Abstract
Aims
Metabolic syndrome (MetS) is associated with insulin resistance (IR) and impaired glucose metabolism in muscle, fat, and other cells, and may induce inflammation and vascular remodeling. Endogenous reparative systems, including adipose tissue-derived mesenchymal stem/stromal cells (MSC), are responsible for repair of damaged tissue. MSC have also been proposed as an exogenous therapeutic intervention in patients with cardiovascular and chronic kidney disease (CKD). The feasibility of using autologous cells depends on their integrity, but whether in MetS IR involves adipose tissue-derived MSC remains unknown. The aim of this study was to examine the expression of mRNA involved in insulin signaling in MSC from subjects with MetS.
Methods
Domestic pigs consumed a lean or obese diet (n=6 each) for 16 weeks. MSC were collected from subcutaneous abdominal fat and analyzed using high-throughput RNA-sequencing for expression of genes involved in insulin signaling. Expression profiles for enriched (fold change>1.4, p<0.05) and suppressed (fold change<0.7, p<0.05) mRNAs in MetS pigs were functionally interpreted by gene ontology analysis. The most prominently upregulated and downregulated mRNAs were further probed.
Results
We identified in MetS-MSC 168 up-regulated and 51 down-regulated mRNAs related to insulin signaling. Enriched mRNAs were implicated in biological pathways including hepatic glucose metabolism, adipocyte differentiation, and transcription regulation, and downregulated mRNAs in intracellular calcium signaling and cleaving peptides. Functional analysis suggested that overall these alterations could increase IR.
Conclusions
MetS alters mRNA expression related to insulin signaling in adipose tissue-derived MSC. These observations mandate caution during administration of autologous MSC in subjects with MetS.
Keywords: mesenchymal stem cells, insulin, metabolic syndrome, mRNA
1. INTRODUCTION
Metabolic syndrome (MetS) is clinically characterized by harmful co-existence of three or more cardiometabolic risk factors that include abdominal obesity, insulin resistance (IR), hypertension, hyperglycemia or dyslipidemia in a single individual1, 2. Aggregation of these factors has profound influence on the development of diabetes mellitus and cardiovascular disease, intensifying the risk by two- and five-fold, respectively3. Nearly 35% of American adults are projected to have MetS, making it a public health concern2. Additionally, MetS confers increased risk for chronic kidney disease (CKD), by leading to renal dysfunction4, microalbuminuria5 and vascular damage in the kidney6. Insulin, an essential hormone involved in glucose homeostasis and lipid metabolism, has been implicated as a common initiator associated with MetS pathogenesis7. Excessive fat expansion during obesity leads to a chronic inflammatory state, in turn disrupting normal adipokine signaling, and may contribute to the diminished insulin response observed in patients with MetS and kidney injury8.
Regenerative cell-based therapies, such as infusion of autologous mesenchymal stem/stromal cells (MSC), may be effective in re-establishing or repairing damaged tissue. This population of self-renewing cells is capable of differentiating into mature cells that comprise the endoderm, mesoderm and ectoderm. With their chemotactic capacities to selectively home to sites of inflammation and deteriorating tissues, MSC have been shown to attenuate the immune response, decrease fibrosis and stimulate angiogenesis through paracrine-mediated activity, which may be beneficial in kidney disease9, 10. MSC can be obtained from an abundance of sources, including hematopoietic bone marrow11, adipose tissue12 and the placenta13. Stem cell frequency and function has been reported to decline with age, influencing degeneration and dysfunction in aging tissues, which may be mediated by impaired insulin signaling14. However, it is unclear if adipose tissue-derived MSC isolated from subjects with MetS have comparable endogenous characteristics in comparison to healthy individuals.
The current study tested the hypothesis that MetS alters expression of mRNAs related to insulin signaling in adipose tissue-derived MSC. To test this hypothesis, we employed high-throughput RNA sequencing (RNA-seq) to comprehensively characterize the expression of mRNAs in MSC obtained from lean and MetS pigs. Our data demonstrate that insulin signaling-related mRNAs enriched in MetS-MSC are primarily involved in cellular processes associated with transcription and cell-cell signaling, whereas those down-regulated in MetS are implicated in regulation of the cell membrane. These alterations in gene expression contribute greatly to our understanding of biological pathways and may have important ramifications for therapeutic applications using autologous MSC.
