cells were then washed free of treatment medium and resuspended with fresh growth medium. Initial experiments examined the kinetics of appearance of mutant cells for up to 10 days following exposure to EMS. Once an appropriate expression time was determined, cells were passed daily (except over the weekend) for a total of 8 days to allow sufficient time for expression of the mutation and turnover of the normal cell-surface complement of GPI-anchored proteins. For passage of the cells during an experiment, between 1.5 ×105 and 1.0 ×106 total cells were passed in order to maintain a sufficient number of mutant cells in culture. For each experiment Pig-a mutant cell frequency data were collected across triplicate treatments per test agent concentration. Furthermore, each chemical was studied in three independent experiments.
Immunofluorescent labeling of the cells for the majority of experiments involved the following steps based on instructions from a prototype In Vitro MutaFlow kit (Litron Laboratories, Rochester, NY). Briefly, 2 × 106 cells were spun down in 15 mL tubes and resuspended in Hank’s Balanced Salt Solution + 1% fetal bovine serum containing optimized concentrations of anti-CD90.2 PE and anti-CD45 APC. The samples were incubated at 4°C in a refrigerator for 10 min then they were washed and resuspended in fresh buffer and kept cold/protected from light until flow cytometric analysis.
For instrument set-up, we originally employed a GPI anchor-deficient L5178Y/Tk+/− clone in conjunction with wild-type cells. Processing an aliquot of these cells alongside the positive/vehicle controls provided a known CD90.2-negative population to ensure proper adjustment of voltage, compensation and setting of gates/regions in the flow cytometer software as described in the prototype kit manual. Experiments which examined the potential effects of non-mutagenic compounds also included EMS as a positive control (250–500 µM) to demonstrate proper execution of the methodology. Mutant cell frequency was determined by interrogation of approximately 1 million cells for CD90.2 fluorescence. Cytotoxicity was based on cell numbers compared to the concurrent solvent control, i.e. relative survival.
Statistical Analyses
All statistical analyses were performed using JMP v12 (SAS, Cary, North Carolina). Since these studies represent our initial method development activities, we show much of the data as individual experiments rather than combining across independent replicates. This was done to better inform the reader about the reproducibility of the various responses across multiple experiments as opposed to combining data and providing summary statistics on groups. Thus, as a means to represent reproducibility of the assay across separate experiments, an initial trend test producing a linear best-fit line was performed for each independent experiment, using JMP’s ANOVA platform. This was followed by Dunnett’s pair-wise test comparing each treatment group to its respective control. To maintain the overall significance level at 0.05, the trend as well as the pairwise differences from the control group are declared statistically significant if P < 0.025. The results from both tests were considered together when making an assessment of activity, thus achieving significance in both tests was considered a positive response. A significant response in only one or none of the tests was considered equivocal or negative respectively.
Method Optimization
In later experiments, the anti-CD45 APC Ab was replaced with a FITC-conjugated version. This allowed for the inclusion of the cell viability dye DRAQ7 in the final resuspension buffer. Further process optimization included exploring the use of an Isotype Control antibody in place of the CD90.2-specific Ab when labeling a sample of vehicle-treated cells for use in instrument set-up. This eliminated the need to maintain a natively Pig-a mutant cell line as described above.
Mutation Sequencing
In order to address the links between the phenotype-based flow cytometric assays and bona fide mutation in the Pig-a gene, gene sequencing was performed. Mutagenized clones were generated from a population of L5178Y/Tk+/− cells that were treated with EMS for 24 hours before being washed free and cultured over 10 days to allow for sufficient expression of mutation. After this period of time, aliquots of the cells exposed to various EMS concentrations were labeled as described above and analyzed to determine mutant frequency. Highly-purified mutant cells from the cultures were then obtained by immunomagnetic separation. Briefly, the cells were labeled with both CD45-FITC and CD90.2-PE Abs as described above. The cells were then incubated with anti-PE superparamagnetic microbeads (Miltenyi Biotec) which bind to the PE molecule conjugated to the CD90.2 Ab. The cell suspensions are then passed through a MACS MS column (Miltenyi Biotec) seated in an OctoMACS magnetic separator. This process retains any cells with surface expression of GPI-anchored CD90.2 protein on the column and allows GPI anchor-deficient cells to pass through. The column purification was repeated three more times with fresh columns for each sample. An aliquot of the samples were taken for verification of separation by flow cytometry and the remaining mutant-enriched eluates were subjected to limiting dilution and approximately 30 cells per plate were distributed across individual wells of nine 96 well plates. Once sufficient outgrowth was achieved over the course of an additional 10 days, clones were re-assessed for GPI anchor phenotype by immunolabeling/flow cytometry and confirmed GPI-negative cells were frozen as pellets (~1 million cells per vial) and sent to the National Center for Toxicological Research (NCTR) for further processing.
