Abstract
Background
The Phosphatase and tensin homolog deleted on chromosome TEN (Pten) is implicated in a broad range of developmental events and diseases. However, its role in neural crest and craniofacial development has not been well illustrated.
Results
Using genetically engineered mouse models, we showed that inactivating Pten specifically in neural crest cells causes malformation of craniofacial structures. Pten conditional knockout mice exhibit perinatal lethality with overgrowth of craniofacial structures. At cellular level, Pten deficiency increases cell proliferation rate and enhances osteoblast differentiation. Our data further revealed that inactivating Pten elevates PI3K/AKT signaling activity in neural crest derivatives, and confirmed that attenuation of PI3K/AKT activity led to decreased neural crest cell proliferation and differentiation both in vitro and in vivo.
Conclusions
Our study revealed that Pten is essential for craniofacial morphogenesis in mice. Inactivating Pten in neural crest cells increases proliferation rate and promotes their differentiation towards osteoblasts. Our data further indicate that Pten acts via modulating PI3K/AKT activity during these processes.
Keywords: Pten, PI3K/Akt, neural crest cells, proliferation, differentiation
Introduction
Neural crest cells (NCCs) are a unique cell population and play an important role during vertebrate development. They are originated as ectodermal cells at the border of neural plate at the dorsal neural tube and transform into mesenchymal cells by epithelial-mesenchymal transformation (EMT). Subsequently, NCCs migrate extensively from the dorsal region to the ventral destinations, where they differentiate into a broad range of cell types and contribute to organogenesis (Selleck and Bronner-Fraser, 1995).
Craniofacial morphogenesis is a highly orchestrated process and is dependent on NCCs (Chai and Maxson, 2006). Lineage studies reveal that cranial NCCs contribute predominantly to the craniofacial mesenchyme (Chai et al., 2000; Jiang et al., 2000; Yoshida et al., 2008). During craniofacial development, NCCs give rise to cartilage, bone, glial cells, peripheral neurons, pigment cells and connective tissues. NCC proliferation and differentiation are under rigorous regulation of a genetic network composed of multiple growth factors, receptors and their intracellular effectors (Knecht and Bronner-Fraser, 2002; Sauka-Spengler and Bronner-Fraser, 2008; Cordero et al., 2011).
The Phosphatase and Tensin homolog deleted on chromosome TEN (Pten) was discovered as a tumor suppressor as its mutation was frequently identified in human tumors (Li and Sun, 1997; Steck et al., 1997). Pten is a lipid phosphatase with a primary role in antagonizing the phosphatidylinositol-3-kinase (PI3K) signaling pathway, which is implicated in multiple processes including cell survival, proliferation, and differentiation (Steelman et al., 2004). Upon stimulation by growth factors, PI3K family members phosphorylate the second messenger phosphatidylinositol 4,5-bisphosphate (PIP2) into phosphatidylinositol 3,4,5-trisphosphate (PIP3), which activates downstream signaling cascades. Pten modulates PI3K signaling activity by converting PIP3 to PIP2 during this process. Besides its primary role of PI3K signaling inhibitor, Pten is also implicated in autophagy signaling of cancer cells (Arico et al., 2001; Errafiy et al., 2013).
Pten also plays a crucial role for embryonic development (Di Cristofano et al., 1998; Salmena et al., 2008). Pten null mutant mice die by E7.5, and heterozygotes develop phenotypes in the prostate, skin and colon, indicating an essential role for Pten in these tissues (Di Cristofano et al., 1998). In addition, accumulating evidence has demonstrated a role for Pten in development of retina, brain, neural stem cells and other tissues (Veleva-Rotse and Barnes, 2014). In the developing craniofacial tissues, previous studies showed that phosphorylated Pten is expressed in palate and tooth mesenchymal cells, as well as in oral and dental epithelial cells (Cho et al., 2008; Kero et al., 2016). However, the role of Pten in neural crest and craniofacial development still remains to be fully elucidated.
In the present study, we showed that Pten regulates neural crest development and craniofacial morphogenesis in mice. Interestingly, inactivating Pten in neural crest cells elevates Akt phosphorylation level in both trigeminal ganglia and craniofacial skeletons, but causes distinct phenotypes. In the trigeminal ganglia, inactivating Pten increases the rate of cellular proliferation, while in the premaxilla, Pten deficiency enhances both cell proliferation and osteoblast differentiation. On the other hand, inhibiting PI3K/AKT signaling activity in primary cells rescues Pten mutant phenotype. We further confirm these findings in Pdgfra PI3K signaling mutant mice. In summary, our study showed that Pten regulates neural crest proliferation and differentiation via modulating PI3K/AKT signaling pathway during mammalian craniofacial development.