2. METHODS
2.1 Experimental Animals and MSC Collection
Approval for this study was granted by the Mayo Clinic Animal Care and Use Committee. After a 16-week observation, twelve 7-month old domestic pigs were assessed. Development of an experimental MetS model was induced by consumption of a high-cholesterol, high-carbohydrate diet15 (5B4L, protein 16.1%, ether extract fat 43.0% and carbohydrates 40.8%, Purina Test Diet, Richmond, IN) in six randomly selected pigs, while the remaining animals were fed regular pig chow (lean) (13% protein, 2% fat, 6% fiber, Purina Animal Nutrition LLC, MN). All animals received water ad libitum. After 16 weeks of diet, animals were euthanized by a terminal intravenous injection of sodium pentobarbital (100mg/kg IV, Sleepaway®, Fort Dodge Laboratories, Fort Dodge, IA). Blood samples and subcutaneous abdominal adipose tissue (5–10 g) were collected from all pigs. Briefly, adipose tissue was enzymatically digested in collagenase-H for 45 minutes, filtered, and expanded in culture for 3 weeks to isolate MSC. Cells were kept in advanced MEM medium supplemented with 5% platelet lysate (PLTmax, Mill Creek Life Sciences, Rochester, MN) at 37°C with 5%CO2. After the 3rd passage primary MSC were characterized by immunofluorescent staining for MSC-specific surface markers and used for transcriptome analysis.
2.2 RNA Sequencing and Bioinformatics Analysis
RNA libraries were prepared according to the manufacturer’s instructions (TruSeq RNA Sample Prep Kit v2, Illumina). Flow cells were loaded with 8–10 pM sequence libraries in order to generate cluster densities of 7,000,000/mm2. The Illumina cBot and cBot Paired-end cluster kit version 3 protocol was used for this analysis. Cells were sequenced on an Illumina HiSeq 2000 using TruSeq SBS kit version 3 and HCS v2.0.12 data collection software. The MAPRSeq v.1.2.1 system, the Bioinformatics Core standard tool, TopHat 2.0.616 and featureCounts17 software were used for further data analysis. Gene expression was standardized to 1 million reads and corrected for gene length (reads per kilobasepair per million mapped reads, RPKM).
To elucidate whether insulin regulates gene expression during MetS, we used the GeneCards® database (http://www.genecards.org/) to screen genes associated with insulin signaling. From this group of genes, search mRNAs with a fold change >1.4 were defined as enriched and a fold change <0.7 was classified as down-regulated in MetS pigs compared to lean. In both enriched and down-regulated mRNAs, a Fisher’s exact test yielding a p-value ≤0.05 was considered statistically significant and included in our analysis. mRNA lists were uploaded into DAVID 6.7 database (https://david-d.ncifcrf.gov/) to determine their functional significance. Additionally PANTHER (http://pantherdb.org/) and Morpheus (https://software.broadinstitute.org/morpheus/) software was utilized to analyze cellular localization and expression of mRNAs between groups, respectively.
2.3 mRNA validation by quantitative-polymerase chain reaction (qPCR) and Statistically Analysis
RNA isolation, cDNA synthesis, and qPCR using the ΔΔCt method was used to evaluate random gene expression levels of enriched mRNAs (NR1H4, TFAP2B and KCND2) and downregulated mRNAs (CXCL10, MSI1 and NTRK2) in MetS-MSC. All primers were from ThermoFisher Scientific (NR1H4: APMFXH4; TFAP2B: ss03373551; KCND2: ss3376943; CXCL10: 333391846; MSI1: APNKR32 and NTRK2: ss3373489). Gene expression was normalized to GAPDH. All data is represented as mean ± standard error (SE). Analysis was accomplished by using JMP 10.0 Software. Differences in mean values between lean and MetS animals were assessed using Wilcoxon signed-rank test at a statistically significant level p<0.05.
3. RESULTS
3.1 Characterization of MetS model
We have previously shown that after 16 weeks of diet, MetS pigs had progressive increases in body weight, blood pressure and lipid levels in comparison to lean animals, and developed IR15. This feeding protocol confirmed successful induction of our MetS pig model.
3.2 Overall mRNA expression in Lean-MSC and MetS-MSC
Our previous studies have demonstrated that MSC produced employing our lab protocols stain positive for CD44, CD73, CD90 and CD105 surface markers and negative for CD14, CD34 and CD4518, 19. Additionally, we demonstrated that adipose tissue-derived MSC cultured from lean and obese pigs execute tri-lineage differentiation12.