Genomic DNA was extracted from the supplied clones using a QiaAmp DNA micro kit (Qiagen, Germantown, MD) and three fragments of the mouse Pig-a gene covering five coding regions of the gene were amplified for each clone in separate reactions [Revollo et al., 2017]. The fragments belonging to each clone were then pooled together in a single sample. The resulting samples were processed using a modification of the Nextera DNA library preparation kit (Illumina, San Diego, CA) and oligonucleotides for dual-indexing as in Nextera XT v2 kit (Illumina). Briefly, 1 µL of each sample containing 0.5–1.0 ng water-diluted amplicon DNA was fragmented with 1.5 µL of Nextera enzyme/buffer mix (1:5 ratio) at 55°C for 10 minutes. A PCR mixture was added to each sample to a final volume of 25 ul and final concentrations of oligos for dual-indexing of 0.25 uM, after which the samples were subjected to the following PCR conditions: 72°C×3min + 95°C×5 min + (95°C × 30sec + 58°C × 30sec + 72°C × 30sec) × 8 cycles. The indexed libraries were then pooled into a single multiplexed library. The multiplexed library was purified with a Qiagen PCR purification kit, and sized with Agencourt AMPure XP beads (Beckman Coulter, Brea, CA). Sequencing was carried out in an Illumina NextSeq500 sequencer using a paired-end dual-indexing sequencing protocol. Sequencing reads were aligned against the mouse Pig-a reference sequence (http://www.ncbi.nlm.nih.gov/nuccore/NC_000086.7?report=genbank&from=164419685&to=164433916) with BWA software [Li and Durbin, 2009] and mutations were called with LoFreq software [Wilm et al., 2012] resulting in an individual variant call file for each sample.
Results and Discussion
Method Development
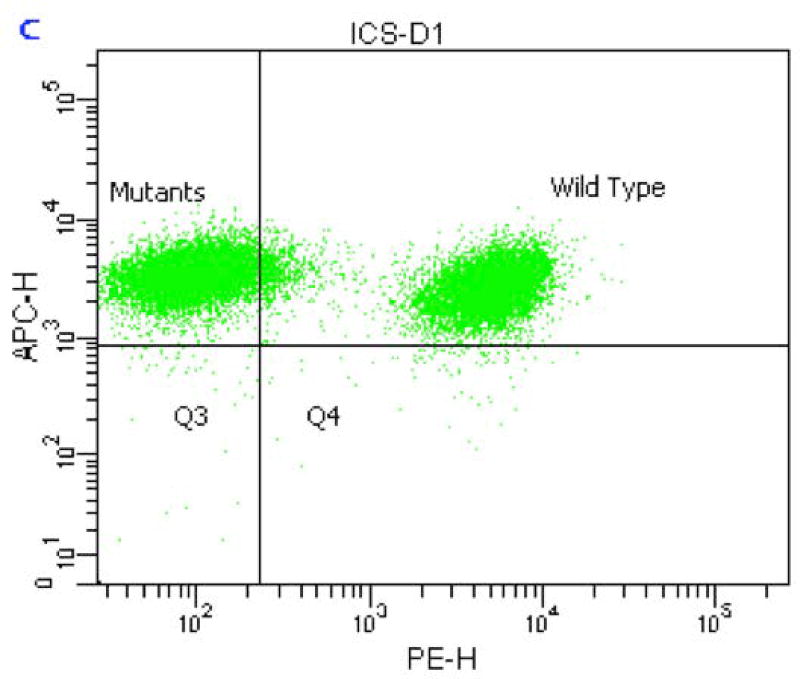
The initial staining procedure utilized a combination of CD45-APC and CD90.2-PE and instrument settings were optimized to enable sufficient resolution of the CD90.2-deficient population from wildtype. The quadrant in Figure 1 was positioned to achieve a minimum resolution of 99% of the mutant cell population. This sample was created by processing a 1:1 mix of wildtype L5178Y/Tk+/− cells and the GPI anchor-deficient clone.
Figure 1.
Bivariate plot of an Instrument Calibration Sample (ICS) comprised of a 1:1 mix of wildtype and natively mutant L5178Y/Tk+/− cells. Note the positioning of the vertical demarcation line of the quadrant that ensures approximately 99% of the mutant cells are captured within the upper left region.
Mutant population biology and kinetics
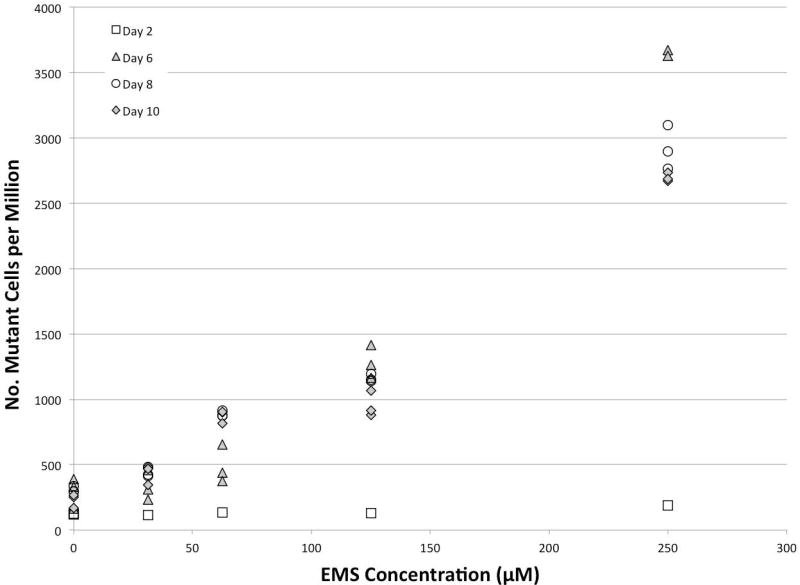
As a means to investigate the kinetics of expression of a mutant cell population, we examined the frequency of CD90.2-negative cells over time from 2 to 10 days post-exposure. Figure 2 shows the lack of any response at 2 days along with a clustering of the maximal response occurring between 6 and 10 days. For practical purposes a total expression time of 8 days (1 day exposure + 7 additional days in culture) was selected for the remaining experiments.
