Abstract
Imaging approaches that track biological molecules within cells are essential tools in modern biochemistry. Lipids are particularly challenging to visualize, as they are not directly genetically encoded. Phospholipids, the most abundant subgroup of lipids, are structurally diverse and accomplish many cellular functions, acting as major structural components of membranes and as signaling molecules that regulate cell growth, division, apoptosis, cytoskeletal dynamics, and numerous other physiological processes. Cells regulate the abundance, and therefore bioactivity, of phospholipids by modulating the activities of their biosynthetic enzymes. Thus, techniques that enable monitoring of flux through individual lipid biosynthetic pathways can provide key functional information. For example, the choline analogue propargylcholine (ProCho) can report on de novo biosynthesis of phosphatidylcholine by conversion to an alkynyl lipid that can be imaged following click chemistry tagging with an azido fluorophore. We report that ProCho is also a substrate of phospholipase D enzymes—which normally hydrolyze phosphatidylcholine to generate the lipid second messenger phosphatidic acid—in a trans-phosphatidylation reaction, generating the identical alkynyl lipid. By controlling the activities of phosphatidylcholine biosynthesis and phospholipase D enzymes, we establish labeling conditions that enable this single probe to selectively report on two different biosynthetic pathways. Just as nature exploits the economy of common metabolic intermediates to efficiently diversify biosynthesis, so can biochemists in interrogating such pathways with careful probe design. We envision that ProCho’s ability to report on multiple metabolic pathways will enable studies of membrane dynamics and improve our understanding of the myriad roles that lipids play in cellular homeostasis.
Phospholipids form the most abundant class of eukaryotic lipids, serving as major structural components of cellular membranes and critical signaling agents.1 These diverse functions necessitate vastly different scales, rates, and subcellular localizations of phospholipid biosynthetic reactions. For example, abundant phospholipids involved in membrane structure are produced at rates matching cell growth, division, and organelle biogenesis.2 In contrast, synthesis of low-abundance signaling lipids is tightly regulated.3,4
Among eukaryotic phospholipids, the most abundant are those containing a choline headgroup.1 These lipids, which include phosphatidylcholine (PC), ether phosphatidylcholine (ePC), and sphingomyelin (SM), are structural components of membrane bilayers. Additionally, PC and ePC are metabolic precursors to signaling lipids like lysophosphatidylcholine and phosphatidic acid (PA) via phospholipase A5 and D6 enzymes, respectively. PC is biosynthesized via the Kennedy pathway,7 wherein the primary alcohol choline is activated to cytidine 5′-diphosphocholine and then coupled to diacylglycerol (Figure 1A). Salic has exploited the plasticity8–10 of the PC biosynthetic machinery by using alkyne-bearing11 and azide-bearing12 choline analogues to produce bioorthogonally functionalized PC analogues within live cells (Figure 1B). Subsequent tagging with fluorophores or biotin via click chemistry enabled visualization of these analogues within cells.
Figure 1.
