Abstract
We proposed an unsupervised hybrid method - Intelligent Word Embedding (IWE) that combines neural embedding method with a semantic dictionary mapping technique for creating a dense vector representation of unstructured radiology reports. We applied IWE to generate embedding of chest CT radiology reports from two healthcare organizations and utilized the vector representations to semi-automate report categorization based on clinically relevant categorization related to the diagnosis of pulmonary embolism (PE). We benchmark the performance against a state-of-the-art rule-based tool, PeFinder and out-of-the-box word2vec. On the Stanford test set, the IWE model achieved average F1 score 0.97, whereas the PeFinder scored 0.9 and the original word2vec scored 0.94. On UPMC dataset, the IWE model’s average F1 score was 0.94, when the PeFinder scored 0.92 and word2vec scored 0.85. The IWE model had lowest generalization error with highest F1 scores. Of particular interest, the IWE model (trained on the Stanford dataset) outperformed PeFinder on the UPMC dataset which was used originally to tailor the PeFinder model.
Keywords: Information extraction, word embedding, pulmonary embolism, report annotation
Graphical abstract
1. Introduction
Radiology is central to modern healthcare, providing detailed clinical information for disease detection, staging and treatment planning while also playing an important role in monitoring and predicting outcomes. Radiology reports are composed of unstructured free-text, and conversion into a computer manageable representation for large scale analysis requires strategies for efficient and automated information extraction. Natural language processing (NLP) tools are designed to convert unstructured text into coded data which may enable automatic identification and extraction of information from radiology text reports for a variety of clinical applications, including diagnostic surveillance, cohort building, quality assessment, labels for computer vision data, and clinical decision support services.
Despite the advantages, NLP remains an underutilized technique for large-volume radiology report data extraction in both research and clinical practice environments due to high development costs and lack of generalizabiltiy of models. Many of the best performing NLP methods are Dictionary-based [1] or Rule-based analysis [2], which, while accurate for a specific task, requires tremendous manual effort to tune the methods for a particular case-study/dataset. Recently, deep learning has provided researchers with tools to create automated classification models without requiring hand-crafted feature engineering which is adapted widely for medical images [3, 4, 5]. However, the deep learning methods have yet to show similar performance gains on information extraction from free text radiology reports. A challenge to applying deep learning methods to information extraction in text is modeling ambiguity of free text narrative style for clinical reports, lexical variations, use of ungrammatical and telegraphic phases, and frequent appearance of abbreviations and acronyms.
We propose a hybrid method - Intelligent Word Embedding (IWE) that combines semantic-dictionary mapping and neural embedding technique for creating context-aware dense vector representation of free-text clinical narratives. Our method leverages the benefits of unsupervised learning along with expert-knowledge to tackle the major challenges of information extraction from clinical texts, which include ambiguity of free text narrative style, lexical variations, use of ungrammatical and telegraphic phases, arbitrary ordering of words, and frequent appearance of abbreviations and acronyms. Ideally, the transformation of large volume of free-text radiology reports into dense vectors may serve to unlock rich source of information for solving a variety of critical research challenges, including diagnostic surveillance, cohort building, and clinical decision support services. In this study, we will exploit the embedding created by the IWE method to generate annotation of a large multi-institutional cohorts of chest CT radiology reports based on various level of categorizations of PE.
In the targeted case-study, the first important determinant is whether the patient has a PE or not, which informs medical care and treatment decisions; however it is possible that the patient has had prior imaging that diagnosed PE and subsequent imaging may demonstrate an unchanged, diminished, or otherwise chronic PE, in which case medical treatment may change based on whether the PE had responded to prior therapy. Finally it is controversial whether subsegmental PE requires treatment at all, and is not felt to have the same clinical implications as central PE, and thus this category holds valuable importance for clinical decision making [6, 7].
We formulated annotation of the radiology reports in terms of three different PE categorical measures (PE present/absent, PE acute/chronic, PE central/subsegmental) as separate classification task. Note that a given report cannot have labels of ‘PE acute/chronic’ or ‘PE subsegmental only/central’ without the label of ‘PE present’. Our formulation is mainly influenced by the fact that the performance of the ‘PE positive’ label alone and drawing conclusions in comparison to other NLP classifiers has significant value as the primary clinical state based on the imaging study. The characteristics of ‘PE acute’ vs ‘PE chronic’ or ‘PE subsegmental’ vs ‘PE central’ location, while important, are each inherently more challenging and have less clinical impact compared to the fundamental disease state and conflating these labeling tasks would provide less information about individual label performance for this exploratory evaluation.
We benchmark the performance of IWE model against a state-of-the-art rule-based solution PeFinder [8] and out-of-the-box word2vec model [9, 10] using radiology reports from two major academic institutions: Stanford and University of Pittsburgh medical center. The proposed embedding produced high accuracy (average F1 score Stanford dataset - 0.97, UPMC dataset - 0.94) for three different categorical measures of PE despite the fact that the reports were generated by numerous radiologists of differing clinical training and experience. Besides, the IWE model trained on the Stanford dataset, and used to create embeddings from UPMC dataset, beat the PeFinder model which was originally developed on the UPMC dataset. IWE model also improved upon the out-of-the-box word2vec and showed more generalizability on heterogeneous datasets. We also explored the visualization of vectors in low dimensional space while retaining the local structure of the high-dimensional vectors, to investigate the legitimacy of the semantic and syntactic information of words and documents. In the following sections, we detail the methodology (Sec. 3), present the results (Sec. 4) and finally conclude by mentioning core contributions, limitations and future research directions (Sec. 5).
2. Related works
MedLEE (Medical Language Extraction and Encoding System) in an example of traditional NLP approach in medical domain which relies on controlled vocabulary and grammatical rules in order to convert free-text into a structured database [11, 12]. Dang et al. processed 1059 radiology reports with Lexicon Mediated Entropy Reduction (LEXIMER) to identify the reports that include clinically important findings and recommendations for subsequent action [13]. A core limitation of such rule-based systems is that all the kinds of entities and relations need to be pre-specified, and it requires enormous amount of manual effort to initiate such systems if the number of such entities and relations that need to be extracted is significantly large. Moreover, extension of such systems, even for a similar case-study, needs nearly equal amount of manual work.