Results
Pten is essential for mouse neural crest development and craniofacial morphogenesis
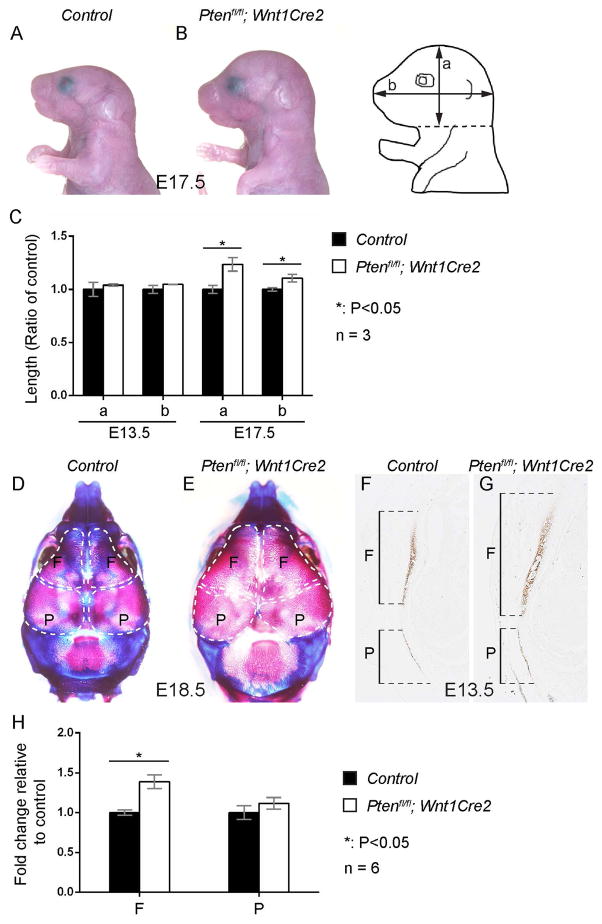
To investigate the role of Pten in neural crest development and craniofacial morphogenesis, we generated neural crest-specific Pten conditional knockout mice Ptenfl/fl; Wnt1Cre2 by crossing Ptenfl/fl (Groszer et al., 2001) with Wnt1Cre2 transgenic line (Lewis et al., 2013). Ptenfl/fl; Wnt1Cre2 mice exhibit 100% perinatal lethality with enlarged heads (Fig 1A–B; n=3). Morphometric analysis revealed that Ptenfl/fl; Wnt1Cre2 embryo heads are comparable to their littermate controls at E13.5 (Fig 1C, n=3). At E17.5, Ptenfl/fl; Wnt1Cre2 embryo heads are 15±3% larger along the dorsal-ventral axis (a) and 34±5% larger along the anterior-posterior axis (b), while the length of their trunks is comparable to that of the control (data not shown, n=3). Skeletal preparations confirmed that Ptenfl/fl; Wnt1Cre2 embryos exhibit an enlarged skull at E18.5 (Fig 1D–E, n=3). Morphometric analysis of the E18.5 skulls revealed that neural crest derived frontal bone of mutant embryos is 39±8% (n=6, P<0.05) larger that that of littermate controls, and mesoderm derived parietal bone of mutant embryos are 12±7% larger than controls (n=6, p<0.05). However, at E13.5, only mutant frontal bones are larger than the control (1.39±0.08 fold, n=6, P<0.05), while mutant parietal bones and the control show comparable length (1.08±0.02 fold, n=6, P>0.05). These results indicate that Pten plays an autonomous role in neural crest-derived frontal bone development, and the enlarged parietal bone at E18.5 is likely secondary to malformation of other structures.
Fig 1. Inactivating Pten in NCCs leads to an enlarged skull.
(A, B) Lateral view of the littermate control (A) and Ptenfl/fl; Wnt1Cre2 (B) at E17.5. (C) Morphometric quantification and statistical analysis of the control and Ptenfl/fl; Wnt1Cre2 embryos along the dorsal-ventral axis (a) and anterior-posterior axis (b) at E13.5 and E17.5 (n=3). (D, E) Dorsal view of skeletal preparations of littermate control (D) and Ptenfl/fl; Wnt1Cre2 (E) at E18.5. (F, G) Immunostaining with anti-Sp7 antibody on transverse sections of the control and Ptenfl/fl; Wnt1Cre2 embryos at E13.5. (H) Quantification and statistical analysis of the frontal bone and parietal bone length of F and G. F, frontal bone. P, parietal bone.
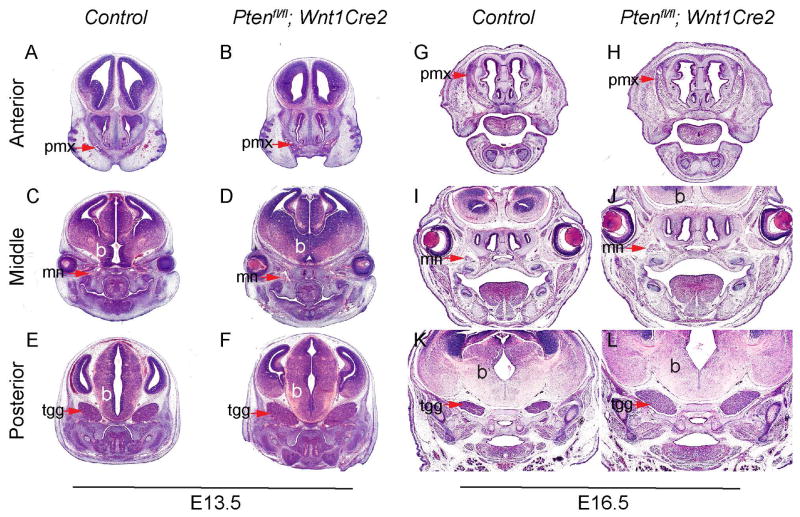
To reveal the details of Ptenfl/fl; Wnt1Cre2 craniofacial phenotype, we performed histological analysis on mutant embryos and littermate controls at E13.5 and E16.5. Hematoxylin/Eosin staining on frontal sections of E13.5 embryos showed that Ptenfl/fl; Wnt1Cre2 and the control are comparable at the anterior level (Fig 2A, B). At the middle and posterior level, the midbrain of the mutant is larger than that of control (Fig 2C–F), consistent with previous studies that report Pten is important for brain development (Groszer et al., 2001; Marino et al., 2002; Yue et al., 2005; Fraser et al., 2008; Lehtinen et al., 2011). Interestingly, we observed that the trigeminal ganglion (tgg) is enlarged in the mutant (Fig 2E, F). At E16.5, the mutant premaxilla appears larger than the control (Fig 2G, H), and the regions of maxillary nerves (mn) and tgg are enlarged in the mutant embryos when compared to the littermate controls (Fig 2I–L).
Fig 2. Ptenfl/fl; Wnt1Cre2 embryos exhibit overgrowth of craniofacial tissues.
H&E staining on frontal sections of littermate control and Ptenfl/fl; Wnt1Cre2 at E13.5 (A–F) and at E16.5 (G–L). Arrows point to pmx in A, B, G and H, to mn in C, D, I and J, and to tgg in E, F, K and L. b, brain; mn, maxillary nerve; pmx, premaxilla; tgg, trigeminal ganglion.