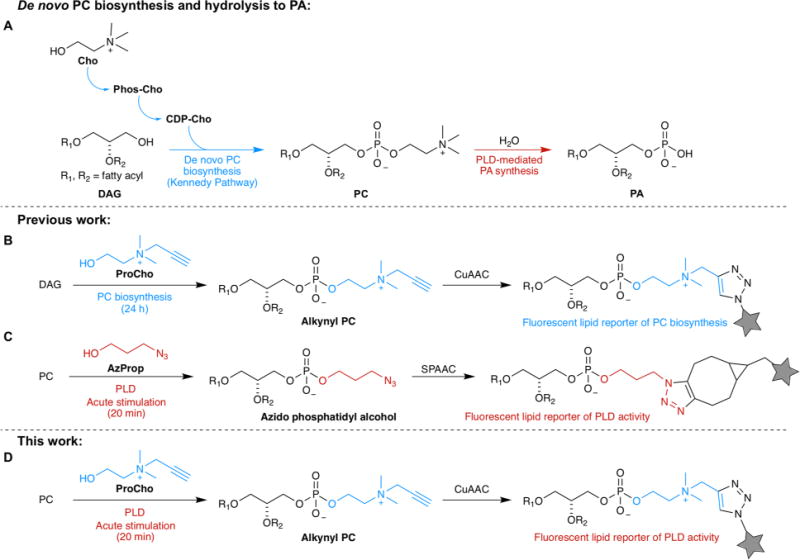
Strategy for using propargylcholine (ProCho) as a reporter for PLD activity and for PC biosynthesis. (A) Abbreviated biosynthetic pathways for phosphatidylcholine (PC) synthesis and phospholipase D (PLD)-mediated phosphatidic acid (PA) production. (B) Established strategy for using ProCho as a reporter for PC biosynthesis.11 (C) Established strategy for using small, linear, clickable primary alcohols such as 3-azido-1-propanol (AzProp) as reporters for PLD activity.16,17 (D) Strategy for repurposing ProCho as a reporter of PLD activity by controlling labeling conditions as indicated. Other abbreviations: Cho, choline; Phos-Cho, phosphocholine; CDP-Cho, cytidine 5′-diphosphocholine; DAG, diacylglycerol; CuAAC, copper-catalyzed azide–alkyne cycloaddition; SPAAC, strain-promoted azide–alkyne cycloaddition.
In contrast to the steady levels of PC biosynthesis, lipid second messengers such as PA are kept at low levels, and their production is tightly regulated spatiotemporally. PA is synthesized via several pathways but is maintained at very low concentrations.6 Production of PA pools for acute signaling purposes occurs by phospholipase D (PLD) and diacylglycerol kinase enzymes in response to upstream signaling from G protein-coupled receptors, tyrosine kinases, and intracellular GTPases and kinases.13
PLD enzymes use PC as the substrate for PA production, acting as hydrolases and releasing choline (Figure 1A). In a convenient enzymological quirk, PLDs also accept primary alcohols such as butanol or ethanol as the nucleophile for PC cleavage.14 Here, PLDs catalyze a transphosphatidylation reaction, producing phosphatidylalcohols rather than PA. This long-appreciated promiscuity of PLDs has enabled methods for quantifying PLD activity using labeling with radioisotopes14 or stable isotopes,15 with detection of phosphatidylalcohols by thin layer chromatography or mass spectrometry, respectively. We have demonstrated that PLDs accept alkynyl16 and azido17 alcohols to generate “clickable” phosphatidylalcohols whose localization can be visualized in cells following click chemistry-based fluorophore tagging in a method termed IMPACT (imaging phospholipase D activity with clickable alcohols via transphosphatidylation) (Figure 1C).
While developing IMPACT, we focused our efforts on linear, aliphatic clickable alcohols, whose structures resembled that of butanol. However, some microbial and plant-derived PLDs accept larger and hydrophilic primary alcohols for trans-phosphatidylation,18 raising the question of whether the larger and charged alcohol propargylcholine (ProCho), developed to report on PC biosynthesis,11 might also be a PLD substrate. We reasoned that if PLDs accept ProCho in transphosphatidylations, then we could use a single alcohol to interrogate two biosynthetic pathways operating on different time scales by controlling labeling time and stimulation (Figure 1D).
Herein, we validate ProCho as a chemical reporter of either PLD activity or choline-containing phospholipid biosynthesis. To explore the viability of using ProCho as a PLD reporter in cells, we first tested its utility as a PLD substrate in vitro using purified Arachis hypogaea (peanut) PLD. Reaction of dioleoylphosphatidylcholine with ProCho in the presence of A. hypogaea PLD, followed by lipid extraction and tagging with an azido-rhodamine 110 derivative (Az488) via copper-catalyzed azide–alkyne cycloaddition (CuAAC),19 resulted in a lipid-derived peak on fluorescence-coupled high-performance liquid chromatography (HPLC) (Figure 2A), which can be identified as the transphosphatidylation product as it was not present in a mock reaction without the enzyme.