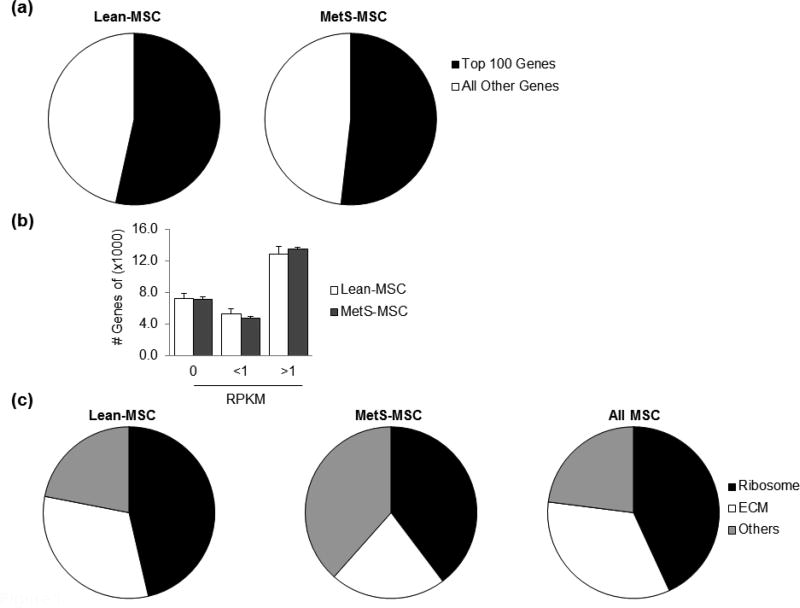
Following RNA-seq analysis, our database yielded a total of 25,476 mapped genes, with the 100 most highly expressed accounting for approximately 50% in both lean- and MetS-MSC (Fig. 1). Of all transcripts, approximately 45% were expressed at levels >1 RPKM, 30% at 0 RPKM, and 20% at <1 RPKM with similar proportions in lean- and MetS-MSC (Fig. 1). The most abundant mRNAs (expressed at >1000 RPKM) encoded mostly ribosomal and extracellular matrix (ECM) proteins (Fig. 1).
Figure 1. Genes in porcine MSC.
(a) Pie chart showing the distribution of genes in lean- and MetS-MSC. The top 100 genes expressed in both lean- and MetS-MSC represent over half of mapped transcripts. (b) The number of genes showing RNA reads of 0, <1 and >1 were similar in lean- and MetS-MSC. Nearly 45% of annotated genes are expressed at levels >1 RPKM. (c) Functional analysis of lean- and MetS-MSC (DAVID 6.7) revealed that mRNAs expressed at >1000 RPKM encode proteins associated with ribosomal and ECM proteins.
3.3 Enriched mRNAs in MetS-MSC
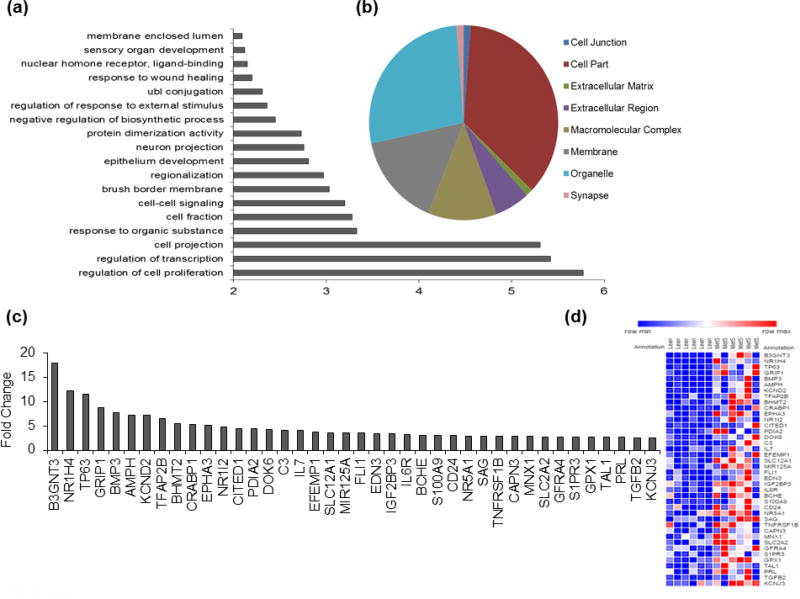
A total of 3,052 mRNAs associated with insulin signaling were identified, of which we identified 168 insulin signaling related mRNAs enriched in MetS-MSC. Of these protein-encoding genes, the majority was localized intracellularly in the nucleus and involved in diverse functions within the cell, including cellular proliferation, transcription, signaling between cells and binding of ligands to nuclear hormone receptors (Fig. 2). Fold changes of the top 40 enriched mRNAs (Fig. 2) included CITED1, an adipocyte differentiation marker; NR1H4, NR1I2, GRIP1 and CRABP1, which play a role in mediating hepatic gluconeogenesis; as well as TFAP2B, an transcriptional factor associated with reduced insulin sensitivity.
