SUMMARY
A novel bacterium was isolated from the subgingival plaque of a patient with periodontal disease. Bacterial strain BA112T is a facultative gram-positive coccus. It metabolizes alanine, arginine, glycine, histidine, leucine, proline, serine, and tyrosine, but does not appear to utilize carbohydrates. Urease, esculin, indole, catalase, and nitrate reduction tests were all negative. Major cellular fatty acids were C18:0, C12:0, C16:0, C18:1 w9c, and C20:0. The genome was sequenced and is 2.4 Mbp in length and has 64% GC content. Based on phylogenetics of the 16S rRNA sequence and concatenated alignments of 37 conserved proteins, BA112T belongs to the family Actinomycetaceae but is located on a branch of the tree without currently named members. Based on our phenotypic and phylogenetic studies, we propose that BA112T is the first known representative of a new genus, for which the name Peptidiphaga gingivicola gen. nov., sp. nov. is proposed. The type strain is BA112T.
Keywords: Actinobacteria, genome, cultivation, phylogenetics
INTRODUCTION
Periodontitis results from interactions between mammalian host cells and complex subgingival bacterial communities. Studies using molecular methods have demonstrated that many of the bacterial species that inhabit the oral cavity remain to be cultivated 1,2 and have provided evidence for the association of previously uncultivated phylotypes with disease 3–6. A better understanding of the biology of these uncultivated bacteria is essential to our understanding of the biofilm community dynamics in both health and disease. This study describes a novel bacterium isolated from the subgingival plaque of a patient with periodontal disease. In a previous investigation this bacterium was more commonly found in subjects with periodontitis than in periodontally healthy subjects 4. Based on the phylogenetic and phenotypic results presented below, we propose that this isolate, referred to in this paper as BA112T, is sufficiently unique to warrant a new genus and species designation, Peptidiphaga gingivicola gen. nov., sp. nov.
METHODS
Sample collection and plating
Subgingival plaque was obtained with paper points, which were transported to the lab in Liquid Dental Transport Medium (Anaerobe Systems, Morgan Hill, CA). The patient provided informed consent via a signed statement before participation. The human subjects protocol was approved by the Ohio State University Institutional Review Board (protocol 2007H0064). After a 104–fold dilution in prereduced BHI, samples were plated on Brucella blood agar (Anaerobe Systems) at 37°C in an atmosphere of air and 5% CO2. Single colonies were subcultured to ensure purity.
Culture identification
16S rRNA gene sequencing was performed to identify colonies of interest. For DNA isolation, colonies were collected from blood agar plates and incubated at 56°C for at least two hours with 10% proteinase K in Buffer ATL (Qiagen, Valencia, CA), and then homogenized for 1 minute with 0.25 g of 0.1 mm glass beads in a BioSpec Products BeadBeater. Genomic DNA was then isolated using the QIAamp DNA Mini Kit (Qiagen). The 16S rRNA gene was amplified by PCR using the semi-universal bacterial primers A17 (5′-GTT TGA TCC TGG CTC AG-3′) (E. coli positions 11–27) and 317 (5′-AAG GAG GTG ATC CAG CC-3′) (E. coli positions 1551–1537) (Biosynthesis, Lewisville, TX), as described previously 7. After purification with the QIAquick® PCR Purification kit (Qiagen), the PCR product was sequenced by the Sanger method, using an ABI Prism cycle sequencing kit (BigDye Terminator Cycle Sequencing) and an ABI 3730 instrument.
Whole genome sequencing and assembly
We processed the genomic DNA for whole genome sequencing by fragmentation in a Covaris S2 ultrasonicator (Covaris, Woburn, MA) and library preparation with the NEBNext Ultra Kit (New England Biolabs, Ipswich, MA) and sequenced it in the Illumina HiSeq 2500 with 100 bp paired end reads. The 17.1 million reads were trimmed with Trimmomatic 0.33 8, and assembled with SPAdes 3.5 9. After removing artifactual low coverage contigs we retained 6 contigs. We checked the assembly by visual inspection of the read alignments with IGV 10. This assembly included a complete copy of the rRNA operon within a contig. Based on the level of coverage and patterns of mapping of read pairs, it appeared that there were two additional copies of the rRNA repeat that connected other fragments. Additionally there was a pattern of mismatching of read pairs that indicated a misassembly in which one large contig was likely to be inserted within another. There were two further apparent misassemblies, again detected by read pair mapping anomalies, that were apparently due to collapsed repeats. We were able to generate a probable assembly for those regions using Bandage 11. We therefore were able to generate a three contig assembly for the Peptidiphaga genome, with each contig ending in a copy of the ribosomal operon. The orientation of the contigs is known but the relative order is uncertain and we have not been able to generate sufficient high molecular weight genomic DNA to discern this arrangement. We checked the genome with REAPR 12 and although it flagged some possible anomalous regions, none of them seemed to be misassemblies by visual inspection of the mapped reads. The model of the genome we produced aligned extensively with the genome of Actinomyces oral taxon 848 (NCBI RefSeq accession NZ_ACUY00000000.2) that had been independently sequenced by others using 454 technology. A set of 12 pairs of PCR primers was used to amplify between unique flanking regions and the rRNA operons and across the corrected misassembly regions and all gave expected size fragments.