Figure 2.
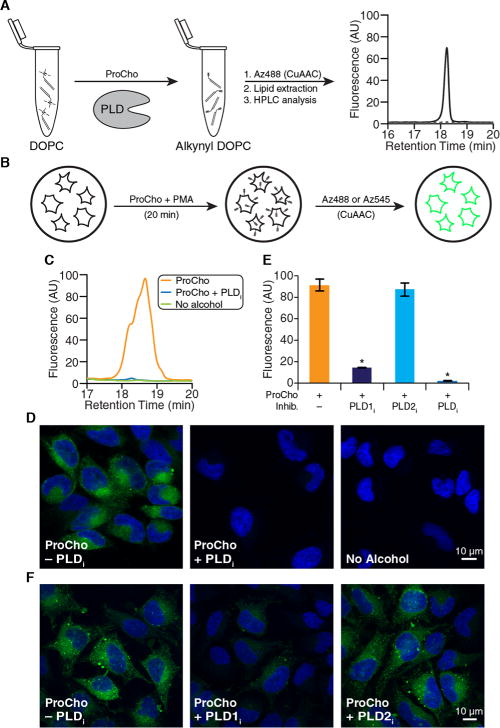
ProCho can report on acutely stimulated PLD activity. (A) HPLC analysis of in vitro transphosphatidylation of dioleoylphosphatidylcholine (DOPC) with ProCho in the presence (solid line) or absence (dotted line) of A. hypogaea PLD, following CuAAC tagging with Az488. (B) Schematic for labeling cellular PLD activity. (C–F) HeLa cells were treated with PLD inhibitors [FIPI (750 nM), VU0359595 (250 nM), or VU0364739 (350 nM)] for 30 min, followed by ProCho (100 μM) and PMA (100 nM) for 20 min. (C and E) Lipid extracts were subjected to CuAAC with Az488 and HPLC analysis. (D and F) Cells were fixed, tagged with Az545 via CuAAC, and imaged by confocal microscopy: green for CuAAC-derived fluorescence and blue for DAPI. (E) Quantification (area under curve) of labeled extracts tagged with Az488 via CuAAC and analyzed by HPLC. n = 3; *p < 0.05. Error bars indicate the standard error of the mean. Abbreviations: PLDi, FIPI; PLD1i, VU0359595; PLD2i, VU0364739; PMA, phorbol 12-myristate 13-acetate.
Having determined that ProCho is a PLD transphosphatidylation substrate in vitro, we evaluated whether human PLDs would accept ProCho within cells (Figure 2A). HeLa cells were treated with 100 μM ProCho, and PLD activity was acutely stimulated using phorbol 12-myristate 13-acetate (PMA) as an agonist to induce rates of PLD-mediated PA production greatly exceeding those of the Kennedy pathway.20 The labeling was performed in the absence or presence of 5-fluoro-2-indolyl des-chlorohalopemide21 (FIPI), a potent inhibitor of PLD1 and PLD2, the two PLD isozymes responsible for PA synthesis via PC hydrolysis. Cellular lipid extracts were prepared and tagged with Az488 via CuAAC (Figure 2B). In the absence of FIPI, a clear HPLC signal was observed that was not present in negative control samples either with FIPI or without ProCho (Figure 2C).
To visualize cellular PLD activity with ProCho, we treated HeLa cells with ProCho and PMA, but rather than proceeding with lysis and lipid extraction, we fixed the cells with paraformaldehyde to preserve their organelle and membrane morphology and tagged them with an azido-tetramethylrhodamine conjugate (Az545) via CuAAC, followed by confocal microscopy. The labeling was sensitive to FIPI and localized to several intracellular membranes, similar to IMPACT with alkynyl and azido alcohols (Figure 2D).