Figure 2. Enriched insulin signaling-related mRNAs in MetS-MSC.
(a) DAVID 6.7, functional annotation analysis of enriched mRNAs in MetS-MSC demonstrated that the majority of mRNAs are involved in cellular processes. (b) Cellular localization of enriched mRNAs using PANTHER analysis identified most mRNAs are located in the nuclear region of the cell. (c) Top 40 enriched mRNAs with the highest fold change (MetS-MSC/Lean-MSC). (d) The heat map of mRNAs displays a visual interpretation of the relative expression of the 40 up-regulated mRNAs in MetS-MSC, where an increase in expression is signified in red and decrease in blue.
3.4 Down-regulated mRNAs in MetS-MSC
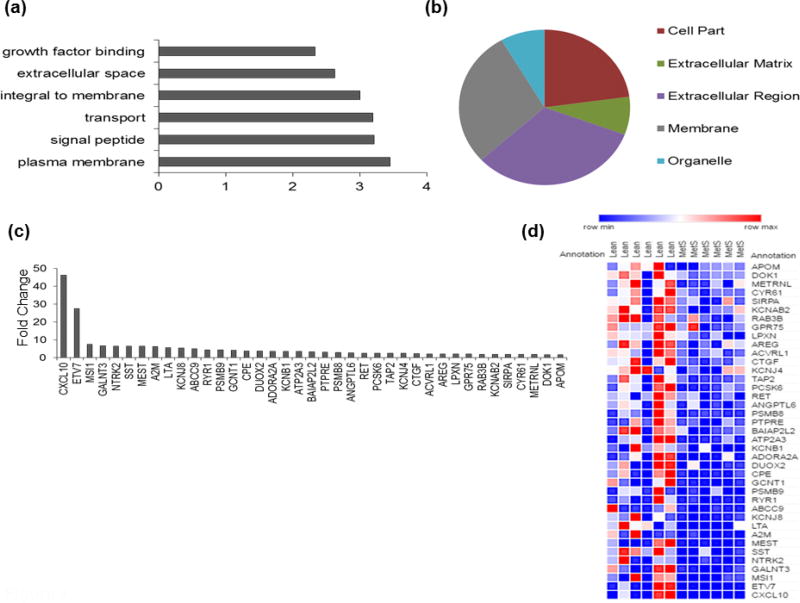
Similar analysis identified 51 insulin signaling-related down-regulated mRNAs in adipose tissue derived-MSC in MetS animals. Many of these down-regulated mRNAs are housed in the extracellular region and membrane of the cell (Fig. 3). Functional analysis showed these mRNAs to be engaged in biological functions including transport, growth factor binding and signaling of peptides (Fig. 3). Primarily, among these mRNAs with the highest fold change (Fig. 3), many are responsible for cleaving peptides and associated with calcium signaling.
Figure 3. Down-regulated insulin signaling-related mRNAs in MetS-MSC.
(a) Downregulated mRNAs in MetS-MSC were mainly involved in functions associated with the membrane. (b) Cellular localization of down-regulated mRNAs revealed primarily the extracellular region and membrane of the cell. (c) Many down-regulated mRNAs with the highest fold change (Lean-MSC/MetS-MSC) account for proteases and proteins involved in calcium flux. (d) Heat map of the top 40 down-regulated mRNAs showing a decreased abundance in MetS-MSC.