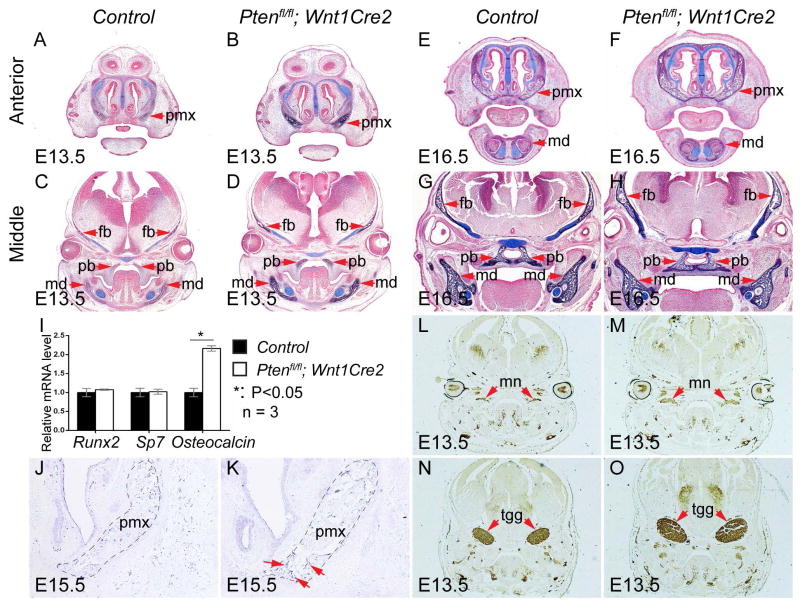
Cranial neural crest cells give rise to the bones, cartilages, and neurons. To identify the cell types of mutant tissues showing abnormality, we conducted alkaline phosphatase-Alcian blue (AP-AB) staining on control and mutant embryos. Our results confirmed elevated AP activity in mutant premaxilla, frontal bones, mandible and palatine bones (Fig 3A–H), and the cartilage formation was not altered. Real time PCR results showed that osteoblast markers Runx2 and Sp7 exhibit comparable expression levels in the control and mutant, while expression level of Osteocalcin is significantly increased in the mutant embryos (Fig 3I). Increased Osteocalcin expression was further confirmed by section in situ hybridization in the mutant pmx at E15.5 (Fig 3J, K), suggesting that inactivating Pten promotes osteoblast maturation. Immunostaining for neurofilaments revealed increased neuronal formation in mutant mn and tgg at E13.5 (Fig. 3L–O). These results indicate that Pten controls osteogenesis and neuron formation during craniofacial morphogenesis.
Fig 3. Pten deficiency causes enhanced osteoblast differentiation and neurogenesis.
(A–H) AP-AB staining on frontal sections of littermate control and Ptenfl/fl; Wnt1Cre2 embryos at E13.5 (A–D) and at E16.5 (E–H). Sections were counterstained with nuclear fast red. (I) Quantification result of osteogenesis markers expression of the control and Ptenfl/fl; Wnt1Cre2 mRNA samples prepared from E13.5 embryos (n=3). (J, K) In situ hybridization with Osteocalcin antisense probe on the frontal sections of the control (J) and Ptenfl/fl; Wnt1Cre2 (K) embryos at E15.5. (L–O) Immunostaining with anti-Neurofilament-L antibody on frontal sections of littermate control (L, N) and Ptenfl/fl; Wnt1Cre2 (M, O) embryos at E13.5. fb, frontal bone; md, mandible; mn, maxillary nerve; pb, palatine bone; pmx, premaxilla; and tgg, trigeminal ganglion.
Pten transcripts are expressed in craniofacial tissues in developing mouse embryos
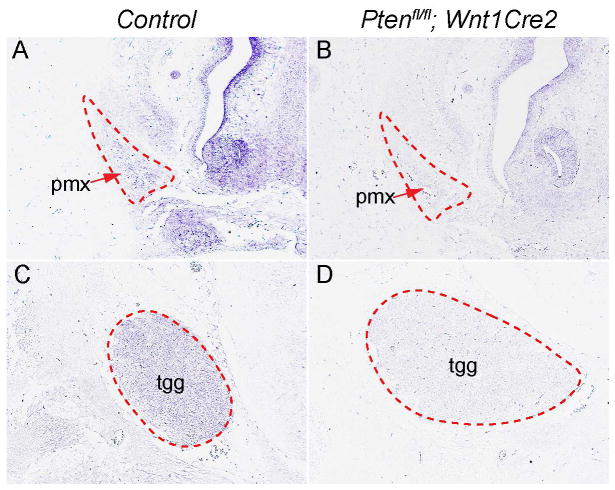
Next we examined the expression pattern of Pten transcripts in the developing craniofacial tissues. In situ hybridization results revealed that Pten mRNA is expressed in the premaxilla and tgg of the control embryos (Fig. 4A, C). In Ptenfl/fl; Wnt1Cre2 mice, Pten expression was efficiently deleted in both tissues (Fig 4B, D). These data indicated an autonomous role for Pten in regulating premaxilla and tgg development.
Fig 4. Expression pattern of Pten mRNA in developing craniofacial structures.
(A–D) In situ hybridization using Pten antisense probe on frontal sections of wild type (A, C) and of Ptenfl/fl; Wnt1Cre2 (B, D) embryos at E13.5. pmx, premaxilla; tgg, trigeminal ganglion.