To establish that the signal was PLD-derived and to determine which of the two PLD isoforms is operative under this acute stimulation, we performed the labeling in the presence of isoform-selective inhibitors of PLD122 (VU0359595, or PLD1i) and PLD223 (VU0364739, or PLD2i). PLD1i treatment caused a loss of most of the fluorescent lipid signal, as quantified by HPLC, while PLD2i treatment had no significant effect (Figure 2E). The fact that the majority of the labeling derives from PLD1 is consistent with PMA’s mode of action via protein kinase C stimulation.13 The sensitivity of the PMA-stimulated, ProCho-derived labeling to PLD1i but not PLD2i was confirmed via microscopy following CuAAC tagging with Az545 (Figure 2F).
Until now, our studies had involved short ProCho labeling (20 min), allowing for maximal PMA-stimulated PLD activity. In developing IMPACT, however, we found that an alternative protocol, longer treatments of cells with 3-azido-1-propanol (AzProp) or hexyn-1-ol (2–3 h) in the absence of PMA, also resulted in clickable phosphatidylalcohol production.17 Under such conditions, these lipid reporters were the product of unstimulated, or basal, levels of PLD activity. Here, we have established that PLDs accept ProCho as a substrate. Thus, we were eager to investigate whether, when we replicated exactly the labeling protocol of Salic (ProCho labeling for 24 h), basal PLD activity was responsible for a substantial portion of the alkynyl PC analogues that were produced.
We thus treated HeLa cells with 100 μM ProCho for 24 h, generated lipid extracts, and tagged alkynyl lipids with Az488 via CuAAC (Figure 3A). Quantification of tagged lipids generated in the presence or absence of FIPI revealed no significant difference (Figure 3B,C), demonstrating that endogenous PLDs insignificantly contribute to alkynyl lipid production under this labeling protocol. In addition, this result was verified by fixation of cells labeled with ProCho for 24 h, followed by CuAAC tagging with Az545 and confocal microscopy analysis revealing that overall cellular fluorescence remained unchanged by the addition of FIPI to the cells (Figure 3D).
Figure 3.
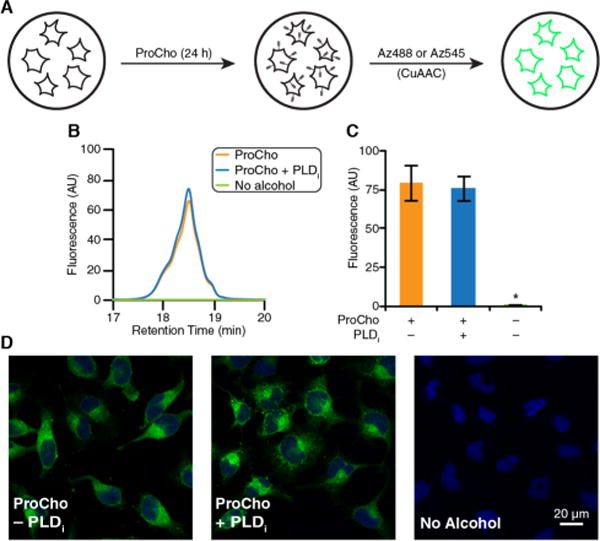
Long-time scale labeling with ProCho results in PLD-independent formation of alkynyl lipids. (A) Schematic of extended HeLa cell labeling with ProCho (100 μM). (B and C) Cells were labeled with ProCho as in panel A, and lipid extracts were generated, tagged with Az488 via CuAAC, and analyzed by HPLC. Shown are representative HPLC traces (B) and quantification (area under curve) (C). (D) Cells were labeled with ProCho as in panel A, fixed, tagged with Az545 via CuAAC, and imaged by confocal microscopy: green for CuAAC-derived fluorescence and blue for DAPI. n = 3; *p < 0.05. Error bars indicate the standard error of the mean.