3.5 Validation of Insulin Signaling mRNAs in MSC
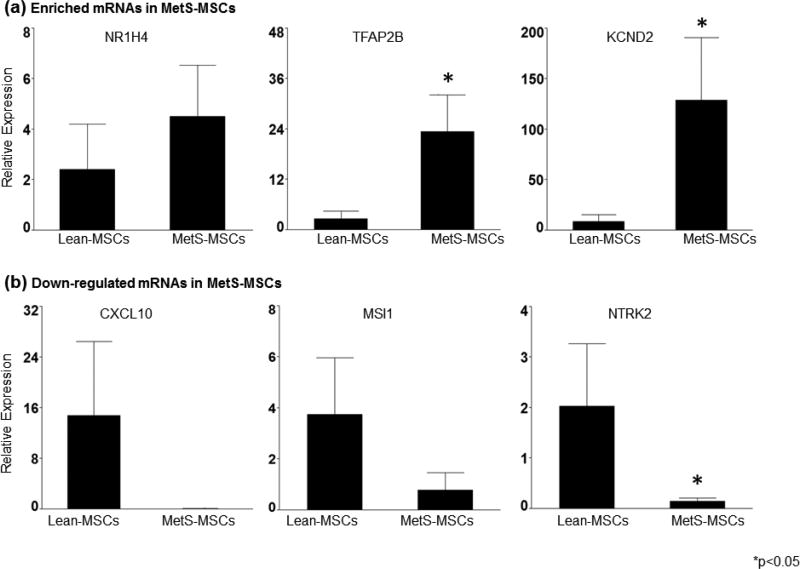
Various bioinformatic tools employed during this study revealed an array of differentially expressed mRNAs in adipose-tissue derived MSC in pigs. Gene expression of enriched and down-regulated mRNAs was subsequently validated using qPCR. The expression of NR1H4, TFAP2B and KCND2 was intensified in MetS-MSC, while CXCL10, MSI1 and NTRK2 were suppressed in these cells (Fig. 4). The similarities in gene expression pattern support our computational analyses.
Figure 4. Validation of candidate mRNAs in Lean- and MetS-MSC by qPCR.
Differential expression of selected up-regulated (a) and down-regulated (b) candidate mRNAs demonstrated agreement with the RNA-seq analysis, where NR1H4, TFAP2B and KCND2 were up-regulated in MetS-MSC and CXCL10, MSI1 and NTRK2 were depleted in these MSCs. * p<0.05, vs. Lean-MSC
4. DISCUSSION
In this study, we evaluated the mRNA expression profiles of adipose tissue-derived MSC in lean and diet-induced MetS pigs. Through a comprehensive sequencing analysis, we compared the enrichment and suppression of mRNA content that may modulate insulin signaling in MSC. It is well recognized that impaired insulin signaling eventually advances to IR and contributes to metabolic derangement. Furthermore, diminished response of normal insulin handling is the most important pathogenic factor which initiates the development of MetS7. Given the paucity of data regarding insulin signaling in stem cells, we aimed to determine whether insulin signaling potential is impaired in MSC isolated from MetS subjects.
Our initial analysis identified 168 insulin-associated mRNAs upregulated in MetS pigs MSC and 51 down-regulated in these cells. The distinct quantifiable differences between lean and MetS groups signify the variances of insulin response in adipose tissue-derived MSC. Of those enriched mRNAs, a subset has been recognized for their participation in triggering IR. For example, sphingolipids are involved in obesity-induced IR, so that exogenous stimulation of palmitate in obese mice upregulates IL-6 production through activation of sphingosine 1-phosphate receptor-3 (S1PR3) in skeletal muscle20. Additionally, sphingosine 1-phosphate expression is dependent on activation of nuclear receptor, peroxisome proliferator-activated receptor-α (PPARα). Likewise, our studies established an increase in S1PR3 in MetS-MSC, as well as upregulated gene expression of the receptor of IL-6. Moreover, our findings revealed an enrichment of two members of the nuclear hormone receptor family (NR1H4 and NR1I2) and various proteins associated with nuclear hormone receptor activation (GRIP1 and CRABP1). Polymorphisms in the TFAP2B gene have been shown to impact insulin sensitivity in healthy male adolescents21. In our dataset, we also observed a nearly 1.5-fold increase in the expression of β1, 3-N-acetylglucosaminyltransferase-3 (B3GNT3). While the exact role of this gene has not been fully explored, in cancer its expression has been correlated to decreased ERK and AKT signaling22.