In addition to traditional dictionary-based and rule-based NLP techniques, various combinations of NLP pipelines and Machine learning methods have been proposed [14, 15] that do not demand substantial manual effort and can be retrained without reprogramming for any domain. Sohn et al used tokenizer combined with machine learning to identify patients with abdominal aortic aneurysms [16]. Nguyen et al.[17] combined traditional supervised learning methods with Active Learning for classification of imaging examinations into reportable and non-reportable cancer cases.
However, the performance of machine learning models heavily depends on finding meaningful vector space projections of the unstructured texts. In most approaches, documents are represented by a simple sparse bag-of-words (BoW) representations which face several challenges in the clinical domain: (i) scalability - BoW encode every word in the vocabulary as one-hot-encoded vector, but clinical vocabulary may potentially run into millions; (ii) semantics of the words - the vectors corresponding to same contextual words are orthogonal; (iii) word orderings - BoW models also don’t consider the order of words in the phrase.
There is now an emerging trend with deep learning that adopts a distributed representation of words by constructing a so-called neural embedding of each word or document. The word2vec model introduced by Mikolov et al. [9, 10] is the most popular approach for providing semantic word embeddings. One of the biggest challenges with word2vec is how to handle unknown or out-of-vocabulary (OOV) words and morphologically similar words. This can particularly be an issue in domains like medicine where synonyms and related words can be used depending on the preferred style of radiologist, and words may have been used infrequently in a large corpus. If the word2vec model has not encountered a particular word before, it will be forced to use a random vector, which is generally far from its ideal representation. Our proposed method - Intelligent Word Embedding (IWE) that can efficiently handle OOV words by combining neural embedding with the semantic dictionary mapping.
3. Material and Methods
3.1. Dataset
3.1.1. Cohorts
Stanford dataset
With the approval from the Stanford Institutional Review Borad (IRB), we obtained radiology reports from Stanford medical center for contrast-enhanced CT examinations of the chest performed between January 1,1998 and January 1,2016. Using radiology procedure codes, a total of 117,816 CT examinations of the chest with contrast reports were selected for our analysis. All examinations were de-identified in a fully HIPAA- compliant manner and processing of data was approved by the IRB.
Two experienced radiologists performed annotation of total 4512 randomly selected reports. Three binary labels were assigned to individual reports which was defined according to three categorical measures of PE: (1) PE present/absent; (ii) PE acute/chronic; (iii) PE central/subsegmental only. If a PE was definitely present in the report it was annotated as positive for PE present, or else annotated as negative. Chronicity was labeled as either acute or chronic based on the text description. In the setting of acute on chronic, or “mixed” chronicity, the report was labeled as acute to reduce the false negative rate. The “subsegmental only” label was used in cases where the PE was described as subsegmental and did not include more central locations.
Interrater reliability was estimated as Cohen’s Kappa Score and the raters were highly consistent for the first two categories of determining PE present and PE Acute with kappa scores of 0.959 and 0.969 respectively. Significant disagreement (kappa score of 0.664) was observed when looking at PE subsegmental label. A senior radiologist resolved all conflicting cases manually for preparing the ground truth labels.
UPMC dataset
We obtained 858 reports from University of Pittsburgh medical center that were originally used to develop PeFinder classifiers. The reports were all de-identified in a fully HIPAA-compliant manner. The annotations were defined according to two categorical measures of PE: (1) PE present/absent; (ii) PE acute/chronic. Three medical students independently annotated the reports with five distinct states and binary annotations for each document were obtained from the user annotations as follows: probably positive and definitely positive were collapsed to positive; probably negative, indeterminate, and definitely negative were considered negative; after collapsing annotations to binary values, the authors generated labels for each report by a majority vote of the annotators [8].
3.1.2. Synopsis of the cohorts
In this study, we utilized the ‘Impression’ section of the radiology reports to classify them according to PE categorical measures since the PE assessment categories are often only reported in impression section of the reports. We implemented a python-based section segmentation algorithm - Report Splitter, to recognize section headings and to use them to segment ‘Impression’ section from both Stanford and UPMC dataset. Despite the fact that the purpose of impression section is to provide a high-level summary of the clinical findings, it is not trivial to recognize the PE assessment descriptions, due the high ambiguity in the expression and variations in syntax. In Table 1, we present the synopsis of the Stanford and UMPC dataset according to report-level, sentence-level and word-level statistics which again reflects a large diversity between the style of impression sections. For instance, the number of words in the impression section of the reports ranged from 11 to 3015 in the UPMC dataset while for Stanford dataset in varies from 11 to 2116. Same observation holds for the sentence length. More importantly, 117,387 unique words are noticed only in the Stanford dataset when all the reports belongs to contrast-enhanced CT examinations of the chest performed in the same institution, which makes the automated report classification task even more challenging.
Table 1.
Statistics of the reports
| Features | Stanford dataset (117,816 reports) | UPMC dataset (859 reports) | |
|---|---|---|---|
| Report-level statistics | Maximum number of words | 2116 | 3015 |
| Minimum number of words | 2 | 11 | |
| Average word count | 666 | 412 | |
| Sentence-level statistics | Maximum number of words | 269 | 125 |
| Minimum number of words | 2 | 2 | |
| Average word count | 33 | 66 | |
| Word-level statistics | Unique word count | 117387 | 4676 |
3.1.3. Sample distribution
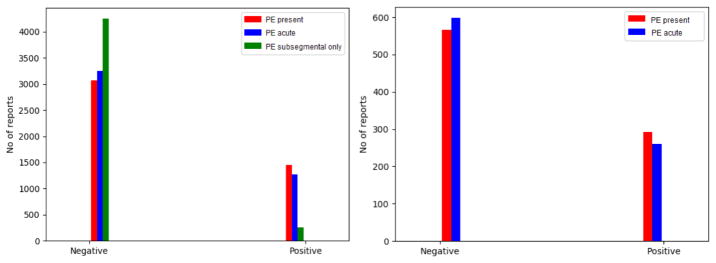
Figure. 1 presents the sample distribution within the annotated Stanford and UPMC dataset according to the PE categorical measures. As seen from the figure, ~ 68% of the samples in both Stanford and UPMC cohorts represents PE absent, and are also labeled as ‘negative’ for PE acute/chronic class label. Only ~ 10% of the samples in the Stanford cohort are labeled as ‘positive’ for the PE subsegmental category. The percentage of imbalance in our dataset can be considered as the representative sample of real clinical scenarios.