Phylogenetics
The NCBI refseq sequence database was searched for genomes from the genera Actinobaculum, Actinotignum, Arcanobaculum, Trueperella, Mobiluncus, and Actinomyces. Because of the large number of species in genus Actinomyces, we limited those to species found in the oral cavity. We extracted 16S rRNA gene sequences with Rnammer 13 from complete genomes or downloaded them directly. 16S alignments were performed by SSU-ALIGN 0.1 14, while concatenated protein alignments were extracted from each genome with the command “phylosift all –isolate –besthit” (phylosift 1.0.1) and the alignments in the “concat.updated.1.fasta” files for each genome were combined to generate the multiple alignment. Masked alignments were converted from Stockholm format with BioPython 15, inspected in Mesquite 3.04 16, trimmed to E. coli positions 29 and 1389, manually adjusted if indicated, and output in the required formats. The CIPRES science gateway 17 was used to run RAxML-HPC BlackBox (8.2.6) 18 and MrBayes 3.2.6 19 to generate phylogenetic trees. Average amino acid identity (AAI) between species was determined using CompareM 20.
RESULTS
Colonies of BA112T became visible after about 4 days on Brucella blood agar, and were round, raised, smooth, off-white, and shiny, with a slight indentation on top. They ranged from about 0.2–0.9 mm in diameter. On subculture, the isolate grew equally well in an anaerobic atmosphere (85% N2, 10% H2, 5% CO2), or in air/5% CO2. Growth in ambient air was poor. After about 5 days, colonies became waxy and solid. Gram stains, performed with a kit from BD (Franklin Lakes, NJ), showed gram-positive cocci, many in small groupings of 2–5 cells. To assess the temperature-dependence of growth, BA112T was grown anaerobically in Peptone-Yeast Broth with starch (PY-S, Anaerobe Systems, Morgan Hill, CA). Growth rate was maximal between 33° and 40°C, with no growth at either 23° or 45°. The pH optimum, assessed in Brain-Heart Infusion broth (BHI, BD), was 6.0–7.0. Growth was prevented by incubating freshly inoculated broth at 80°C for 10 min., indicating that no spores were formed.
Biochemical characterizations of substrate utilization and enzyme activity were performed using the API test kits rapid ID 32Strep, rapid ID 32A, Coryne, and ZYM. Cells metabolized alanine, arginine, glycine, histidine, leucine, proline, serine, and tyrosine. Phenylalanine use was variable. Tests were also positive for acetoin production, acid phosphatase, alanyl-phenylalanyl-proline arylamidase, and naphthol-AS-BI-BD-phosphohydrolase. Leucyl-glycine arylamidase activity was variable. The following substrates were not utilized: arabinose, fucose, mannose, D-arabitol, glucose, glycogen, lactose, maltose, mannitol, ribose, saccharose, xylose, gelatin, glutamic acid, melezitose, melibose, pullulane, raffinose, sorbitol, tagatose, trehalose, valine. Urease and esculin were negative, nitrate was not reduced, and indole was not produced. Catalase was negative. The following tests were also negative: A- and B-galactosidase, A- and B-Glucosidase, chymotrypsin, fucosidase, alkaline phosphatase, A- and B-mannosidase, arginine dihydrolase, B-galactosidase-6-phosphate, B-glucuronidase, cystine arylamidase, esterase, esterase lipase, glycyl-tryptophan arylamidase, hippurate hydrolysis, lipase, Methyl-BD-Glucopyranoside Acidification, N-Acetyl-B-Glucosaminidase, pyrazinamidase, pyrolidonyl arylamidase, and trypsin. Tests that distinguish BA112T from the Actinobaculum species and their phylogenetic relative, Trueperella (Arcanobacterium) abortisuis, are shown in Table 1. Complete results for BA112 T and A. suisT can be found in Supplemental Table S1. Results for A. suisT in this study were consistent with those previously published, except where noted.