Inactivating Pten in neural crest cells increases cell proliferation rate of pmx osteoprogenitors and tgg
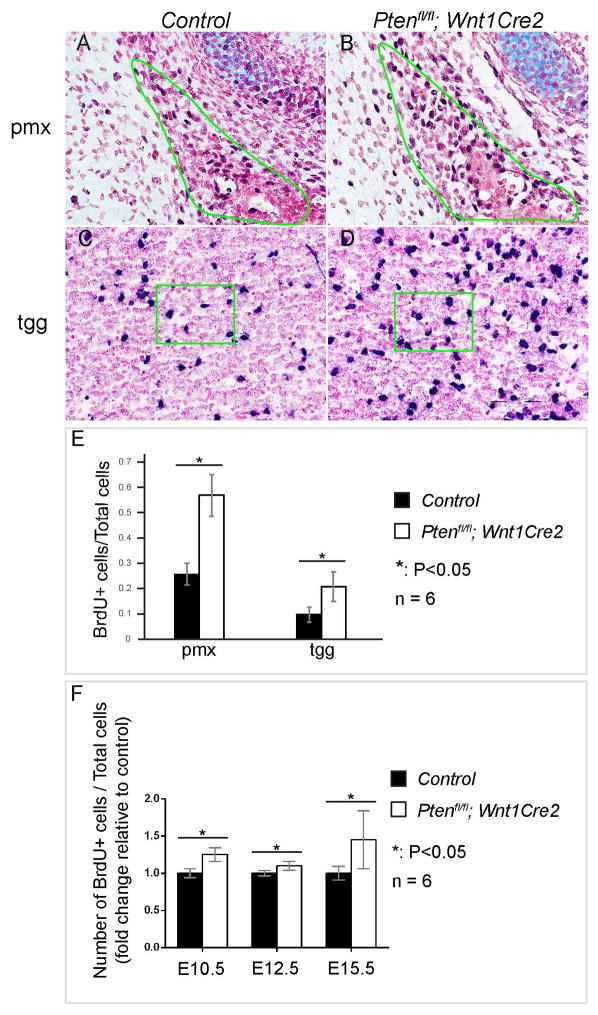
To understand the cellular mechanisms underlying Ptenfl/fl; Wnt1Cre2 craniofacial phenotype, we assayed the cell proliferation rate, with a focus on the pmx osteoblasts and tgg neurons. BrdU labeling assay showed that at E13.5, 26±4% of pmx cells and 10±3% of tgg cells are proliferating in the control embryos; in the mutant, 57±8% of pmx cells and 21±6% of tgg cells are proliferating (Fig 5A–E, n=6, P<0.05). Further analysis showed that mutant pmx cells shows higher proliferation rate than control at E10.5 (1.3±0.1 fold), E12.5 (1.1±0.1 fold) and E15.5 (1.5±0.4 fold) (n=6, P<0.05). These results indicate that Pten controls cell proliferation rate in these neural crest derived tissues.
Fig 5. Pten is essential to regulate proliferation of osteoprogenitors and tgg cells.
(A–D) BrdU labeling on frontal sections of littermate control (A, C) and Ptenfl/fl; Wnt1Cre2 (B, D) at E13.5, with a focus on pmx (A, B) and tgg (C, D). Slides were counterstained with nuclear fast red to facilitate quantification. (E) Quantification result and statistical analysis of percentage of BrdU+ cells in control and Ptenfl/fl; Wnt1Cre2 tissues. (F) Quantification and statistical analysis of the percentage of BrdU+ cells in Ptenfl/fl; Wnt1Cre2 and littermate control pmx at E10.5, E12.5 and E15.5. The result is presented as fold change relative to the control. pmx, premaxilla; tgg, trigeminal ganglion.
Inactivating Pten enhances AKT signaling activity in neural crest derivatives
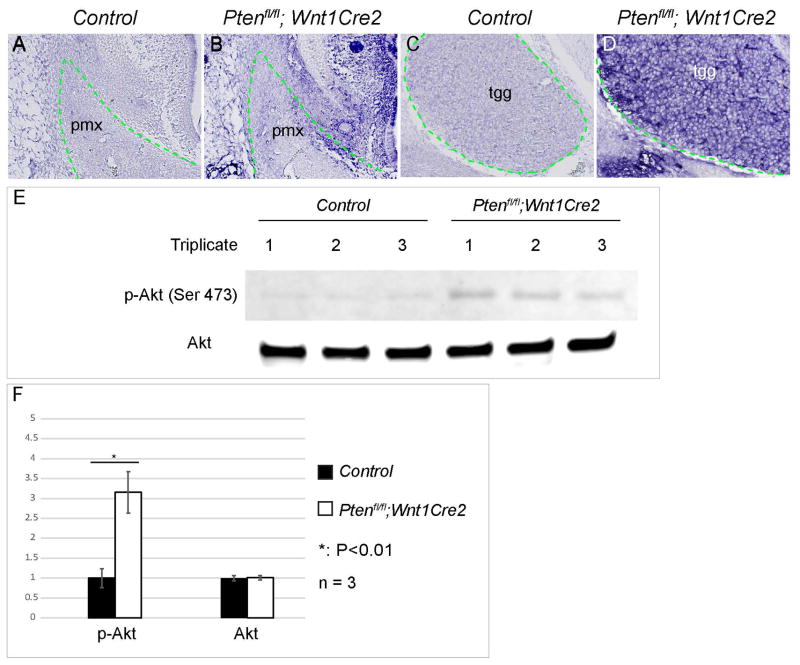
To examine Pten downstream signaling pathways in neural crest development, we assayed both autophagy and PI3K/AKT signaling activity. Our data showed that expression of autophagy markers LC3-B and ubiquitin were not altered in mutant craniofacial structures (data not shown). Previous studies showed that in multiple cell types, Pten functions via antagonizing PI3K/AKT signaling pathway, which has been identified as an important player of craniofacial development (Fantauzzo and Soriano, 2015). Therefore, we examined if PI3K/AKT signaling activity is altered in Ptenfl/fl; Wnt1Cre2 embryos. Immunohistochemistry results showed that expression of phosphorylated AKT is maintained at basal level in the control embryos, but is significantly increased in Ptenfl/fl; Wnt1Cre2 pmx and tgg (Fig 6A, B, C and D). Western blot confirmed a 3.2±0.5 fold increase of phosphorylated AKT expression in the mutant pmx lysates (Fig 6E and F, n=3). Our data showed that Pten regulates the phosphorylation of AKT in vivo during craniofacial morphogenesis, and indicated that Pten might control NCC development via AKT signaling.
Fig 6. Pten deficiency increases PI3K/AKT signaling activity in neural crest cells.