Figure 2.
Mutant cell manifestation time in L5178Y/Tk+/− cells exposed to ethyl methanesulfonate (EMS). Cells were exposed for 24 hours and then washed free of treatment medium. No effects were observed 2 days after exposure and stable expression was seen between 6 and 10 days post-exposure.
The potential for Pig-a gene mutation to result in resistance to apoptosis has been raised by several groups in the literature associated with the disease paroxysmal nocturnal hemoglobinurea (PNH) [Brodsky et al., 1997; Marsh and Elebute 2003]. However, there are other PNH investigators that suggest there is no alteration in sensitivity to apoptosis mediated by GPI anchor deficiency [Ware et al., 1998; Bastisch et al., 2000]. Given these observations, the potential role of a pre-existing mutant cell population experiencing some preferential growth advantage due to challenging the cells with toxic agents was explored. Figure 3 shows the cytotoxicity responses of both wild-type L5178Y/Tk+/− cells and the GPI anchor-deficient mutant clone to various compounds that act via genotoxic or non-genotoxic mechanisms. In all cases, the dose-response profiles of the two cell populations behaved similarly. This supports the conclusion that there was no growth or survival advantage for the mutant cells that would result in higher mutant frequencies relative to the wild-type cell population. This is also consistent with the in vivo literature that characterizes Pig-a mutation in bone marrow stem cells as “neutral” in that it does not provide any selection for or against the mutant cell population in the whole animal [Dertinger et al., 2014].
Figure 3.
Cytotoxicity curves for wild-type or mutant L5178Y/Tk+/− cells exposed to the nongenotoxic agent carbonyl cyanide m-chlorophenyl hydrazone (CCCP) or the genotoxicants vinblastine, etoposide or camptothecin. Solvent control for these experiments was DMSO. Similar shaped curves suggest comparable sensitivities to toxicant challenge between the two cell lines. Relative cell counts were determined via flow cytometry-based volumetric cell counting.
Response to chemical agents
Once the basic experimental conditions were established by the initial experiments, an investigation of the responses of L5178Y/Tk+/− cells to various genotoxic and non-genotoxic compounds was performed. The panels in Figure 4 depict the effects of genotoxic agents on both mutant frequency and cytotoxicity as determined by relative survival. For all of these studies we sought to achieve exposures that resulted in at least 50% relative cell survival as the top passing concentration in order to ensure adequate exposure of the cells to the study compounds. This level of cytotoxicity is provisional and needs additional testing to confirm the utility of this criterion. The overwhelming majority of the data were normally distributed as indicated by Shapiro-Wilk test of the residuals. Combined trend and pair-wise testing was performed on each separate replicate experiment. Table 1 represents the statistical analyses performed on each individual experiment. As described in the Materials and Methods section, both a linear trend test and pairwise testing was performed and the combined significance of these tests served to make the positive, negative or equivocal call. Thus both tests had to reach statistical significance in order for the experiment to be considered positive and if one test was significant it was considered equivocal. Table 1 shows that the mutagenic agents elicited statistically significant elevations in CD90.2-deficient cells. One out of three CSP experiments was considered a negative result.
Figure 4.
Graphs summarize the frequency of CD90.2-negative cells and cytotoxicity results of three independent replicate experiments examining exposure of L5178Y/Tk+/− cells to various genotoxic agents. Cells were exposed for 24 hours at which time cytotoxicity assessment and removal of test article occurred, then an additional 7 days in culture provided time for expression of a stable mutant frequency.
Table 1.
Results of statistical analyses for mutation frequency data in L5178Y/Tk+/− cells.