Previously, we had established that IMPACT labeling with AzProp for 2 h results in phosphatidylalcohol production that is entirely dependent upon basal PLD activity. As an additional control, we treated cells with AzProp (100 μM) for 24 h by replacing ProCho with AzProp in the Salic protocol, again in the presence and absence of FIPI, to determine whether a longer labeling would result in the production of phosphatidylalcohols by non-PLD-mediated processes (e.g., Kennedy pathway enzymes). We found that this protocol resulted in azidolipid production that is entirely sensitive to FIPI (Figure S1), demonstrating that AzProp is an exclusive substrate of PLDs, unlike ProCho.
Having established the ability to monitor two distinct lipid biosynthetic pathways with a single reporter, we next turned to HPLC-coupled, high-resolution electrospray ioniziation–time-of-flight mass spectrometry-based lipidomics to verify the identities of the tagged lipids and analyze the distribution of lipid tails in the naturally occurring and ProCho-bearing lipids produced under each labeling regime. Treatment of HeLa cells with ProCho (100 μM) for 24 h resulted in alkynyl PC species with a distribution and relative abundance of acyl tail composition similar to those observed for natural PC in both cells treated with ProCho or natural PC in a control population that received no alcohol treatment (Figure S2A,B).
We also examined incorporation of ProCho into ePCs, a related class of phosphatidylcholines bearing an ether-linked fatty tail.24 We detected both natural and ProCho-modified ePCs and observed a similar level of agreement between alkynyl ePC and natural ePC species, though with a lower apparent level of incorporation of the unnatural ProCho headgroup into these lipids (Figure S3A,B). Largely, these data mirrored lipidomics analysis of ProCho labeling (100–500 μM for 24 h) by Salic.11
The previously published data were then compared to those collected from cells labeled acutely with ProCho and PMA for 20 min, i.e., under a protocol using ProCho as a reporter of PLD activity. The distribution of alkynyl PC species resulting from PLD-mediated transphosphatidylation (Figure S2C) resembles that of natural PC from either those cells (Figure S2C) or cells receiving only PMA stimulation and no alcohol (Figure S2D). We also detected molecular masses corresponding to alkynyl ePCs from this PLD-selective ProCho labeling protocol, suggesting that this enigmatic class of lipids may be substrates of cellular PLDs (Figure S3C,D). Collectively, these mass spectrometry studies demonstrate that ProCho can report on two different lipid biosynthetic pathways and that the distribution of ProCho-containing lipids roughly mirrors that of the corresponding choline-containing lipid precursors.
Finally, we performed two-color imaging experiments using AzProp and ProCho under two different labeling conditions to validate the localization of the acutely stimulated, PLD-derived, ProCho signal and to simultaneously image PC biosynthesis and sites of PLD activity within cells. First, acute labeling with both AzProp and ProCho concomitant with PMA stimulation for 20 min, followed by sequential tagging of azidolipids by SPAAC with a BODIPY-cyclooctyne conjugate25 (BODIPY-BCN) and then of alkynyl lipids by CuAAC with Az545, resulted in substantial colocalization of the fluorescent signals (Figure 4A,B). This high degree of colocalization was expected given that both alcohols are substrates of the same PLD isoforms under this labeling protocol. Second, we labeled cells with ProCho for 24 h to mark PC biosynthesis and subsequently labeled cells acutely with AzProp in the presence of PMA for 20 min to report on stimulated PLD activity. Again, following SPAAC and CuAAC tagging, we observed substantial colocalization (Figure 4C,D). This result fits well with the understanding that PC is the major substrate for cellular PLDs,6 whose activity is expected to correlate strongly with PC localization.
Figure 4.