(A–D) Immunostaining with anti phospho-AKT antibody on frontal sections of littermate control embryos (A, C) and Ptenfl/fl; Wnt1Cre2 (B, D) at E13.5. (E) Western blot using anti phospho-AKT antibody (p-Akt Ser473) and craniofacial lysates from littermate control and Ptenfl/fl; Wnt1Cre2 embryos at E13.5. (F) Quantification result and statistical analysis of western blot data on E. pmx, premaxilla; tgg, trigeminal ganglion.
Inhibiting AKT signaling rescues Pten mutant cell proliferation and differentiation
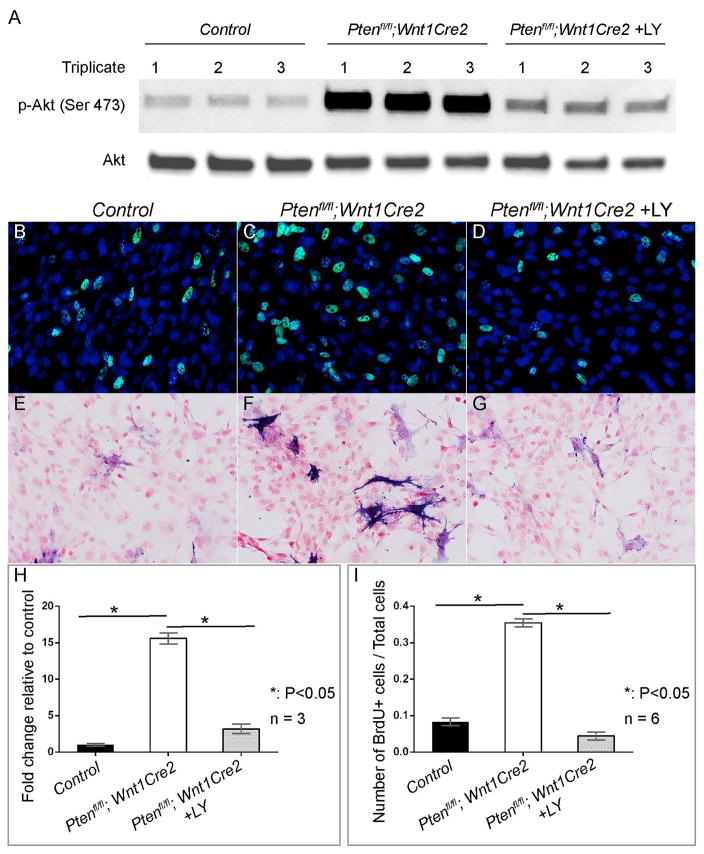
To examine the role of AKT signaling in neural crest cells, we isolated the premaxilla tissues from E13.5 embryos and used them for primary cell culture. Western blot results showed that expression level of phosphorylated AKT of the mutant cells is significantly higher than the control (15.6±0.3 fold change), and treating mutant cells with AKT signaling inhibitor LY294002 (LY) decreases phosphorylated AKT expression (4.9±0.2 fold change relative to the mutant) (Fig 7A, B, H). BrdU labeling and AP staining showed that LY treatment inhibits cell proliferation rate (from 35±1% to 4%±1%) (Fig 7B–D, I; n=3) and AP activity (Fig 7E–G) of mutant cells. These results indicate that AKT signaling plays an important role in neural crest cell proliferation and differentiation, and Pten acts via AKT signaling to regulate neural crest cells.
Fig 7. Inhibiting PI3K/AKT signaling rescues Pten mutant neural crest cells phenotype in vitro.
(A) Western blot of phospho-AKT and total AKT in primary pmx cells prepared from littermate control and Ptenfl/fl; Wnt1Cre2 embryos at E13.5. (B–D) BrdU labeling of primary pmx cells of littermate control (B), Ptenfl/fl; Wnt1Cre2 (C) and Ptenfl/fl; Wnt1Cre2 treated with 10uM LY for 30 minutes (D). BrdU+ cells are labeled with green fluorescence and total nuclei are stained with DAPI (blue). (E–G) AP staining of primary E13.5 pmx cells of littermate control (E), Ptenfl/fl; Wnt1Cre2 (F) and Ptenfl/fl; Wnt1Cre2 treated with 10uM LY for 8 hours (G). AP positive cells are blue and nuclei are counterstained with nuclear fast red. (H) Quantification and statistic analysis of western blot result of (A). (I) Quantification and statistic analysis of BrdU labeling results of (B–D).
Attenuation of PI3K signaling affects neural crest cell proliferation and osteogenesis in vivo
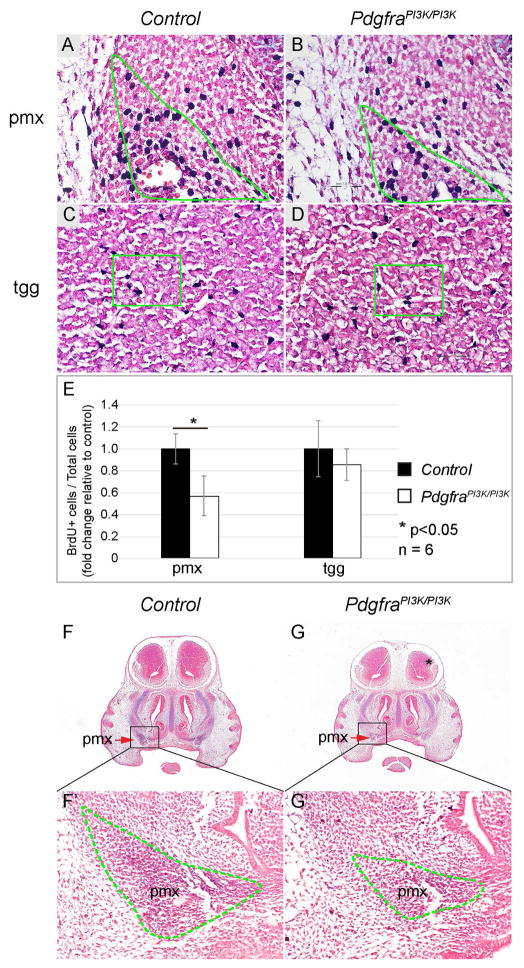
To examine if AKT mediates Pten regulation of neural crest development in vivo, we analyzed PdgfraPI3K/PI3K embryos, in which the ability of Pdgfra to activate PI3K signaling is abrogated (Klinghoffer et al., 2002). BrdU labeling showed that cell proliferation rate of pmx oestoprogenitors is decreased in PdgfraPI3K/PI3K embryos compared to the control (57±18%) (Fig 8A, B and E, n=4, P<0.05). On the other hand, cell proliferation rate of tgg is not significantly altered in PdgfraPI3K/PI3K mice (86±14%) (Fig 8C, D and E, n=6, P>0.05). AP activity in the pmx of PdgfraPI3K/PI3K embryo at E13.5 is attenuated compared to the control (Fig 8F, F′, G, and G′). These results confirmed that PI3K signaling is critical to maintain neural crest cell proliferation and osteoblast differentiation in vivo.