Figure 1.
Distribution of PE categorical measure: Stanford dataset (4512 reports) on left and UPMC dataset (858 reports) on right
3.2. Intelligent word embedding (IWE) model
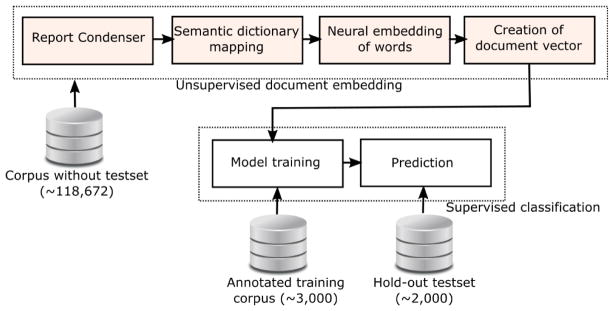
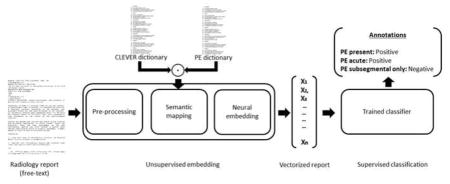
Figure. 2 presents the high-level model schema of Intelligent word embedding (IWE) where two complimentary approaches - (i) Semantic dictionary mapping, and (ii) word2vec, are combined together for creating dense vector representation of individual clinical reports. Finally, a supervised classification model is trained to learn the mapping between the vectors of the training set and ground truth labels for predicting the annotation of test cases. The majority of the pipeline is unsupervised, and only the classification block needs manually labeled data.
Figure 2.
Schema of Intelligent word embedding (IWE) approach
3.2.1. Report Condenser
We implemented a python-based text processor, Report Condenser which transformed all 117,816 report impressions through a series of pre-processing steps to focus only on the significant concepts of the free-text reports, that would enhance the semantic quality of the resulting word embeddings. First, it cleansed the texts by normalizing the texts to lowercase letters and removing words of following types: general stop words (a, an, are,...,be, by,...,has, he,...,etc), words with very low frequency (< 50), unwanted terms and phrases (e.g. medicolegal phrases, headers, etc). Following removal steps, Report Condenser searched the updated corpus to identify frequently appearing pairs of words based on pre-defined threshold value of occurrence (> 5000) and condensed them into a single word to preserve useful semantic units for further processing. Some examples of the concatenated words are: ‘bilater pulmonari’ → ‘bilater_pulmonari’, ‘mass effect’ → ‘mass_effect’, ‘lung nodule’ → ‘lung_nodule’.
3.2.2. Semantic-dictionary mapping
We use a lexical scanner that recognizes corpus terms which share a common root or stem with pre-defined terminology, and map them to controlled terms. In contrast with traditional NLP approaches, this step does not need any sentence parsing, noun-phrase identification, or co-reference resolution. We used dictionary style string matching where we directly search and replace terms, by referring to the dictionary. We adopted multilevel semantic mapping methodology. First, we used the more general publicly available CLEVER terminology [18] to replace common analogies/synonyms for creating more semantically structured texts. We mainly focused on the terms that describe family, progress, risk, negation, and punctuations, and normalized them using the formal terms derived from the terminology. For instance, {’no’, ‘absent’, ‘inadequate to rule out’ .. } → ‘NEGEX’, {’suspicion’, ‘probable’, ‘possible’} → ‘RISK’, {’increase’, ‘invasive’, ‘diffuse’, ..} → ‘QUAL’. The common-term dictionary contains on total 800 unique terms.
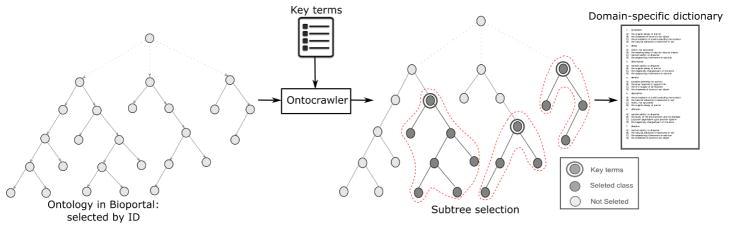
In the second level of the mapping, we built a domain specific dictionary for Pulmonary embolism (PE) case study by using three distinct bio-portal ontologies [19] - SNOMEDCT, MEDDRA, and RadLex. The main idea is to reduce the variations of radiological terms that are often used by the clinician while reporting pulmonary embolism cases. We collected 44 unique domain-specific terms from the radiologists. We created a SPARQL based ontology crawler that search for the domain specific key terms remotely on a bio-portal ontology specified by the unique ID, and grabs all the sub-classes and synonyms of the domain-specific terms from the targeted ontology. Figure 3 presents the functioning of the ontology crawler in a higher-level where, the crawler extracted 3 sub-trees given 3 key-terms(red dashed). Finally, the crawler automatically resolves the redundancy and creates a focused dictionary for “Pulmonary embolism (PE)” case-study.
Figure 3.