Table 1.
Phenotypic characteristics* that distinguish BA112T from other related species. 1, BA112 T; 2, A. suisT; 3, A. massiliense; 4, A. schaalii; 5, A. urinale; 6, T. abortisuis. Symbols: +, positive; −, negative; v, variable.
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| β-glucuronidase | − | + | − | + | + | |
| Alkaline phosphatase | − | + | − | − | − | |
| Alanyl-phenylalanyl-proline arylamidase | + | + | + | − | ||
| Pyroglutamic acid arylamidase | − | + | + | − | ||
| Urease | − | + | − | − | v | − |
| α-glucosidase | − | + | + | + | − | + |
| Glucose | − | +# | + | + | + | + |
| Ribose | − | − | + | + | + | + |
| Maltose | − | −# | + | + | + | |
| Mannitol | − | +# | − | − | − | |
| Lactose | − | − | − | − | + | |
| Xylose | − | − | + | + | − | − |
| Glycogen | − | + | + | − | − | + |
Analysis of cellular fatty acid composition (FAME) was performed by Microbial ID (Newark, DE) using the MIDI Sherlock® Microbial Identification System. The major fatty acid components were: C18:0 (41.57%), C12:0 (16.04%), C16:0 (11.86%), C18:1 1 9c (7.75%), and C20:0 (7.17%). Lesser amounts of the following were found: C10:0 (3.53%), C14:0 (1.64%), C18:1 1 7c (1.64%), and C17:0 (1.23%). Summed features were C18:2 1 6,9c/C18:0 ante (3.91%) and C16:1 1 7c/C16:1 6c (1.01%). This differs from the Actinobaculum and Actinomyces genera, in which C16:0 and unsaturated C18 fatty acids predominate 21,22 (and CCUG culture collection information).
The 1,438 bp 16S rRNA sequence obtained was compared to 16S sequences in the GenBank database using the BLASTN program available through the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/blast/). The closest matches were the uncultured Actinobaculum sp. clone 7BB627 (FJ976277) and the unnamed cultivated isolate Actinomyces oral taxon 848 (GQ131416), both at 99% similarity. The closest named species, Actinobaculum massiliense, (Accession number AF487679), had a similarity value of 90%, indicating that BA112T is a novel species 23.
We subsequently performed whole genome sequencing with the Illumina technology and performed de novo genome assembly as described in Methods. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession LVZK00000000. Version LVZK01000000 is described in this paper. The raw data is associated with BioProject PRJNA317166. We were able to derive a three contig genome with the aid of assembly graph inspection, pair read mapping and designed PCR probes. The remaining ambiguity was associated with the order of three unique regions between rRNA repeats, and one base within the 23S rRNA genes (nucleotide 1563) which 72% of reads identify as C and 27% T. It is likely that there are 2 alleles of the 23S rRNA gene. We calculated average nucleotide identity (ANI) and aligned fraction (AF) of complete genomes with tools on the IMG-ER web site 24, comparing BA112T with related sequenced genomes. BA112T had 98.94 % ANI and 0.92 AF when compared with the unnamed sequenced isolate “Actinomyces sp. oral taxon 848 F0322” (NCBI RefSeq NZ_ACUY00000000.2). It has been suggested that thresholds of 96.5% ANI and 0.6 AF define bacterial species 25, therefore by this criterion these two isolates represent members of the same species. All other related genomes fell well below these thresholds with the closest being Actinobaculum sp. oral taxon 183 F0552 at 76.57% ANI and 0.53 AF.