Fig 8. Attenuation of PI3K/AKT signaling affects NCCs proliferation and differentiation during craniofacial development.
(A–D) BrdU labeling on frontal sections of littermate control (A, C) and PdgfraPI3K/PI3K (B, D) embryos at E13.5, with a focus on the pmx (A, B) and tgg (C, D). Slides were counterstained with nuclear fast red to facilitate quantification. (E) Quantification and statistical analysis of percentile of BrdU+ cells in the control and PdgfraPI3K/PI3K tissues. The result is presented as fold change relative to the control. (F, G) AP-AB staining on frontal sections of littermate control (F) and PdgfraPI3K/PI3K (G) embryos at E13.5. (F′, G′) Magnification of defined area in F and G. pmx, premaxilla; tgg, trigeminal ganglion.
Discussion
Originally discovered as a tumor suppressor gene, Pten is now recognized as an important player in development of multiple tissues. Previous studies assayed Pten expression in developing orafacial tissues (Cho et al., 2008; Kero et al., 2016), but it remains unclear if Pten is required for normal craniofacial morphogenesis. Our study showed that Pten plays critical roles in craniofacial development by regulating neural crest cell proliferation and differentiation. Our data also indicates that Pten might function via inhibiting PI3K/AKT signaling during mouse craniofacial development.
Pten is important for brain development. The brain overgrowth phenotype we found in Ptenfl/fl; Wnt1Cre2 mice is consistent with previous reports in Pten conditional knockout using other Cre lines expressed in embryonic brains (Groszer et al., 2001; Marino et al., 2002; Yue et al., 2005; Fraser et al., 2008; Lehtinen et al., 2011). The craniofacial phenotype of Ptenfl/fl; Wnt1Cre2 mice could thus be intrinsic, or secondary to the brain malformation, or combination of both. In situ hybridization showed that Pten transcripts are detected in the osteoprogenitors support the notion that Pten plays a primary role in these cells development (Fig 4). In addition, our data revealed that both cell proliferation and AKT phosphorylation were altered in the pmx bones of Ptenfl/fl; Wnt1Cre2 embryos (Fig 5 and Fig 6). Although these findings cannot exclude the effect of brain overgrowth on craniofacial phenotype in the mutant, it highlights the autonomous role for Pten in neural crest and craniofacial development.
There are two pathways to form a bone. In long bones, the mesenchymal cells form a cartilaginous template for osteoblasts through endochondral ossification pathway. In calvarial and partial facial bones, osteoprogenitors differentiate into osteoblasts via the intramembranous pathway. Pten has been implicated in both these processes. During endochondral ossification, Pten was found to be essential for chondrocyte patterning and differentiation in the growth plate of long bones, as well as proliferation of osteoblast progenitors (Ford-Hutchinson et al., 2007; Yang et al., 2008; Hsieh et al., 2009; Guntur et al., 2011). In intramembranous ossification, inactivation of Pten using osteocalcin-Cre led to increased calvarial bone density (Zhang et al., 2002). Neural crest cells give rise to the entire facial skeleton. Our data showed that deletion of Pten in neural crest cells elevated proliferation rate and enhanced differentiation in osteoprogenitors of pmx (Fig 3 and Fig 5). However, we did not observe an obvious phenotype in the facial cartilages (Fig 3A–H).
Platelet derived growth factor receptors (PDGFRs) are major regulators of PI3K signaling in craniofacial development (Klinghoffer et al., 2002). PdgfraPI3K/PI3K mice exhibit decreased proliferation of neural crest cells during craniofacial development (He and Soriano, 2013; Fantauzzo and Soriano, 2014; Fantauzzo and Soriano, 2015), and PDGFAA treatment stimulates neural crest cell proliferation and differentiation via PI3K/AKT pathway (Vasudevan et al., 2015). All of the studies supported the notion that PI3K signaling is critical for craniofacial development. Our data showed that Ptenfl/fl; Wnt1Cre2 mice exhibited a reversed phenotype to PdgfraPI3K/PI3K in proliferation and differentiation of neural crest-derived osteoprogenitors (Fig 3, 5 and 8). These results confirmed the essential role for PI3K in craniofacial development. Interestingly, cell proliferation rate in Ptenfl/fl; Wnt1Cre2 tgg is increased significantly but not affected in that of PdgfraPI3K/PI3K mice. This result suggests that PDGFRa might play a dispensable role in the regulation of PI3K/AKT signaling and cell proliferation in tgg or Pten might act via other pathways to control tgg development.
Experimental Procedures
Animals
All animal experimentation was approved by the Institutional Animal Care and Use Committee of Tulane University. Tg(Wnt1-cre)2Sor (Lewis et al., 2013), referred to as Wnt1Cre2; Ptentm1Hwu (Groszer et al., 2001), referred to as Pten+/fl; and Pdgfratm5Sor (Klinghoffer et al., 2002), referred to as PDGFRα+/PI3K, were maintained on a C57BL/6J/129S4 mixed genetic background. Vaginal plug of the females was checked daily in the morning. Noon on the day when vaginal plug was found was considered embryonic day (E) 0.5. Genotyping was done by PCR on tail lysates following protocol described in the references.