Ontocrawler pipeline
The PE dictionary contains total 44 unique terms and on average 5–8 are mapped with each unique terms. For instance, all the equivalent terms of pulmonary embolism are formalized as: {’pulmonary embolism’, ‘pulmonary embolus’, ‘lungenembolie’, ...} → ‘pe’. Our semantic dictionary mapping step considerably reduced the size of our vocabulary (~40%) by preserving the true semantics of terms, thereby making the words in the vocabulary more frequent. Therefore, the automatic dictionary tailoring using ontology crawler provide a solution to efficiently utilize multiple large-scale ontologies while increasing the overall processing performance.
3.2.3. Unsupervised embedding
The corpus of pre-processed reports was used to create vector embeddings for words in a completely unsupervised manner using a word2vec predictive model. The Semantic dictionary mapping step (Section 3.2.2) not only considerably reduced the size of our vocabulary by mapping the words in corpus to the controlled terms derived from the domain specific dictionary for Pulmonary embolism (PE) and CLEVER, but also decreased the probability of OOV word encounter. The idea behind this is that the context of controlled terms formalized in the knowledge-base should capture the true semantics and can facilitate information extraction from the reports. Therefore, the reports are pre-processed using Report Condenser and Semantic dictionary mapping to balance the text consistency with less term variation which facilitates the application of word2vec directly to parse radiology reports.
The model probes the finer structure of the word vector space by representing each word as a distribution of weights across several hundred dimensions. So instead of a one-to-one mapping between an element in the vector and a word, the representation of a word is spread across all of the elements in the vector. It also captures the semantic regularities of words. We first constructed a vocabulary from our pre-processed corpus, and then learned vector representations of words in the vocabulary. One word is considered per context, which means the model will predict one target word given one context word. The loss function of prediction model is: , where wo is the output word, vwo′ is its output vector, h is the average of vectors of the context words, and V is the entire vocabulary.
We used both Hierarchical Softmax as well as Negative Sampling for training word embedding model and we found based on the experiments that Negative Sampling to be faster and better training algorithm for this case-study. Mikolov et al. [10] also advocated Negative Sampling as the method that results in faster training and better vector representations for frequent words. The cost function of Negative Sampling is: E = −log σ(vwo′.h) − Σwj∈ωneg log σ(−vwj′.h), where ωneg is the set of negative samples, wo is the output word, vwo′ is its output vector and h is the average of vectors of the context words.
We explored all possible combinations of the following configurations to train the word2vec model: (1) the dimension of word vectors: (100, 200, 300, and 700); and (2) the size of the context window: (10, 20, 30, and 50). Using 10-fold cross validation on the training dataset (Stanford dataset), we found that the optimized performance was achieved with the skip-gram architecture, vector dimension of 300 and a window size of 30.
3.2.4. Document vector creation
The document vectors were created by a word averaging approach that simply averages the word vectors created through the trained model. Each document vector was computed as: , where Vdoc is the set of words in the report and vw refers to the word vector of word w. We also experimented with 2-step Doc2vec model that, first, modifies the word2vec algorithm to unsupervised learning of continuous representations for larger blocks of text, and then we retrain the model with lower learning rate (10 times smaller than original learning rate) on a smaller subset of labeled data. But the initial experiments showed that accuracy of unsupervised word averaging approach for the targeted learning task performed better than the 2-step semi-supervised Doc2Vec approach.
3.2.5. Classification
Ideally, IWE generated document embeddings can be utilized to train any Parametric classifiers (Logistic Regression) as well as Non-Parametric classifiers (Random Forests, Support Vector Machines, K-Nearest Neighbors (KNN)) to fulfill various type of classification tasks. For this study, we experimented with binary logistic regression models (Lasso) with 10 fold cross validation on the same training dataset as described in Sec. 3.1 and report the performance on the test sets. We train three separate Lasso models for predicting PE present/absent, Acute/chronic, Central/Subsegmental using the same vector embeddings as input. Out of the 4512 annotated reports in the Stanford corpus, 3512 reports were randomly selected for training and remaining 1000 selected as test sets. The classification models trained on the Stanford dataset have been directly applied on the UPMC dataset (858 reports) for testing. Note that in the current study only the impression section of the radiology reports has been considered.
4. Results
4.1. Validation of the embedding
We explored the semantic quality of the embedding generated by our IWE method in two different ways. First, we find similar word clusters in a totally unsupervised manner to verify the positioning of synonyms (and related words). This can show at the very low scale that if our vector embedding is able to preserve legitimate semantics of the natural words and clinical terms. Second, we visualize the document vectors to fulfill the purpose of analyzing the proximity of documents that have different levels of PE categorization. If the documents corresponding to the same class (risk) appear close to each other and form clusters, we can infer that our embedding carries substantial signals which can be useful to boost the performance of any standard classifiers.
Word embedding
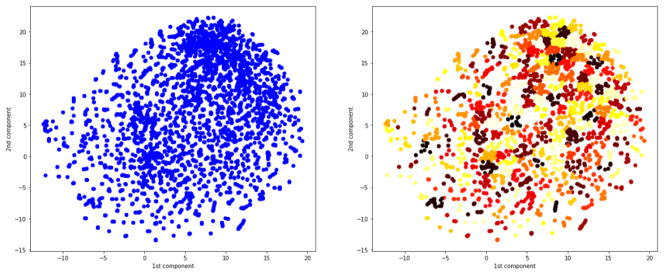
For projecting the high dimensional trained embeddings of words and documents in 2D, we used t-Distributed Stochastic Neighbor Embedding (t-SNE) method [20] since it is able to capture much of the local structure of the high-dimensional data, while also revealing global structure. Figure. 4 (on the left) shows the 2D visualization of the complete vocabulary constructed using the t-SNE approach where each data point represents a word. To investigate the semantic correctness of the word embedding, we performed clustering of the space using a modified version of k-means - k-means++, where randomized seeding technique has been adopted to determine the starting centroids of the clusters. The optimal number of clusters - ‘k’, has been determined by Silhouette analysis [21] where ‘k’ is selected according to the highest separation distance between the resulting clusters. For our vocabulary of size 3650 words, we tried the number of cluster within the range [10, 1000] and the highest silhouette measure was 0.62 for k = 200 which depict a reasonably strong clustering of the space. Interestingly, most of the clusters contain semantically similar words. We present a few representative clusters in Table 2 where terms related to ‘Cancer’, ‘Cardiac’, ‘Skeletal’, ‘Location’, ‘Effsusion’, ‘Procedure’ are clustered together without even inclusion of any prior knowledge. This clustering outcome illustrates that our word embedding was able to preserve the semantics of the relevant terms in an unsupervised manner.