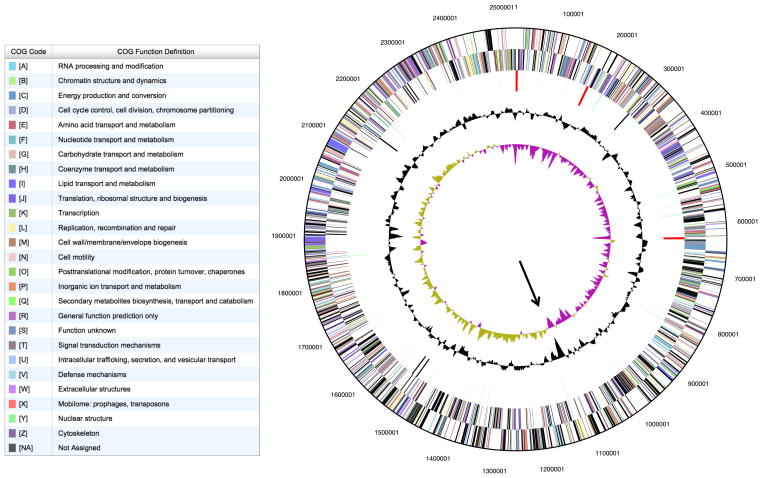
The genome was annotated using the IMG-ER system and was found to comprise 2,524,828 bp with a GC content of 64.4% (Figure 1). There were 2065 predicted protein coding sequences and 60 RNA encoding genes including three ribosomal operons as discussed above. There are two regions with CRISPR repeats and CRISPR associated proteins. The observed lack of growth on carbohydrate substrates was not easily explained by the genome sequence as the organism contains genes for glycolysis and sugar transport. It also has genes for an F1F0 type membrane ATPase and an NADH:ubiquinone oxidoreductase together with fumarate reductase and a nitrite reductase which may be involved in anaerobic respiration. The genome also appears to code for a type VII secretion system as also found in Mycobacteria and other gram positive organisms 26, however in BA112T the WXG100 genes that encode the putative secreted substrates are located in a separate genomic location.
Fig. 1.
Circular map of a model of the genome of BA112T. From outside to inside, the tracks represent: (1) Protein coding genes on the forward strand, colored by COG as indicated (2) Protein coding genes on the reverse strand (3) RNA coding genes with rRNAs red (4) GC content (5) GC skew. The arrow points to the putative origin, determined through GC skew and the presence of dnaA and rpmH genes 31. Note that the position of the unique contigs between the rRNA repeats has not been determined and that they might be switched from the picture, although the orientation should be maintained.
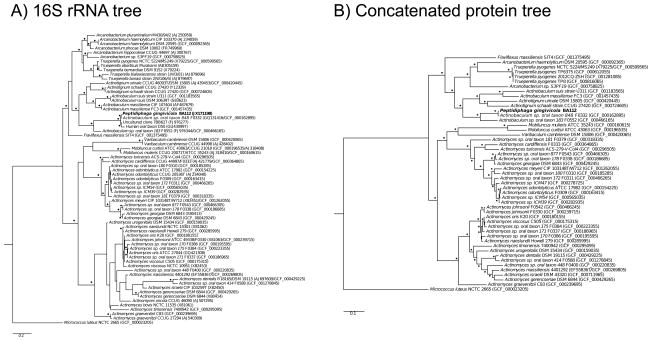
We compared the genomic sequence of BA112T to the genomes from related bacteria to clarify the phylogenetic relationships among organisms in three ways. The first used 16S rRNA genes from type strains and strains with sequenced genomes aligned with SSU-ALIGN 14 (Figure 2A), while the second used a concatenated alignment of 37 conserved proteins that was generated by the PhyloSift program as suggested by Rinke et al. 27,28 (Figure 2B). Figure 2 shows trees generated by the RAxML maximum likelihood method 18. The trees predicted by either protein or 16S rRNA alignments were essentially the same, as were additional trees calculated with the MrBayes Bayesian inference method (Supplemental Figure 1) 19. Finally we determined the AAI values between BA114 and related species using compareM. These clustered in ranges that agree with the tree topology. AAI between BA114 and Actinotignum schaalii, Actinotignum urinale, Actinotignum timonense, Actinobaculum suis, and Actinobaculum massiliense ranged from 59.68% to 61.21%. AAI between BA114 and Actinobaculum sp. oral taxon 183 was 70.93%, while AAI between BA114 and Actinomyces sp. oral taxon 848 was 98.51%.
Fig. 2.
Dendrograms based on A) 16S rRNA sequences and B) 37 concatenated protein sequences showing the phylogenetic relationship between BA112 T, the type strains of closely related species, strains with sequenced genomes, and a few closely related amplified sequences. 16S rRNA gene sequences were obtained via the accession numbers found on the List of Prokaryotic Names with Standing in Nomenclature website (http://www.bacterio.cict.fr/), or extracted from genomes. The best fitting tree was calculated with the RAxML black box program. Branches with bootstrap numbers above 70% are marked with black dots. The sequence of Micrococcus luteus NCTC 2665 was used as the outgroup. Bar = 0.1 sequence divergence.