Skeletal preparations
E18.5 embryos were dissected in PBS followed by removal of the skin and internal organs. After incubation in 95% ethanol overnight at room temperature, skeletons were stained with staining solution (0.005% alizarin red, 0.015% alcian blue and 5% glacial acetic acid in 70% ethanol) for 2 days at 37°C. The skeletons were then rinsed briefly in 95% ETOH at room temperature, and then cleared with 1% KOH overnight. Samples were then equilibrated in gradient washes of glycerol/KOH mixture until glycerol concentration reached 80%.
Histology and AP-AB staining
Timed embryos were harvested in ice-cold PBS. Embryos were decapitated and heads were fixed in 4% paraformaldehyde (PFA) in PBS for 18 – 24 hours at 4°C. Following dehydration with graded washes of ethanol, samples were embedded in paraffin and processed for 10μm frontal sections and subjected to standard Hematoxylin/Eosin staining or Alkaline Phosphatase and Alcian Blue (AP-AB) double staining. For AP-AB staining, sections were deparaffinized in Histo-clear followed by graded ethanol and incubated in 0.03% nitro-blue tetrazolium chloride (NBT) and 0.02% 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (BCIP) to detect AP activity. Slides were then rinsed in water and dipped in 1% alcian blue 8GX (A5268, Sigma) in 0.1N HCl to label cartilage, and counterstained with 0.1% nuclear fast red solution.
In situ hybridization and immunohistochemistry
In situ hybridization was carried out as described previously (He and Soriano, 2013). Pten antisense RNA probes was generated using primers: 5-TTACAGTTGGGCCCCGTACA-3 and 5- TAAAACACCCACACAATGACAAGA-3. For immunohistochemistry, samples were sectioned at 10μm and subjected to standard protocols using primary antibodies to anti-phospho-Akt (1:25, Cell Signaling Technology 9271), anti-Neurofilament-L (1:100, Cell Signaling Technology 2837), HRP-conjugated secondary antibody (1:100, Cell Signaling Technology 7074), anti-Sp7(1:200, Abcam ab94744) and AP-conjugated secondary antibody (1:100, Novus Biologicals, NB7157).)
Quantitative PCR
Expression of Runx2, Sp7 and Osteocalcin mRNA were analyzed by real-time quantitative RT-PCR. The premaxilla and adjacent medial nasal tissues were dissected from E13.5 embryos in ice cold PBS and total RNA was extracted using the RNeasy Mini Kit (Qiagen 74104). First strand cNDA was synthesized using 1μg of total RNA, oligo-dT primers and SuperScript III RT (Invitrogen 18080). Quantitative PCR was performed on Bio-Rad iCycler using SYBR super mix kit (Bio-Rad 170-8880). The following protocol was used: step 1: 95°C for 10 min; step 2: 95°C for 10 s; step 3: 55° for 30 s; repeat to step 2 39× (40 cycles in total). PCR products were run on agarose gel to confirm correct amplicon size. β-actin expression was used as an internal control. The primers are as follows: mouse Runx2, 5′-CCCAGCCACCTTTACCTACA-3′ and 5′-TATGGAGTGCTGCTGGTCTG-3′; mouse Sp7, 5′-AGTTCACCTGCCTGCTCTGT-3′ and 5′-GGAGCTGGAGACCTTCCTCT-3′; mouse osteocalcin, 5′-AAGCAGGAGGGCAATAAGGT-3′ and 5′-ACTTGCAGGGCAGAGAGAGA-3′; mouse β-actin, 5′-GCAAGTGCTTCTAGGCGGAC-3′ and 5′-AAGAAAGGGTGTAAAACGCAGC-3′.
Cell proliferation assay
For mice, BrdU was administered into pregnant female mice by intraperitoneal (I.P.) injection at 30 ug/g body weight. Embryos were dissected after 1 hour of injection, fixed in Carnoy’s solution for 1 hour, dehydrated through graded ethanol washes, cleared in Histo-clear and embedded in paraffin. Frontal sections were cut at 10μm. Immunostaining for BrdU was performed using a Bromodeoxyuridine (BrdU) Labeling and Detection Kit (Roche, 11299964001). At least three embryos for each genotype and each stage were analyzed. BrdU labeled cells were counted in the pmx area in mutant and control samples. Data from three continuous sections of each embryo were collected for statistical analysis.
For cells, primary cells were incubated with 10 ug/ml BrdU for 30 minutes and were washed in PBS. The cells were then fixed in 4% PFA followed by incubation in 1N HCl for 10 minutes. Subsequently, the cells were blocked with 10% donkey serum for 1 hour, followed by incubation with anti-BrdU antibody (1:200, Novus Biologicals, NBP2-14890) for 1 hour and Alexa FluoR 448 donkey anti-rabbit IgG secondary antibody (Abcam, ab150073). DAPI was used for nuclei staining. BrdU labeled cells were counted in comparable areas in mutant and control samples. Data collected from at least three independent experiments were subject for statistical analysis using student’s t-test. Statistical data are presented as mean ± SEM, and subjected to double tailed Student’s t-tests.
Western blot
E13.5 embryos were dissected in ice-cold PBS. With the brain removed, the embryonic head was homogenized by sonication in ice-cold NP-40 buffer with proteinase inhibitors including protease inhibitor cocktail (Roche, 11836153001), 1 mM PMSF, 10 mM NaF, 1 mM Na3VO4 and 25 mM β-glycerophosphate. Tissue lysates were then centrifuged at 12000 rpm at 4°C. Supernatants were collected and protein concentrated was determined by BCA assay. Western blot was carried out using the primary antibodies from Cell Signaling Technology: Anti-β-actin (8457), anti-phospho-Akt (9271), and the secondary antibody from LI-COR Biosciences: IRDye 680RD (925-68071). An Odyssey CLx imaging system was used for result recording. ImageJ software was used to quantify the western blot result.