These findings may have important implications for stem cell biology. Insulin is an essential hormone with diverse roles throughout the body, including glucose uptake, cellular proliferation and growth, and lipid synthesis. The alterations in expressed mRNA associated with insulin signaling observed in the present study may suggest significant implications for using MSC for autologous therapies. MSC obtained from subjects with MetS could potentially have injurious modifications linked to cellular processes, such as transcription, cell proliferation, and diminished responses associated with transport and binding of growth factors. Importantly, altered expression of genes involved in insulin signaling may affect MSC integrity, since recent reports suggest that insulin transduction regulates aging in stem cells14. Furthermore, these changes may have adverse effects on the communication of MSC with neighboring cells and on their capacity for endogenous tissue repair. Similarly, their potency and utility for autologous delivery may be impeded.
Several limitations of this study need to be considered. The present study investigated the early alterations in insulin signaling associated protein-encoding genes during MetS in a porcine model fed an atherogenic, high-fat diet for 16 weeks. Our model displayed several characteristics related to human MetS; however, progressive modifications that occur over time in patients with prolonged MetS might be underestimated, due to the short duration of disease. Furthermore, functional changes of MSC in vivo are not captured in this analysis. However, previously we observed a senescent phenotype and an increase in TNF-α expression in cultured MetS MSC12, suggesting that changes in gene expression are functionally meaningful.
In summary, we observed that MSC obtained from pigs with MetS have modified mRNA expression involved in insulin signaling. Hence, careful assessment of modifications in MSC in patients is imperative. Autologous cell-based therapies have proven to be valuable in organ repair and slowing disease progression, yet chronic long-lasting pathologies may negatively influence the biological properties and cellular functions linked with MSC regenerative potential. Future studies are needed to develop approaches to improve the therapeutic efficacy of autologous MSC or identify alternative treatment modalities.
Acknowledgments
This study was partly supported by NIH grants: DK73608, HL123160, DK104273, DK102325, DK007013 and UL1-TR000135.
List of Abbreviations
- B3GNT3
β1, 3-N-acetylglucosaminyltransferase-3
- CKD
Chronic Kidney Disease
- ECM
Extracellular matrix
- IR
Insulin resistance
- MSC
Mesenchymal stem/stromal cells
- MetS
Metabolic syndrome
- PPARα
Peroxisome proliferator-activated receptor-α
- RNA-seq
RNA sequencing
- S1PR3
Sphingosine 1-phosphate receptor-3
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST
None
Research idea and study design: SMC, XYZ, AE, LOL; experiment and data acquisition: SMC, XZ, AE, HT; data analysis/interpretation: SMC, XYZ, AE, HT, LOL; statistical analysis: SMC; supervision or providing intellectual content of critical importance to the work described: XYZ, AE, AL, AJVW, LOL; mentorship: AL, AJVW, LOL. Each author contributed important intellectual content during manuscript drafting or revision and accepts accountability for the overall work by ensuring that questions pertaining to the accuracy or integrity of any portion of the work are appropriately investigated and resolved.
References
- 1.Alberti KG, Zimmet P, Shaw J. Metabolic syndrome--a new world-wide definition. A Consensus Statement from the International Diabetes Federation. Diabet Med. 2006;23:469–80. doi: 10.1111/j.1464-5491.2006.01858.x. [DOI] [PubMed] [Google Scholar]
- 2.Roberts CK, Hevener AL, Barnard RJ. Metabolic syndrome and insulin resistance: underlying causes and modification by exercise training. Compr Physiol. 2013;3:1–58. doi: 10.1002/cphy.c110062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isomaa B, Almgren P, Tuomi T, Forsen B, Lahti K, Nissen M, Taskinen MR, Groop L. Cardiovascular morbidity and mortality associated with the metabolic syndrome. Diabetes Care. 2001;24:683–9. doi: 10.2337/diacare.24.4.683. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Muntner P, Hamm LL, Jones DW, Batuman V, Fonseca V, Whelton PK, He J. The metabolic syndrome and chronic kidney disease in U.S. adults. Ann Intern Med. 2004;140:167–74. doi: 10.7326/0003-4819-140-3-200402030-00007. [DOI] [PubMed] [Google Scholar]