Figure 4.
On left all word embeddings generated by IWE (vocabulary size - 3650 words) and visualized in two dimensions using t-SNE; On right clustering of the word embedding space using K-means++.
Table 2.
Clustered explored from IWE space using K-means++
| Clusters | Words |
|---|---|
| Cluster 1: Cancer | ‘carcinoma’, ‘metastas’, ‘metastasi’, ‘mass’, ‘malign’, ‘adenocarcinoma’, ‘lymphoma’, ‘tumor’, ‘lymphadenopathi’, ‘carcinomatosi’, ‘adenopathi’, ‘neoplasm’, ‘cancer’, ‘lymphomat’, ‘metastat’, ‘metastat_diseas’, |
| Cluster 2: Cardiac | ‘ventricl’, ‘heart’, ‘pulmonari_arteri’, ‘atrium’, ‘ventricular’, ‘atrial’ |
| Cluster 3: Skeletal | ‘boni’, ‘lytic’, ‘vertebr_bodi’, ‘sclerot’, ‘skeleton’, ‘bone’, ‘lucent’, ‘spine’, ‘sclerosi’, ‘osseous’ |
| Cluster 4: Location | ‘right_lower’, ‘left_lower’, ‘left_upper’, ‘right_upper’, ‘upper’, ‘lower’ |
| Cluster 5: Effusion | ‘pleural_effus’, ‘bilater_pleural_effus’, ‘left_pleural_effus’, ‘effus’, ‘right_pleural_effus’ |
| Cluster 6: Hemorrhage/infection in lungs | ‘hemorrhag’, ‘layer’, ‘air’, ‘pneumoperitoneum’, ‘space’, ‘wound’, ‘hemoperitoneum’, ‘empyema’, ‘pneumothorac’, ‘pneumomediastinum’, ‘hemothorax’, ‘blood’, ‘abscess’, ‘hydropneumothorax’, ‘pneumothorax’, ‘hemithorax’, ‘bronchopleur’, ‘pigtail’, ‘fluid’, ‘intraperiton’, ‘bleed’, ‘hematoma’, ‘pocket’ |
| Cluster 7: Suspicious | ‘concern’, ‘suspici’, ‘worrisom’ |
| ....... ....... |
|
| Cluster 200: Procedure | ‘procedur’, ‘right_upper_lobectomi’, ‘left_upper_lobectomi’, ‘right_lower_lobectomi’, ‘right_middl_lobectomi’, ‘left_lower_lobectomi’, ‘transplant’, ‘mastectomi’, ‘therapi’, ‘therapeut’, ‘lumpectomi’,’thyroidectomi’, ‘pneumonectomi’, ‘nephrectomi’, ‘colectomi’, ‘lobectomi’, ‘hemicolectomi’, ‘treatment’, ‘oper’, ‘posttreat’, ‘chemotherapi’, ‘cholecystectomi’, ‘adrenalectomi’, ‘orchiectomi’, ‘surgic’, ‘radiotherapi’,’radiation’, ‘hepatectomi’, ‘pleurodesi’ |
Report embedding
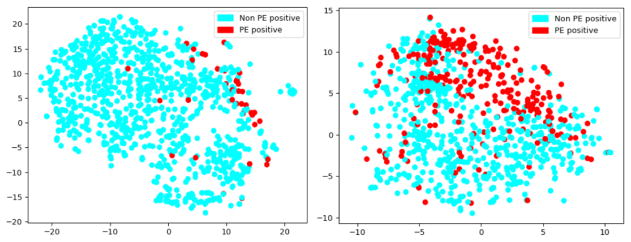
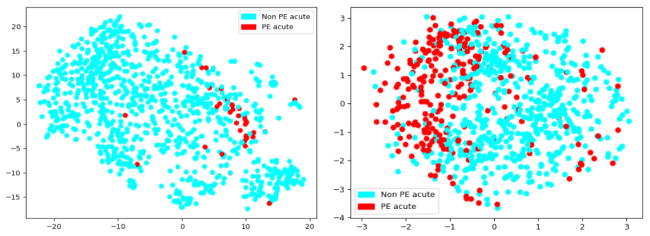
In the next level of validation, we used one time execution of the t-SNE method with the perplexity of the conditional probability distribution as 30 for projecting the high-dimensional document vector created by IWE model into 2D space. In Figure. 5 and 6, we visualize the subsequent vector embeddings of the whole reports from Stanford and UPMC dataset derived by the IWE model. These visualizations have been created only on the test cases (Stanford - 1000 and UPMC - 858) and the coloring has been shown based on the ground truth annotations defined by the radiologists. The figures show that the embeddings created from IWE models were able to preserve meaningful signal for distinguishing the reports annotated with varying risk PE factors, and formed natural clusters in the embedding space. Though this is only a two-dimensional projection of the original high dimensional document vector, the results clearly exhibit that the embeddings could be very informative to automatically classify the reports using any standard classifier. It is important to note that the document embeddings generated by the IWE model is completely unsupervised (see Figure 2), yet it was able to preserve the semantics of the reports at varying level of PE categorizations.
Figure 5.
Unsupervised IWE reports embedding projected in 2D highlighting the label PE positive - Stanford test set (on left) and UPMC dataset (on right)
Figure 6.