| Trend line p-value* | Pair-wise testing results* | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Compound | Exp. 1 | Exp. 2 | Exp. 3 | Exp. 1 | Exp. 2 | Exp. 3 |
| 4-Nitroquinoline oxide | 0.0001 | 0.0002 | 0.0001 | 4,5 | 5 | 5 |
| Cisplatin | 0.0140 | 0.6266 | 0.0001 | 5 | - | 5 |
| 1,3-Propane sultone | 0.0001 | 0.0006 | 0.0001 | 4,5 | 5 | 5 |
| Ethylnitrosourea | 0.0001 | 0.0001 | 0.0001 | 3,4,5 | 4,5 | 4,5 |
| Ethyl methanesulfonate | 0.0001 | 0.0001 | 0.0001 | 4,5 | 5 | 4,5 |
| Methyl methanesulfonate | 0.0001 | 0.0002 | 0.0006 | 5 | 5 | 5 |
|
| ||||||
| Phenformin hcl | 0.0082 | 0.0768 | 0.1972 | - | - | - |
| Cycloheximide | 0.0832 | 0.4845 | 0.0144 | - | - | - |
| Dexamethasone | †0.0024 | 0.8896 | 0.0176 | - | - | 2,4,5 |
| Diethanolamine | 0.2185 | 0.5788 | 0.0448 | - | - | - |
| d-Mannitol | 0.6276 | 0.3820 | 0.5686 | - | - | - |
significance level set at p ≤0.025 for each of the combined tests and significant effects were noted by italics
numbers correspond to treatment group: 1–5 = low to high
significant trend, but in negative direction
Figure 5 depicts the responses to non-genotoxic compounds. Despite the clear cytotoxicity experienced by these exposures, there were no corresponding changes in mutant cell frequency compared to vehicle controls for the majority of experiments performed. One experiment conducted with PHE yielded an equivocal result due to a positive trend test (Table 1). A separate study with DEX also resulted in an equivocal call due to pair-wise testing showing the second, fourth and fifth treatment groups to be statistically different from control. Given the lack of a clear dose response and the negative results from the two other experiments which examined DEX, it does not appear that this compound elicited a robust and reproducible elevation in CD90.2-deficient cells.
Figure 5.
Graphs summarize the frequency of CD90.2-negative cells and cytotoxicity results of three independent replicate experiments examining exposure of L5178Y/Tk+/− cells to various nongenotoxic agents. Cells were exposed for 24 hours at which time cytotoxicity assessment and removal of test article occurred, then an additional 7 days in culture provided time for expression of a stable mutant frequency.
Method Improvements
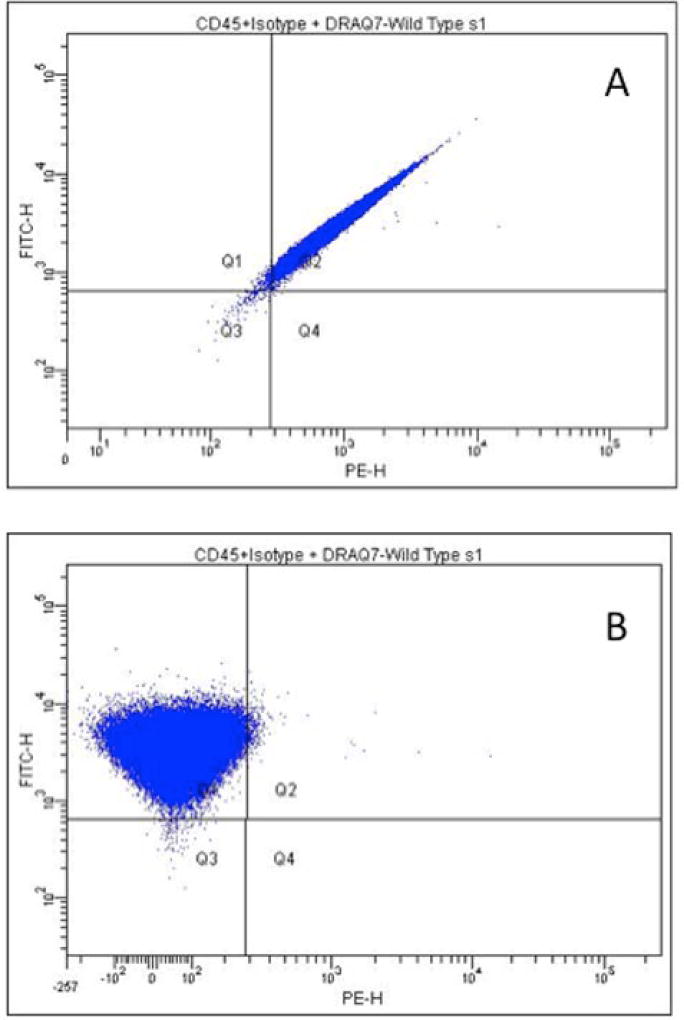
Cytotoxicity-based criteria for selection of the top concentration in dose-response studies is clearly important for the satisfactory performance of a mutagenicity assay. The inclusion of the viability dye DRAQ7 permits the elimination of cells with compromised membranes from analysis, thus focusing the determination of mutant frequency on healthy, live cells only. As described by Rees et al. [2017], this reduces the occurrence of potential artifacts, due to overt cytotoxicity, from influencing detection of mutagenicity. In order to include the DRAQ7 dye in the labeling protocol, we had to modify the other antibody-fluorochrome conjugates used for labeling the cells. Thus for the new method we still use the CD90.2-PE Ab, but replaced the CD45-APC with a CD45-FITC Ab. Figure 6 shows the entire gating scheme for the optimized protocol. Panel B specifically shows the resolution of DRAQ7-positive cells in a population of cells exposed to ENU. These cells labeled as dead and dying are readily identified based on their fluorescent signature. This allows for more accurate identification of “healthy single cells” in the light scatter plot shown in Fig. 6 (Panel A) since the green-labeled population can be eliminated from the region and any subsequent plots that examine this population will only report on healthy cells.
Figure 6.
Complete gating scheme for the optimized protocol showing the identification of DRAQ7-positive events (dead cells) as an indicator of dead and dying cells (Plot B). Note the ability to distinguish the DRAQ7-positive cells (green) in the region used to capture “Healthy single cells” shown in Plot A.