- 5.Hoehner CM, Greenlund KJ, Rith-Najarian S, Casper ML, McClellan WM. Association of the insulin resistance syndrome and microalbuminuria among nondiabetic native Americans. The Inter-Tribal Heart Project. J Am Soc Nephrol. 2002;13:1626–34. doi: 10.1097/01.asn.0000015762.92814.85. [DOI] [PubMed] [Google Scholar]
- 6.Alexander MP, Patel TV, Farag YM, Florez A, Rennke HG, Singh AK. Kidney pathological changes in metabolic syndrome: a cross-sectional study. Am J Kidney Dis. 2009;53:751–9. doi: 10.1053/j.ajkd.2009.01.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrannini E, Haffner SM, Mitchell BD, Stern MP. Hyperinsulinaemia: the key feature of a cardiovascular and metabolic syndrome. Diabetologia. 1991;34:416–22. doi: 10.1007/BF00403180. [DOI] [PubMed] [Google Scholar]
- 8.Bluher M. Clinical relevance of adipokines. Diabetes Metab J. 2012;36:317–27. doi: 10.4093/dmj.2012.36.5.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernardo ME, Locatelli F, Fibbe WE. Mesenchymal stromal cells. Ann N Y Acad Sci. 2009;1176:101–17. doi: 10.1111/j.1749-6632.2009.04607.x. [DOI] [PubMed] [Google Scholar]
- 10.Reinders ME, Fibbe WE, Rabelink TJ. Multipotent mesenchymal stromal cell therapy in renal disease and kidney transplantation. Nephrol Dial Transplant. 2010;25:17–24. doi: 10.1093/ndt/gfp552. [DOI] [PubMed] [Google Scholar]
- 11.Friedenstein AJ, Petrakova KV, Kurolesova AI, Frolova GP. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation. 1968;6:230–47. [PubMed] [Google Scholar]
- 12.Zhu XY, Ma S, Eirin A, Woollard JR, Hickson LJ, Sun D, Lerman A, Lerman LO. Functional Plasticity of Adipose-Derived Stromal Cells During Development of Obesity. Stem Cells Transl Med. 2016;5:893–900. doi: 10.5966/sctm.2015-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.In 't Anker PS, Scherjon SA, Kleijburg-van der Keur C, de Groot-Swings GM, Claas FH, Fibbe WE, Kanhai HH. Isolation of mesenchymal stem cells of fetal or maternal origin from human placenta. Stem Cells. 2004;22:1338–45. doi: 10.1634/stemcells.2004-0058. [DOI] [PubMed] [Google Scholar]
- 14.Signer RA, Morrison SJ. Mechanisms that regulate stem cell aging and life span. Cell Stem Cell. 2013;12:152–65. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pawar AS, Zhu XY, Eirin A, Tang H, Jordan KL, Woollard JR, Lerman A, Lerman LO. Adipose tissue remodeling in a novel domestic porcine model of diet-induced obesity. Obesity (Silver Spring) 2015;23:399–407. doi: 10.1002/oby.20971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 18.Eirin A, Zhu XY, Krier JD, Tang H, Jordan KL, Grande JP, Lerman A, Textor SC, Lerman LO. Adipose tissue-derived mesenchymal stem cells improve revascularization outcomes to restore renal function in swine atherosclerotic renal artery stenosis. Stem Cells. 2012;30:1030–41. doi: 10.1002/stem.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebrahimi B, Eirin A, Li Z, Zhu XY, Zhang X, Lerman A, Textor SC, Lerman LO. Mesenchymal stem cells improve medullary inflammation and fibrosis after revascularization of swine atherosclerotic renal artery stenosis. PLoS One. 2013;8:e67474. doi: 10.1371/journal.pone.0067474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross JS, Hu W, Rosen B, Snider AJ, Obeid LM, Cowart LA. Sphingosine kinase 1 is regulated by peroxisome proliferator-activated receptor alpha in response to free fatty acids and is essential for skeletal muscle interleukin-6 production and signaling in diet-induced obesity. J Biol Chem. 2013;288:22193–206. doi: 10.1074/jbc.M113.477786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nordquist N, Gokturk C, Comasco E, Eensoo D, Merenakk L, Veidebaum T, Oreland L, Harro J. The transcription factor TFAP2B is associated with insulin resistance and adiposity in healthy adolescents. Obesity (Silver Spring) 2009;17:1762–7. doi: 10.1038/oby.2009.83. [DOI] [PubMed] [Google Scholar]
- 22.Ho WL, Che MI, Chou CH, Chang HH, Jeng YM, Hsu WM, Lin KH, Huang MC. B3GNT3 expression suppresses cell migration and invasion and predicts favorable outcomes in neuroblastoma. Cancer Sci. 2013;104:1600–8. doi: 10.1111/cas.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]