Unsupervised IWE reports embedding projected in 2D highlighting the label PE acute - Stanford test set (on left) and UPMC dataset (on right)
4.2. Classification performance
We first created a simple baseline using bag-of-words model (BoW) which represents the radiology reports as the bag (multiset) of its words, disregarding grammar and even word order but keeping multiplicity. For comparative assessment of IWE model, we applied the previously published rule-based PeFinder model [8]. We also performed comparison of IWE with baseline neural embedding where we generated the embedding of the documents using out-of-the-box word2vec model [9, 10] and used logistic regression on top of it. The experiments have been performed on 1,000 classified reports from Stanford dataset and 858 reports from the University of Pittsburgh Medical Center which were used originally to train the PeFinder. Both IWE and standard word2vec model has been trained from scratch on 117,816 CT reports belonging to the Stanford dataset, and on top of the embeddings the same supervised classifier has been trained only on the Stanford training dataset (3512 reports) and tested on both unseen Stanford and UPMC testset.
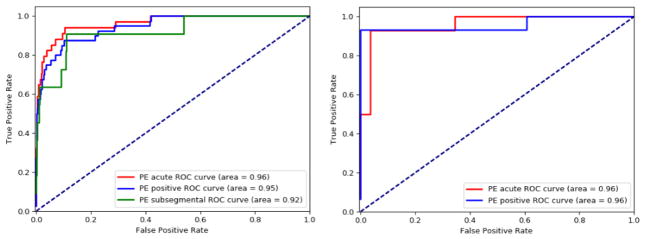
Receiver Operating Characteristic (ROC) curves is a well-accepted method to show the tradeoff between specificity and sensitivity where the models produced a sort of scores for test samples, and presents pairs of specificity and sensitivity values calculated at all possible threshold scores. The ROC curve also provides a single performance measure called the Area under the ROC curve (AUC) score where higher AUC represents better performance. The ROC for IWE model are shown in Figure 7 where we present the curves on both Stanford and UPMC dataset side-by-side. Different colors represent three different PE categorical labels considered in the study. On Stanford test set (ROC on the left), our model achieved AUC 0.96 for PE acute, AUC 0.95 for PE positive, and AUC 0.92 for PE subsegmental. Even without retraining on UPMC test set (ROC on the right), our model maintained the similar performance level with AUC 0.96 for both PE acute and PE positive. However, the interpretation of ROC curves requires a special caution. This is mainly due to the fact that ROC may actually introduce more uncertainty into machine learning classification accuracy comparisons than resolution, and end up showing a very optimized performance. Moreover, it is not possible to draw a fair comparison between the models with ROC curves because the PeFinder model does not output probability scores for its predictions - just binary outcomes.
Figure 7.
ROC Curve for IWE classifier - Stanford Test Set (on left) and UPMC Test set (on right)
Therefore, we decided to evaluate the proposed model’s, BoW, out-of-box word2vec and PeFinder performance in terms of Precision, Recall, and F1 score. In Table 3, we present the simple baseline BoW model’s performance. In Table 4 and 5, we show the performance measures in-terms of precision and recall value for the three comparative models - (i) rule-based - PEFinder, (ii) baseline neural embedding - word2vec, (iii) proposed hybrid model - IWE. UPMC dataset does not have the ground truth labels for PE subsegmental only category, and PeFinder is not engineered for classifying this category. Therefore, we mentioned ‘N/A’ where the performance could not be validated in comparison with the ground truth labels or model could not performed the classification task.
Table 3.
Performance measures of Bag-of-Words model on both Stanford and UPMC dataset
| Simple baseline - Bag-of-Words (BoW) Model | ||||||
|---|---|---|---|---|---|---|
| Labels | Stanford dataset | UPMC dataset | ||||
| Precision | Recall | F1-score | Precision | Recall | F1-score | |
| PE acute | 72.38% | 83.53% | 77.11% | 82.04% | 81.55% | 81.79% |
| PE present | 78.15% | 88.75% | 82.49% | 83.19% | 83.46% | 83.33% |
| PE subsegmental only | 72.82% | 76.92% | 74.70% | N/A | ||
Table 4.
Performance measures on the Stanford dataset
| Comparative models | ||||||
|---|---|---|---|---|---|---|
| Labels | IWE method | Out-of-box word2vec | PEFinder | |||
| Precision | Recall | Precision | Recall | Precision | Recall | |
| PE acute | 97.71% | 97.40% | 96.44% | 93.20% | 91.2% | 91.2% |
| PE present | 97.25% | 96.70% | 96.29% | 89.45% | 87% | 90% |
| PE subsegmental only | 97.81% | 98.90% | 98.11% | 99.05% | N/A | N/A |
Table 5.
Performance measures on the UPMC dataset
| Comparative models | ||||||
|---|---|---|---|---|---|---|
| Labels | IWE method | Out-of-box word2vec | PEFinder | |||
| Precision | Recall | Precision | Recall | Precision | Recall | |
| PE acute | 90.85% | 90.70% | 88.11% | 87.21% | 91% | 95% |
| PE present | 93.03% | 93.02% | 84.81% | 84.88% | 87% | 96% |
As seen from the measures, all three comparative models outperformed simple baseline BoW model for the Stanford as well as for the UPMC test set. The IWE model had the lowest generalization error across most of the dimensions of PE measures among the models. But, as seen from Table 4, precision measures of standard word2vec model trained on the same dataset are close to IWE model, and even closely outperformed IWE model for PE subsegmental category. But, for a completely different UPMC dataset, IWE model performs signifocantly better than word2vec, but PeFinder model achieved slightly higher Recall value for this dataset (Table 5). Therefore, we performed nonparametric statistical signifocant tests for both Precision and Recall score using Fisher’s exact test [22] which calculates an exact p – value based on the sample data (see Table 6). From the results, we can conclude that even if the Recall is higher for the PeFinder on UPMC dataset and Precision of word2vec for Stanford dataset, it is not statistically significant as p – value < 0.001.
Table 6.