The use of an Isotype Control for instrument set-up evolved from our earlier practice of maintaining and preparing a mutant L5178Y/Tk+/− cell clone for each experiment. These natively GPI anchor-deficient cells could be used for determining the position of gates and setting other important instrument parameters when processed through the standard labeling protocol. The disadvantage of using these cells is that any investigator that wishes to employ the method would have to generate or obtain a confirmed mutant clone and continuously propagate it alongside wildtype cells for each experiment.
We examined the use of an Isotype Control antibody as a means to define a negative population of cells that provided similar characteristics to the natively GPI-deficient cell clone we previously used for instrument set-up. To do so we incubated cells with the isotype control Ab in place of the CD90.2 PE Ab, but kept all the other reagents/steps the same. This sample was then used prior to analysis of any experimental samples to adjust voltages and gating associated with the resolution of the CD90.2-negative cell population.
Using the newly modified reagent mix, we examined the fluorescence profiles in the bivariate plots of various dye combinations. The panels in Figure 7 show the before and after profiles for compensation of the FITC signal from the PE channel due to spectral overlap. Additional compensation was not necessary for the APC channel given the separation in the emissions between the fluorochromes. This new reagent combination enables easier, more rapid instrument setup based on the use of the isotype control and reduction in the amount of fluorescence compensation required.
Figure 7.
Bivariate plots showing the uncompensated profile (A) of the FITC vs PE channels and the profile after compensation that serves to remove the FITC component from the PE channel (B).
Pig-a Gene Sequencing
Identification of bona fide Pig-a gene mutation was accomplished by sequencing the DNA from 38 clones developed from CD90.2-deficient cells that were derived from cultures of EMS-treated L5178Y/Tk+/− cells using several rounds of immunomagnetic enrichment. The sequencing approach involved preparation of dual-indexed mini libraries for each individual clone, and subsequent processing of the combined multiplexed library on an Illumina NextSeq sequencer [Revollo et al., 2017]. Figure 8 shows the primer arrangement for generation of the fragments covering the exons of the Pig-a gene containing the open reading frame, along with the depth of the sequencing of individual bases.
Figure 8.
The upper part of the diagram shows a representation of the Pig-a gene and its six exons, along with the positioning of the primers to generate three fragments that cover the coding region of the gene. The lower portion of the diagram shows the depth of coverage of the sequencing for the three fragments of a representative sample.
Table 2 shows each of the individual clones submitted for sequencing and their respective mutation in the Pig-a gene. All fragments of the gene were determined to be present for the analyses. The nature of the mutations, i.e. point mutations, is consistent with the typical mutation spectra reported for EMS [Klungland et al., 1995]. Examinations of the sequence alterations revealed that the mutations resulted in either amino acid changes, stop codons or occurred at splice sites. It should be noted that other genes that participate in the biosynthesis of GPI anchors were not investigated, but in each case the phenotypically identified mutant clones contained a disruptive mutation localized to the Pig-a gene. These data support the link between specific mutation at the Pig-a gene and expression of the GPI anchor-deficient phenotype assessed by flow cytometry.
Table 2.
Mutation sequencing data from 38 individual CD90-deficient clones derived from the culture of L5178Y/Tk+/− cells treated with EMS
| Litron ID | Primary X- chromosome mutation |
Primary mutation frequency |
Amino acid change |
Litron ID | Primary X- chromosome mutation |
Primary mutation frequency |
Amino acid change |
|---|---|---|---|---|---|---|---|
| a-c3 | 164423363 G>A | 0.99 | Splice site | b2–b4 | 164428596 G>A | 0.99 | Gly>Glu |
| a-c5 | 164423346 G>A | 0.99 | Arg>Lys | b2-c4 | 164423087 C>T | 0.99 | Gln>Stop |
| a-d2† | 164428832 G>A | 0.99 | Gly>Arg | b2-c6 | 164428539 G>A | 0.99 | Splice site |
| a-d2 | 164428832 G>A | 0.99 | Gly>Arg | b2-d1 | 164428833 G>A | 0.99 | Gly>Glu |
| a-d3 | 164428004 G>A | 0.99 | Gly>Glu | b2-d2 | 164428833 G>A | 0.99 | Gly>Glu |
| a-d5 | 164428010 G>A | 0.99 | Gly>Glu | b3-a5 | 164428833 G>A | 0.99 | Gly>Glu |
| a1–a5 | 164423003 G>A | 0.99 | Glu>Lys | c1-a1 | 164428617 C>T | 0.99 | Ser>Phe |
| a1-b3 | 164423003 G>A | 0.99 | Glu>Lys | c1-a2 | 164423021 C>T | 0.99 | His>Tyr |
| a1-d1 | 164423109 C>T | 0.99 | Ser>Phe | c1-a3 | 164428932 C>A | 0.99 | Ser>Stop |
| b1-a3 | 164423363 G>A | 0.84* | Splice site | c1-a6 | 164427979 C>T | 0.99 | Gln>Stop |
| b1-a5 | 164423363 G>A | 0.78** | Splice site | c1-b1 | 164428004 G>A | 0.99 | Gly>Glu |
| b1-a6 | 164423363 G>A | 0.99 | Splice site | c1-c1 | 164423363 G>A | 0.99 | Splice site |
| b1–b5 | 164428596 G>A | 0.99 | Gly>Glu | c1–c2 | 164423021 C>T | 0.99 | His>Tyr |
| b1-c1 | 164423363 G>A | 0.99 | Splice site | c1–c6 | 164428617 C>T | 0.99 | Ser>Phe |
| b1-c6 | 164428009 G>A | 0.99 | Gly>Arg | c1-d1 | 164423021 C>T | 0.99 | His>Tyr |
| b1-d2 | 164428596 G>A | 0.99 | Gly>Glu | c1-d4 | 164428832 G>A | 0.99 | Gly>Arg |
| b1-d3 | 164428596 G>A | 0.99 | Gly>Glu | c3-a4 | 164428004 G>A | 0.99 | Gly>Glu |
| b2-a1 | 164428670 C>T | 0.99 | Gln>Stop | c3–c4 | 164427932 G>A | 0.99 | Splice site |
| b2-a4 | 164428009 G>A | 0.99 | Gly>Arg | c3-d2 | 164427979 C>T | 0.99 | Gln>Stop |
| b2-b1 | 164428989 G>A | 0.99 | Trp>Stop | L5178Y | None | --- | None |
A secondary mutation at 164428670 C>T (Gln>Stop, 0.16 frequency) was present in this clone.