DISCUSSION
In analyzing the phylogenetic trees, we note although BA112T forms a clade with the isolate designated as Actinobaculum sp. oral taxon 848, all named Actinobaculum species are located on a well-supported separate branch that also includes Actinotignum.species. Therefore we suggest that Actinbaculum was an incorrect genus designation for oral taxon 848.
Phylogenetically, the closest genera to BA112T are Actinobaculum and the recently named Actinotignum 29, and as mentioned above these are located on a well-supported branch separate from BA112T. The designation as a novel genus is further supported by AAI values of less than 65% with the closest named relatives, as 65% AAI has been suggested as a dividing line for genus 30. By a related argument, it appears that the species currently named Actinobaculum sp. oral taxon 183 might be a second species in the genus with BA112, with the AAI value of 70.93%. At any rate it is clear based on the phylogenetic analysis that BA112T should be assigned to a novel genus and species.
We propose as the genus name Peptidiphaga (Pep.ti.di.pha’.ga. N.L. n. peptidis, -is, peptide; Gr. fem. n. phaga, -ae, eater; N.L. fem. n. Peptidiphaga, peptide eater) with a description as follows: A gram-positive, facultative, non-spore forming coccus. Grows well in anaerobic conditions (85% N2, 5% CO2, 10% H2) and in air-5% CO2, but poorly in ambient air. The type species is Peptidiphaga gingivicola.
We propose as the species name: gingivicola (gin.gi.vi.co’.la. L. fem. n. gingiva, -ae, gum (of the mouth); L. masc. suf. cola, -ae, denotes an inhabitant. N.L. masc. n. gingivicola, an inhabitant of the gum). The species description is based on the study of a single strain. The genome sequence shows high sequence similarity to the unnamed cultivated bacterium HOMD Human Oral Taxon 848. The species description is: Cells are facultative, non-spore forming cocci. Grows well in anaerobic conditions (85% N2, 5% CO2, 10% H2) and in air with 5% CO2, but poorly in ambient air. Cells stain gram-positive and occur in short chains or groups. After 6 days colonies on Brucella blood agar are round, raised, smooth, off-white and shiny, with a small indentation on top. They are waxy and solid, non-hemolytic, and 0.2–0.9 mm in diameter. Temperature and pH optima for growth in tryptic soy broth were 33–40°C and 6.0–7.0. Cells metabolized alanine, arginine, glycine, histidine, leucine, proline, serine, and tyrosine. Phenylalanine use was variable. Tests were also positive for acetoin production, acid phosphatase, alanyl-phenylalanyl-proline arylamidase, and naphthol-AS-BI-BD-phosphohydrolase. Leucyl-glycine arylamidase activity was variable. The following substrates were not utilized: arabinose, fucose, mannose, D-arabitol, glucose, glycogen, lactose, maltose, mannitol, ribose, saccharose, xylose, gelatin, glutamic acid, melezitose, melibose, pullulane, raffinose, sorbital, tagatose, trehalose, valine. Other negative tests were urease, esculin, nitrate reduction, indole, catalase, A- and B-galactosidase, A- and B-Glucosidase, chymotrypsin, fucosidase, alkaline phosphatase, A- and B-mannosidase, arginine dihydrolase, B-galactosidase-6-phosphate, B-glucuronidase, cystine arylamidase, esterase, esterase lipase, glycyl-tryptophan arylamidase, hippurate hydrolysis, lipase, Methyl-BD-Glucopyranoside Acidification, N-Acetyl-B-Glucosaminidase, pyrazinamidase, pyrolidonyl arylamidase, and trypsin. Major fatty acid components were: C18:0 (41.57%), C12:0 (16.04%), C16:0 (11.86%), C18:1 1 9c (7.75%), and C20:0 (7.17%). Lesser amounts of the following were found: C10:0 (3.53%), C14:0 (1.64%), C18:1 1 7c (1.64%), and C17:0 (1.23%). Summed features were C18:2 1 6,9c/C18:0 ante (3.91%) and C16:1 1 7c/C16:1 1 6c (1.01%).
The type strain BA112T, has been deposited in the ATCC (www.atcc.org, BAA-2480) and CCUG (www.ccug.se, CCUG63931) culture collections.
Supplementary Material
Acknowledgments
We gratefully acknowledge the help of Dr. Joshua Daniels and Ms. Nancy Martin at the OSU Veterinary Hospital for the phenotypic analyses, Ms. Jennifer Harris for patient sampling, and Mr. Danny Dayeh for laboratory assistance. We thank Mr. Marion Kruse for assistance with the Latin etymology. This work was supported by grants from the National Institutes of Health NIH/NIDCR DE10467 and DE024463.