Statistical significance of Precision and Recall
| Dataset | Statistical Significance (IWE - PeFinder) | p-value | Statistical Significance (IWE - word2vec) | p-value |
|---|---|---|---|---|
| Stanford | Precision | <0.0001 | Precision | <0.0001 |
| Recall | 0.0018 | Recall | 0.002 | |
| UPMC | Precision | 0.2076 | Precision | 0.1204 |
| Recall | 0.1724 | Recall | 0.2403 |
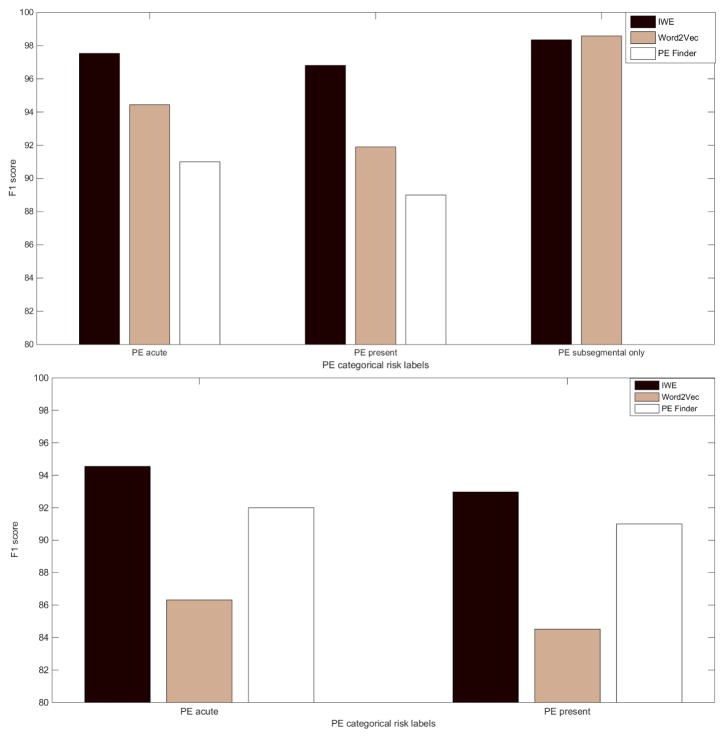
To capture a better view of the performance, we computed the F1 score which is a harmonic mean of precision and recall, for all the models and visualize as bar plot in Fig. 8). The graph clearly shows that IWE model performed consistently better on both Stanford and UPMC data set for all the categorical PE measures. On the Stanford test set, the IWE model achieved average F1 score 0.97, when the PeFinder scored 0.9 and the out-of-the-box word2vec scored 0.94. On UPMC dataset, the IWE model’s average F1 score was 0.94, when the PeFinder scored 0.92 and out-of-the-box word2vec scored 0.85. This is an exciting result since the IWE model outperformed Pefinder which was actually trained as tested on the UPMC dataset. Therefore, it clearly shows that the IWE model is not over-fitted to the Stanford training dataset and can be easily extendable to a completely different organization dataset without retraining. The cross-institutional generalizibility of the IWE method is mainly facilitated due to the efficient integration of neural embedding and semantic dictionary mapping, since it can tackle the major challenges of information extraction from clinical texts, which include ambiguity of free text narrative style, lexical variations, use of ungrammatical and telegraphic phases, arbitrary ordering of words, and frequent appearance of abbreviations and acronyms. Only for PE subsegmental for Stanford testset, F1 score for standard word2vec is slightly higher than IWE, mainly due to the high precision. However, the Fisher’s exact test showed that the difference is not statistically significant.
Figure 8.
Bar plots showing F1 scores in percentage computed on Stanford Test Set(on top) and UPMC Dataset(on bottom) where IWE is represented as dark brown, Out-of-box word2vec as sand, and PEFinder as white colored bar.
5. Discussion
The purpose of this study was to propose an efficient method that can classify free text contrast enhanced chest CT reports based on three different clinically relevant classifications related to the diagnosis of pulmonary embolism - (1) PE present/absent; (ii) PE acute/chronic; (iii) PE central/subsegmental only. We designed a hybrid semi-supervised method termed Intelligent Word Embedding (IWE) that combines word embedding approach proposed by Mikolov et al [10] with domain specific semantic dictionary mapping technique for creating dense vector embedding of the reports. The combination of the neural language model with a semantic dictionary aims to address one of the biggest limitation of word2vec which is the inability to handle unknown or out-of-vocabulary (OOV) words and morphologically similar words. Thanks to the embeddings, we successfully annotated the radiology reports according to the categorization of PE with average F1 score of 0.97% given a small set of annotated reports. Experiments performed in this study showed that the hybrid IWE model outperforms the out-of-the-box word2vec model for the PE risk categorization task while using the same discriminative model on top of the embedding. Moreover, considerable higher performance of the IWE model over the word2vec on the UPMC dataset suggests that the neural embedding model combined with semantic dictionary mapping can be more generalizable for a different organizational dataset. Therefore, such model will also require minimal human effort for task specific customization. To our knowledge, this is a new approach to overcoming the challenge of handling OOV words in word embedding techniques.
We selected the pulmonary embolism case-study mainly because it is one of the most common indications for a chest CT with contrast in the emergency setting and has a high population morbidity and mortality [23]. Extracting the characterization of the location, severity, and timing of the pulmonary embolism diagnosis from the unstructured radiology report would be valuable to precision health and big data initiatives that leverage the clinical record for clinical informatics prediction and risk modeling. In a typical clinical setting, identification of cases requires through manual review of the huge hospital repository and identify exams in order to review the dictated report to determine the characteristics of the resulting exam and diagnosis. Our work has demonstrated that deep-learning based word embedding tool, rather than more traditional labor intensive feature engineering approaches to NLP, can efficiently automate this process and achieve superior performance. Our framework only need a small subset of labeled data to train the discriminative model on top of the dense vector embedding.