This secondary mutation was found as a primary mutation in other clones.
A secondary mutation at 164428810 G>A (Splice site, 0.21 frequency) was present in this clone.
This secondary mutation was not found as a primary mutation in other clones.
Duplicate samples of this clone were submitted, both showed same results.
Conclusions
The data presented here represent our initial proof-of-concept studies into the utility of a Pig-a gene-based mutation assay in vitro. An immunofluorescent labeling protocol was created using L5178Y/Tk+/− cells and optimized to provide information not only on mutant cell frequency, but also cytotoxicity—an important aspect in mutagenicity assays. Additionally, the set-up for this assay was simplified via the use of an isotype control for setting voltage and compensation. Mutation sequencing work was accomplished that demonstrated loss-of-function mutations in the Pig-a gene of cells identified via the phenotype-based flow cytometric methodology. These data support the role of GPI anchor deficiency as a viable and robust reporter of Pig-a gene mutation. Future work will involve the investigation of a larger number of chemicals with a broader range of genotoxic and nongenotoxic mechanisms using the optimized labeling protocol. Examination of additional parameters associated with similar genotoxicity tests such as appropriate cytotoxicity limits, methods for data analysis including formal determination of statistical significance or point of departure. More extensive genome-based studies of the effects of mutagenic compounds with mutation spectra different from EMS and examination of the potential role of other Pig genes in GPI anchor-deficient L5178Y/Tk+/− cells are also planned. Overall, the continued development of in vitro Pig-a gene mutation methods will serve to strengthen the utility of all assays based on this gene and expand the repertoire of methodologies that are available to address mammalian cell mutagenicity.
Acknowledgments
The authors would like to thank Ariel Berg for her early assay optimization work. This work was funded in part by a grant from the National Institute of Health/National Institute of Environmental Health Sciences (NIEHS; grant no. R44ES021973). The contents are solely the responsibility of the authors, and do not necessarily represent the official views of the NIEHS or the US Food and Drug Administration.
Footnotes
AUTHOR CONTRIBUTIONS
JCB, SLA, SDD and CL designed the studies. SLA, CL, PM and JR performed the experiments. JCB and SDD analyzed the data and prepared the manuscript. PM, JR and VD performed all the gene sequencing work and associated data analysis at the NCTR. All authors reviewed and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
JCB, SLA, CL and SDD are employed by Litron Laboratories. Litron has patents covering flow cytometric assessment of Pig-a gene mutation and sells commercial kits based on these procedures under the name MutaFlow®.