Acknowledgments
This work is supported by NIH/NIDCR grant R00DE024617 to F.H.
We thank YiPing Chen for the constructive comments. We are grateful to our laboratory colleagues for their comments and discussion on the manuscript. This work is supported by NIH/NIDCR grant R00DE024617 to F.H.
References
- Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Cho KW, Cho SW, Lee JM, Lee MJ, Gang HS, Jung HS. Expression of phosphorylated forms of ERK, MEK, PTEN and PI3K in mouse oral development. Gene Expr Patterns. 2008;8:284–290. doi: 10.1016/j.gep.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Cordero DR, Brugmann S, Chu Y, Bajpai R, Jame M, Helms JA. Cranial neural crest cells on the move: their roles in craniofacial development. Am J Med Genet A. 2011;155A:270–279. doi: 10.1002/ajmg.a.33702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Errafiy R, Aguado C, Ghislat G, Esteve JM, Gil A, Loutfi M, Knecht E. PTEN increases autophagy and inhibits the ubiquitin-proteasome pathway in glioma cells independently of its lipid phosphatase activity. PLoS ONE. 2013;8:e83318. doi: 10.1371/journal.pone.0083318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantauzzo KA, Soriano P. PI3K-mediated PDGFRalpha signaling regulates survival and proliferation in skeletal development through p53-dependent intracellular pathways. Genes Dev. 2014;28:1005–1017. doi: 10.1101/gad.238709.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantauzzo KA, Soriano P. Receptor tyrosine kinase signaling: regulating neural crest development one phosphate at a time. Curr Top Dev Biol. 2015;111:135–182. doi: 10.1016/bs.ctdb.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford-Hutchinson AF, Ali Z, Lines SE, Hallgrimsson B, Boyd SK, Jirik FR. Inactivation of Pten in osteo-chondroprogenitor cells leads to epiphyseal growth plate abnormalities and skeletal overgrowth. J Bone Miner Res. 2007;22:1245–1259. doi: 10.1359/jbmr.070420. [DOI] [PubMed] [Google Scholar]
- Fraser MM, Bayazitov IT, Zakharenko SS, Baker SJ. Phosphatase and tensin homolog, deleted on chromosome 10 deficiency in brain causes defects in synaptic structure, transmission and plasticity, and myelination abnormalities. Neuroscience. 2008;151:476–488. doi: 10.1016/j.neuroscience.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- Guntur AR, Reinhold MI, Cuellar J, Jr, Naski MC. Conditional ablation of Pten in osteoprogenitors stimulates FGF signaling. Development. 2011;138:1433–1444. doi: 10.1242/dev.058016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Soriano P. A critical role for PDGFRalpha signaling in medial nasal process development. PLoS Genet. 2013;9:e1003851. doi: 10.1371/journal.pgen.1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh SC, Chen NT, Lo SH. Conditional loss of PTEN leads to skeletal abnormalities and lipoma formation. Mol Carcinog. 2009;48:545–552. doi: 10.1002/mc.20491. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Kero D, Cigic L, Medvedec Mikic I, Galic T, Cubela M, Vukojevic K, Saraga-Babic M. Involvement of IGF-2, IGF-1R, IGF-2R and PTEN in development of human tooth germ - an immunohistochemical study. Organogenesis. 2016;12:152–167. doi: 10.1080/15476278.2016.1197460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinghoffer RA, Hamilton TG, Hoch R, Soriano P. An Allelic Series at the PDGFalphaR Locus Indicates Unequal Contributions of Distinct Signaling Pathways During Development. Dev Cell. 2002;2:103–113. doi: 10.1016/s1534-5807(01)00103-4. [DOI] [PubMed] [Google Scholar]
- Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat Rev Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, D’Ercole AJ, Wong ET, LaMantia AS, Walsh CA. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AE, Vasudevan HN, O’Neill AK, Soriano P, Bush JO. The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev Biol. 2013;379:229–234. doi: 10.1016/j.ydbio.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DM, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- Marino S, Krimpenfort P, Leung C, van der Korput HA, Trapman J, Camenisch I, Berns A, Brandner S. PTEN is essential for cell migration but not for fate determination and tumourigenesis in the cerebellum. Development. 2002;129:3513–3522. doi: 10.1242/dev.129.14.3513. [DOI] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Selleck MA, Bronner-Fraser M. Origins of the avian neural crest: the role of neural plate-epidermal interactions. Development. 1995;121:525–538. doi: 10.1242/dev.121.2.525. [DOI] [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- Steelman LS, Bertrand FE, McCubrey JA. The complexity of PTEN: mutation, marker and potential target for therapeutic intervention. Expert Opin Ther Targets. 2004;8:537–550. doi: 10.1517/14728222.8.6.537. [DOI] [PubMed] [Google Scholar]
- Vasudevan HN, Mazot P, He F, Soriano P. Receptor tyrosine kinases modulate distinct transcriptional programs by differential usage of intracellular pathways. Elife. 2015:4. doi: 10.7554/eLife.07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleva-Rotse BO, Barnes AP. Brain patterning perturbations following PTEN loss. Front Mol Neurosci. 2014;7:35. doi: 10.3389/fnmol.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Sun Q, Teng Y, Li F, Weng T, Yang X. PTEN deficiency causes dyschondroplasia in mice by enhanced hypoxia-inducible factor 1alpha signaling and endoplasmic reticulum stress. Development. 2008;135:3587–3597. doi: 10.1242/dev.028118. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Yue Q, Groszer M, Gil JS, Berk AJ, Messing A, Wu H, Liu X. PTEN deletion in Bergmann glia leads to premature differentiation and affects laminar organization. Development. 2005;132:3281–3291. doi: 10.1242/dev.01891. [DOI] [PubMed] [Google Scholar]
- Zhang M, Xuan S, Bouxsein ML, von Stechow D, Akeno N, Faugere MC, Malluche H, Zhao G, Rosen CJ, Efstratiadis A, Clemens TL. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem. 2002;277:44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]