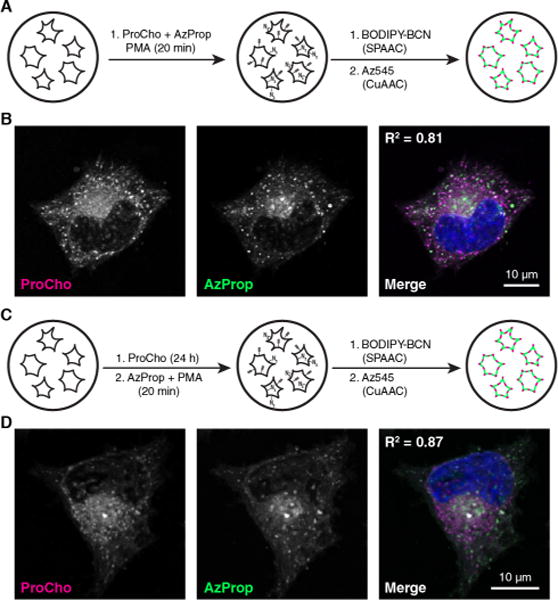
Two-color imaging of PLD activity and PC biosynthesis with orthogonal chemical reporters. (A) Schematic for simultaneous, two-color labeling of PLD activity with AzProp and ProCho. (B) HeLa cells were treated with ProCho (100 μM), AzProp (750 μM), and PMA (100 nM) for 20 min, rinsed, labeled with BODIPY-BCN via SPAAC, fixed, tagged with Az488 via CuAAC, and imaged by confocal microscopy. (C) Schematic of two-color labeling of PC biosynthesis and PLD activity. (D) HeLa cells were treated with ProCho (100 μM) for 24 h to report on PC biosynthesis, rinsed, and then incubated in AzProp (1 mM) for 20 min followed by an additional 20 min incubation with PMA (100 nM) to report on stimulated PLD activity. Cells were then fixed, labeled via SPAAC and CuAAC, and imaged as in panel B. The merge image shows ProCho (magenta), AzProp (green), and DAPI (blue); colocalization of ProCho and AzProp appears white. Pearson correlation coefficients (R2) are provided to aid in interpreting colocalization of the two markers.
In summary, just as nature exploits the economy of common metabolic intermediates to efficiently diversify biosynthetic pathways, so can biochemists in the development of chemical probes to interrogate such pathways. Our results demonstrate that a simple alkynyl choline analogue can serve as a reporter of PMA-stimulated PLD activity or the biosynthesis of choline-containing phospholipids, by careful control of labeling conditions. Future applications may include its use to study flux through these pathways as stimulated by a variety of endogenous and exogenous agents. This property of a single alcohol to report on multiple biochemical pathways based on exposure time and activation state of the cell expands the ability to interrogate the dynamics of lipid synthesis and membrane biogenesis within cells and better understand the many roles that biological membranes and their lipid constituents play in cellular homeostasis. We propose that this dual functionality may extend to other choline analogues and potentially to bioorthogonally tagged metabolic precursors to other classes of lipids, metabolites, and macromolecules.
Supplementary Material
Acknowledgments
The Cornell NMR facility is supported by National Science Foundation Grant MRI-CHE-1531632. The authors thank Alex Brown for providing VU0359595 and VU0364739.
Funding
We thank the National Institutes of Health (R00GM110121 to J.M.B.), the Arnold and Mabel Beckman Foundation (Beckman Young Investigator, J.M.B.), and the National Science Foundation (GRFP, DGE-165-0441, T.W.B.).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b01021.
Figures S1–S3 and Materials and Methods (PDF)
Author Contributions
T.W.B. and F.J.L. performed experiments. T.W.B. and J.M.B. conceived of experiments, interpreted data, and wrote the manuscript.
Notes
The authors declare no competing financial interest.