References
- Ames BN, McCann J, Yamasaki E. Methods for Detecting Carcinogens and Mutagens with the Salmonella/Mammalian-Microsome Mutagenicity Test. Mutation Res. 1975;31:347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- Araten DJ, Krejci O, Ditata K, Wunderlich M, Sanders KJ, Zamechek L, Mulloy JC. The rate of spontaneous mutations in human myeloid cells. Mutat Res. 2013;749(1–2):49–57. doi: 10.1016/j.mrfmmm.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastisch I, Tiede A, Deckert M, Ziolek A, Schmidt RE, Schubert J. Glycosylphosphatidylinositol (GPI)-deficient Jurkat T cells as a model to study functions of GPI-anchored proteins. Clin Exp Immunol. 2000;122(1):49–54. doi: 10.1046/j.1365-2249.2000.01350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky RA, Vala MS, Barber JP, Medof ME, Jones RJ. Resistance to apoptosis caused by PIG-A gene mutations in paroxysmal nocturnal hemoglobinuria. Proc Natl Acad Sci US. 1997;94(16):8756–60. doi: 10.1073/pnas.94.16.8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Eshleman JR, Brodsky RA, Medof ME. Glycophosphatidylinositol-anchored protein deficiency as a marker of mutator phenotypes in cancer. Cancer Res. 2001;61(2):654–8. [PubMed] [Google Scholar]
- Dertinger SD, Phonethepswath S, Franklin D, Weller P, Torous DK, Bryce SM, Avlasevich S, Bemis JC, Hyrien O, Palis J, MacGregor JT. Integration of mutation and chromosomal damage endpoints into 28-day repeat dose toxicology studies. Toxicol Sci. 2010;115(2):401–11. doi: 10.1093/toxsci/kfq070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dertinger SD, Heflich RH. In vivo assessment of Pig-a gene mutation-recent developments and assay validation. Environ Mol Mutagen. 2011;52(9):681–4. doi: 10.1002/em.20685. [DOI] [PubMed] [Google Scholar]
- Dertinger SD, Phonethepswath S, Weller P, Nicolette J, Murray J, Sonders P, Vohr HW, Shi J, Krsmanovic L, Gleason C, Custer L, Henwood A, Sweder K, Stankowski LF, Jr, Roberts DJ, Giddings A, Kenny J, Lynch AM, Defrain C, Nesslany F, van der Leede BJ, Van Doninck T, Schuermans A, Tanaka K, Hiwata Y, Tajima O, Wilde E, Elhajouji A, Gunther WC, Thiffeault CJ, Shutsky TJ, Fiedler RD, Kimoto T, Bhalli JA, Heflich RH, MacGregor JT. International Pig-a gene mutation assay trial: evaluation of transferability across 14 laboratories. Environ Mol Mutagen. 2011;52(9):690–8. doi: 10.1002/em.20672. [DOI] [PubMed] [Google Scholar]
- Dertinger SD, Phonethepswath S, Avlasevich SL, Torous DK, Mereness J, Bryce SM, Bemis JC, Bell S, Weller P, Macgregor JT. Efficient monitoring of in vivo pig-a gene mutation and chromosomal damage: summary of 7 published studies and results from 11 new reference compounds. Toxicol Sci. 2012;130(2):328–48. doi: 10.1093/toxsci/kfs258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dertinger SD, Avlasevich SL, Torous DK, Bemis JC, Phonethepswath S, Labash C, Carlson K, Mereness J, Cottom J, Palis J, MacGregor JT. Persistence of cisplatin-induced mutagenicity in hematopoietic stem cells: implications for secondary cancer risk following chemotherapy. Toxicol Sci. 2014;140(2):307–14. doi: 10.1093/toxsci/kfu078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatehouse D, Haworth S, Cebula T, Gocke E, Kier L, Matsushima T, Melcion C, Nohmi T, Venitt S, Zeiger E. Recommendations for the Performance of Bacterial Mutation Assays. Mutation Res. 1994;312:217–233. doi: 10.1016/0165-1161(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Gollapudi BB, Lynch AM, Heflich RH, Dertinger SD, Dobrovolsky VN, Froetschl R, Horibata K, Kenyon MO, Kimoto T, Lovell DP, Stankowski LF, Jr, White PA, Witt KL, Tanir JY. The in vivo Pig-a assay: A report of the International Workshop On Genotoxicity Testing (IWGT) Workgroup. Mutat Res Genet Toxicol Environ Mutagen. 2015;783:23–35. doi: 10.1016/j.mrgentox.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Kimoto T, Horibata K, Miura D, Chikura S, Okada Y, Ukai A, Itoh S, Nakayama S, Sanada H, Koyama N, Muto S, Uno Y, Yamamoto M, Suzuki Y, Fukuda T, Goto K, Wada K, Kyoya T, Shigano M, Takasawa H, Hamada S, Adachi H, Uematsu Y, Tsutsumi E, Hori H, Kikuzuki R, Ogiwara Y, Yoshida I, Maeda A, Narumi K, Fujiishi Y, Morita T, Yamada M, Honma M. The PIGRET assay, a method for measuring Pig-a gene mutation in reticulocytes, is reliable as a short-term in vivo genotoxicity test: Summary of the MMS/JEMS-collaborative study across 16 laboratories using 24 chemicals. Mutat Res. 2016;811:3–15. doi: 10.1016/j.mrgentox.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Kinoshita T. Biosynthesis and deficiencies of glycosylphosphatidylinositol. Proc Jpn Acad Ser B Phys Biol Sci. 2014;90(4):130–43. doi: 10.2183/pjab.90.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungland A, Laake K, Hoff E, Seeberg E. Spectrum of mutations induced by methyl and ethyl methanesulfonate at the hprt locus of normal and tag expressing Chinese hamster fibroblasts. Carcinogenesis. 1995;16(6):1281–5. doi: 10.1093/carcin/16.6.1281. [DOI] [PubMed] [Google Scholar]