References
- 1.Griffen AL, Beall CJ, Firestone ND, et al. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS One. 2011;6(4):e19051. doi: 10.1371/journal.pone.0019051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dewhirst FE, Chen T, Izard J, et al. The Human Oral Microbiome. J Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colombo AP, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80(9):1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffen AL, Beall CJ, Campbell JH, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44(10):3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82(5):338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 7.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43(8):3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31(20):3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunt M, Kikuchi T, Sanders M, Newbold C, Berriman M, Otto TD. REAPR: a universal tool for genome assembly evaluation. Genome Biol. 2013;14(5):R47. doi: 10.1186/gb-2013-14-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nawrocki EP. Structural RNA Homology Search and Alignment using Covariance Models. St. Louis: Washington University School of Medicine; 2009. [Google Scholar]
- 15.Cock PJ, Antao T, Chang JT, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mesquite: a modular system for evolutinoary analysis [computer program]. Version 3.042015.
- 17.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. Paper presented at: Gateway Computing Environments Workshop; 2010; New Orleans, LA. [Google Scholar]
- 18.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 19.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 20.Parks D. https://github.com/dparks1134/CompareM.
- 21.Hall V, Collins MD, Hutson RA, Falsen E, Inganas E, Duerden BI. Actinobaculum urinale sp. nov., from human urine. Int J Syst Evol Microbiol. 2003;53(Pt 3):679–682. doi: 10.1099/ijs.0.02422-0. [DOI] [PubMed] [Google Scholar]
- 22.Lawson PA, Falsen E, Akervall E, Vandamme P, Collins MD. Characterization of some Actinomyces-like isolates from human clinical specimens: reclassification of Actinomyces suis (Soltys and Spratling) as Actinobaculum suis comb. nov. and description of Actinobaculum schaalii sp. nov. Int J Syst Bacteriol. 1997;47(3):899–903. doi: 10.1099/00207713-47-3-899. [DOI] [PubMed] [Google Scholar]
- 23.Tindall BJ, Rossello-Mora R, Busse HJ, Ludwig W, Kampfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010;60(Pt 1):249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 24.Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25(17):2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 25.Varghese NJ, Mukherjee S, Ivanova N, et al. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015;43(14):6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bottai D, Groschel MI, Brosch R. Type VII Secretion Systems in Gram-Positive Bacteria. Curr Top Microbiol Immunol. 2016 doi: 10.1007/82_2015_5015. [DOI] [PubMed]
- 27.Rinke C, Schwientek P, Sczyrba A, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499(7459):431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 28.Darling AE, Jospin G, Lowe E, Matsen FAt, Bik HM, Eisen JA. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2014;2:e243. doi: 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yassin AF, Sproer C, Pukall R, Sylvester M, Siering C, Schumann P. Dissection of the genus Actinobaculum: Reclassification of Actinobaculum schaalii Lawson et al. 1997 and Actinobaculum urinale Hall et al. 2003 as Actinotignum schaalii gen. nov., comb. nov. and Actinotignum urinale comb. nov., description of Actinotignum sanguinis sp. nov. and emended descriptions of the genus Actinobaculum and Actinobaculum suis; and re-examination of the culture deposited as Actinobaculum massiliense CCUG 47753T ( = DSM 19118T), revealing that it does not represent a strain of this species. Int J Syst Evol Microbiol. 2015;65(Pt 2):615–624. doi: 10.1099/ijs.0.069294-0. [DOI] [PubMed] [Google Scholar]
- 30.Konstantinidis KT, Rossello-Mora R, Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017 doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sernova NV, Gelfand MS. Identification of replication origins in prokaryotic genomes. Brief Bioinform. 2008;9(5):376–391. doi: 10.1093/bib/bbn031. [DOI] [PubMed] [Google Scholar]
- 32.Greub G, Raoult D. “Actinobaculum massiliae,” a new species causing chronic urinary tract infection. J Clin Microbiol. 2002;40(11):3938–3941. doi: 10.1128/JCM.40.11.3938-3941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ülbegi-Mohyla H, Hassan AA, Hijazin M, et al. Characterization of Arcanobacterium abortisuis by phenotypic properties and by sequencing the 16S–23S rDNA intergenic spacer region. Vet Microbiol. 2011;148(2–4):431–433. doi: 10.1016/j.vetmic.2010.08.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.