References
- 1.Vance JE. Traffic. 2015;16(1):1–18. doi: 10.1111/tra.12230. [DOI] [PubMed] [Google Scholar]
- 2.Gibellini F, Smith TK. IUBMB Life. 2010;62(6):414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- 3.Antal CE, Newton AC. Mol Cell Proteomics. 2013;12:3498–3508. doi: 10.1074/mcp.R113.029819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balla T. Physiol Rev. 2013;93(3):1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke JE, Dennis EA. J Lipid Res. 2009;50:S237–S242. doi: 10.1194/jlr.R800033-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selvy PE, Lavieri RR, Lindsley CW, Brown HA. Chem Rev. 2011;111(10):6064–6119. doi: 10.1021/cr200296t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy EP, Weiss SB. J Biol Chem. 1956;222(1):193–214. [PubMed] [Google Scholar]
- 8.Bieber LL, Newburgh RW. J Lipid Res. 1963;4(4):397–401. [PubMed] [Google Scholar]
- 9.Hodgson E, Dauterman WC. J Insect Physiol. 1964;10(6):1005–1008. [Google Scholar]
- 10.Bridges RG, Ricketts J. J Insect Physiol. 1970;16(4):579–593. doi: 10.1016/0022-1910(70)90092-2. [DOI] [PubMed] [Google Scholar]
- 11.Jao CY, Roth M, Welti R, Salic A. Proc Natl Acad Sci U S A. 2009;106(36):15332–15337. doi: 10.1073/pnas.0907864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jao CY, Roth M, Welti R, Salic A. ChemBioChem. 2015;16(3):472–476. doi: 10.1002/cbic.201402149. [DOI] [PubMed] [Google Scholar]
- 13.Bruntz RC, Lindsley CW, Brown HA. Pharmacol Rev. 2014;66(4):1033–1079. doi: 10.1124/pr.114.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang SF, Freer S, Benson AA. J Biol Chem. 1967;242(3):477–484. [PubMed] [Google Scholar]
- 15.Brown HA, Henage LG, Preininger AM, Xiang Y, Exton JH. Methods Enzymol. 2007;434:49–87. doi: 10.1016/S0076-6879(07)34004-4. [DOI] [PubMed] [Google Scholar]
- 16.Bumpus TW, Baskin JM. Angew Chem, Int Ed. 2016;55(42):13155–13158. doi: 10.1002/anie.201607443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bumpus TW, Baskin JM. ACS Cent Sci. 2017;3(10):1070–1077. doi: 10.1021/acscentsci.7b00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto Y, Hosokawa M, Miyashita K. Water. 2009:317–338. [Google Scholar]
- 19.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem. 2002;114(14):2708–2711. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 20.Halenda SP, Rehm AG. Biochem J. 1990;267:479–483. doi: 10.1042/bj2670479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su W, Yeku O, Olepu S, Genna A, Park J, Ren H, Du G, Gelb MH, Morris AJ, Frohman MA. Mol Pharmacol. 2009;75(3):437–446. doi: 10.1124/mol.108.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis JA, Scott SA, Lavieri R, Buck JR, Selvy PE, Stoops SL, Armstrong MD, Brown HA, Lindsley CW. Bioorg Med Chem Lett. 2009;19(7):1916–1920. doi: 10.1016/j.bmcl.2009.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavieri RR, Scott SA, Selvy PE, Kim K, Jadhav S, Morrison RD, Daniels JS, Brown HA, Lindsley CW. J Med Chem. 2010;53(18):6706–6719. doi: 10.1021/jm100814g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honsho M, Fujiki Y. FEBS Lett. 2017;591(18):2720–2729. doi: 10.1002/1873-3468.12743. [DOI] [PubMed] [Google Scholar]
- 25.Alamudi SH, Satapathy R, Kim J, Su D, Ren H, Das R, Hu L, Alvarado-Martínez E, Lee JY, Hoppmann C, Peña-Cabrera E, Ha HH, Park HS, Wang L, Chang YT, Melorose J, Perroy R, Careas S. Nat Commun. 2016;7:11964. doi: 10.1038/ncomms11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