- Krüger CT, Hofmann M, Hartwig A. The in vitro PIG-A gene mutation assay: mutagenicity testing via flow cytometry based on the glycosylphosphatidylinositol (GPI) status of TK6 cells. Arch Toxicol. 2015;89(12):2429–43. doi: 10.1007/s00204-014-1413-5. [DOI] [PubMed] [Google Scholar]
- Krüger CT, Fischer BM, Armant O, Morath V, Strähle U, Hartwig A. The in vitro PIG-A gene mutation assay: glycosylphosphatidylinositol (GPI)-related genotype-to-phenotype relationship in TK6 cells. Arch Toxicol. 2016;90(7):1729–36. doi: 10.1007/s00204-016-1707-x. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JC, Elebute MO. Stem cells in paroxysmal nocturnal haemoglobinuria and aplastic anaemia: increasing evidence for overlap of haemopoietic defect. Transfus Med. 2003;13(6):377–86. doi: 10.1111/j.1365-3148.2003.00465.x. [DOI] [PubMed] [Google Scholar]
- Moore MM, Honma M, Clements J, Bolcsfoldi G, Burlinson B, Cifone M, Clarke J, Clay P, Doppalapudi R, Fellows M, Gollapudi B, Hou S, Jenkinson P, Muster W, Pant K, Kidd DA, Lorge E, Lloyd M, Myhr B, O’Donovan M, Riach C, Stankowski LF, Jr, Thakur AK, Van Goethem F. Mouse Lymphoma Thymidine Kinase Mutation Assay: Meeting of the International Workshop on Genotoxicity Testing, San Francisco, 2005, Recommendations for 24-h Treatment. Mutation. Res. 2005;627(1):36–40. doi: 10.1016/j.mrgentox.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Moore MM, Clive D, Hozier JC, Howard BE, Batson AG, Turner NT, Sawyer J. Analysis of Trifluorothymidine-Resistant (TFTr) Mutants of L5178Y/TK+/− Mouse Lymphoma Cells. Mutation Res. 1985;151(1):161–174. doi: 10.1016/0027-5107(85)90194-0. [DOI] [PubMed] [Google Scholar]
- Morrison V, Ashby J. A preliminary evaluation of the performance of the Muta Mouse (lacZ) and Big Blue (lacI) transgenic mouse mutation assays. Mutagenesis. 1994;9(4):367–75. doi: 10.1093/mutage/9.4.367. [DOI] [PubMed] [Google Scholar]
- Nakamura J, Gul H, Tian X, Bultman SJ, Swenberg JA. Detection of PIGO-deficient cells using proaerolysin: a valuable tool to investigate mechanisms of mutagenesis in the DT40 cell system. PLoS One. 2012;7(3):e33563. doi: 10.1371/journal.pone.0033563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas JA, Carter EW, Albertini RJ. Both PIGA and PIGL mutations cause GPI-a deficient isolates in the Tk6 cell line. Environ Mol Mutagen. 2015;56(8):663–73. doi: 10.1002/em.21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T, Suzuki T, Masumura K. Recent advances in the protocols of transgenic mouse mutation assays. Mutat Res. 2000;455(1–2):191–215. doi: 10.1016/s0027-5107(00)00077-4. [DOI] [PubMed] [Google Scholar]
- Olsen AK, Dertinger SD, Krüger CT, Eide DM, Instanes C, Brunborg G, Hartwig A, Graupner A. The Pig-a Gene Mutation Assay in Mice and Human Cells: A Review. Basic Clin Pharmacol Toxicol. 2017 Sep;121(Suppl 3):78–92. doi: 10.1111/bcpt.12806. [DOI] [PubMed] [Google Scholar]
- Piberger AL, Krüger CT, Strauch BM, Schneider B, Hartwig A. BPDE-induced genotoxicity: relationship between DNA adducts, mutagenicity in the in vitro PIG-A assay, and the transcriptional response to DNA damage in TK6 cells. Arch Toxicol. 2017 Jun 7; doi: 10.1007/s00204-017-2003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees BJ, Tate M, Lynch AM, Thornton CA, Jenkins GJ, Walmsley RM, Johnson GE. Development of an in vitro PIG-A gene mutation assay in human cells. Mutagenesis. 2017;32(2):283–297. doi: 10.1093/mutage/gew059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M, Gollapudi BB, Thybaud V, Kim JH. Need and potential value of the Pig-a in vivo mutation assay-A HESI perspective. Environ Mol Mutagen. 2011;52(9):685–9. doi: 10.1002/em.20687. [DOI] [PubMed] [Google Scholar]
- Walker DM, McDonald JD, Meng Q, Kracko DA, Bauer MJ, Seilkop SK, Walker EL, Henderson RF, Walker VE. Measurement of plasma or urinary metabolites and Hprt mutant frequencies following inhalation exposure of mice and rats to 3-butene-1,2-diol. Chem Biol Interact. 2007;166(1–3):191–206. doi: 10.1016/j.cbi.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Ware RE, Nishimura J, Moody MA, Smith C, Rosse WF, Howard TA. The PIG-A mutation and absence of glycosylphosphatidylinositol-linked proteins do not confer resistance to apoptosis in paroxysmal nocturnal hemoglobinuria. Blood. 1998;92(7):2541–50. [PubMed] [Google Scholar]
- Wills JW, Long AS, Johnson GE, Bemis JC, Dertinger SD, Slob W, White PA. Empirical analysis of BMD metrics in genetic toxicology part II: in vivo potency comparisons to promote reductions in the use of experimental animals for genetic toxicity assessment. Mutagenesis. 2016;31(3):265–75. doi: 10.1093/mutage/gew009. [DOI] [PubMed] [Google Scholar]
- Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40(22):11189–201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]