We also compared the performance of proposed semi-supervised hybrid method to a state-of-the-art rule-based solution for classifying free text contrast enhanced chest CT reports - called PeFinder [8]. The complementary architectures help to draw substantial assessment of two well-known aspects of natural language processing on same contextual analysis. Among the three comparative models that we evaluated in the current study, the proposed IWE model resulted highest accuracy with average F1 score > 0.97 for Stanford dataset (Sec. 3.1) within all dimensions of PE measures (PE positive, PE acute and PE subsegmental). Of particular interest, the IWE model (trained on the Stan-ford dataset) outperformed PeFinder on the University of Pittsburgh Medical Center dataset labeling task which was used originally to tailor the PeFinder model. This demonstrates good generalizability of the proposed approach across institutions.
IWE performed better than PE-Finder in several ways. While the IWE model’s prediction was accurate, the rule-based PE-Finder wrongly classified the reports as PE present when there is mention of historic evidence of pulmonary embolism, e.g. “The previously described pulmonary emboli are no longer visualized.” or embolism occurs in a different anatomical region, e.g. “This is most consistent with a renal infarct, possible from embolism to a small renal artery branch.”. Moreover, the PEFinder also failed to identify non-significant negation claims in the impression section and tagged the report incorrectly as PE absent, e.g. “Stable partially occlusive chronic thrombus in the right main pulmonary artery. No new emboli”. There is no standardized lexicon of all the different words or combinations of words that can represents various aspect of pulmonary embolism, and the type of relations that can be encountered in free text reports are difficult to know in advance. Therefore, it is an impractical task to generate rules for every situation which is one of the main limitations of any rule-based method. From the high accuracy achieved by the IWE model, we can conclude that neural embedding can be utilized as a very powerful tool for extracting semantics of the free-text radiology reports. We suspect that better performance of IWE model occurs mainly due to the significant reduction of vocabulary size and domain specific word embedding steps.
This study has several limitations. First, our model lacks the sensitivity for word order that limits the ability of learning long term and rotated scope of negex term. However, thanks to the semantic mapping and word occurrence analysis, the adjacent negex terms are concatenated with the targeted entity in the pre-processed text before Word2Vec training, e.g. ‘negex_pulmonari’. As a result, the model had the opportunity to learn the representation of the entity and the adjacent negation of the entity without explicitly considering the sentence boundary. The trained IWE model derived negative cosine similarity score(−0.245) for word the ‘pulmonari’ and ‘negex_pulmonari’.
Second, we used free text reports from two large academic medical practices which may have some similarities in radiology report dictation narrative which could be more variable in smaller institutions or non-academic medical practices and may limit the generalizability of the models. Additionally, the dataset included in the study are associated to a very narrow domain, i.e. contrast-enhanced CT examinations of the chest, and thus the variation in the vocabulary of radiology reports is relatively small. We expect that the superiority in the performance of the proposed combination of semantic mapping and neural embedding will vary when multi-topic and multi-institutional free-text reports will be considered; future work may consider applying the IWE model to the whole medical repository of major hospital system for creating dense vector representation of clinical notes of different domains (e.g. oncology, pathology), given appropriate domain-ontologies. This can help to develop an “Intelligent Clinical Data” platform that can normalize free text into a dense vector representation, while aggregating it with discreet data from EMRs and diagnostic information systems via its transformation interfaces.
The IWE method can be extended to a different domain with minimal human effort, only given two vital inputs - domain specific key terms and ontology identifiers. However, a completely different domain other than radiology may need also some manual tuning in the Report condenser and Ontocrawler, since data quality and domain-ontology schema may vary significantly among domains. In addition, training the discriminative model on top of the embedding needs human labeled data for performing the task specific annotation. Future work will be to discover the domain specific key terms from the free-text corpus for the domain-specific taxonomy creation and unsupervised clustering of the vector space for identifying meaningful annotation of the reports.
6. Conclusions
In conclusion, our deep learning approach to natural language processing for classifying free text radiology reports demonstrates high fidelity compared to state-of-the-art rule-based method and appears to be generalizable across institutions for the clinically relevant categorization related to the diagnosis of pulmonary embolism. Automated information extraction from radiology reports with deep learning may have many applications in machine vision work by providing accurate labeling of medical images on a large scale from the radiology report. Further, these techniques may make the valuable diagnostic information in radiology report text available at a large scale to be used in models that evaluate imaging utilization, used as part of clinical decision support models, used to predict outcomes, and used as a valuable tool to evaluate ordering provider imaging yield rates. This information can be useful in an array of applications from large scale retrospective research to focused clinical applications.
Highlights.
Proposed an unsupervised method that combines neural embedding method with a semantic dictionary mapping for creating a dense vector representation of unstructured radiology reports.
Applied to generate embedding of chest CT radiology reports from two healthcare organizations and utilized the vectors to semi-automate report categorization based on diagnosis of pulmonary embolism (PE).
Resulted lowest generalization error with highest F1 scores and outperformed state-of-the-art rule-based system – PEFinder.
The method can be extended to a different domain with minimal human effort, only given two vital inputs - domain specific key terms and ontology identifiers.
Acknowledgments
The authors would like to thank Brian Chapman, Ph.D. of the University of Utah for his generous contribution of report data and the PeFinder model code. This work was supported in part by grants from the National Cancer Institute, National Institutes of Health, U01CA142555, 1U01CA190214, and 1U01CA187947, and Stanford Child Health Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical text analysis and knowledge extraction system (ctakes): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dublin S, Baldwin E, Walker RL, Christensen LM, Haug PJ, Jackson ML, Nelson JC, Ferraro J, Carrell D, Chapman WW. Natural language processing to identify pneumonia from radiology reports. Pharmacoepidemiology and drug safety. 2013;22(8):834–841. doi: 10.1002/pds.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho J, Lee K, Shin E, Choy G, Do S. Medical image deep learning with hospital pacs dataset. arXiv preprint arXiv:1511.06348. [Google Scholar]
- 4.Hua K-L, Hsu C-H, Hidayati SC, Cheng W-H, Chen Y-J. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets and therapy. :8. doi: 10.2147/OTT.S80733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anavi Y, Kogan I, Gelbart E, Geva O, Greenspan H. A comparative study for chest radiograph image retrieval using binary texture and deep learning classification. Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE; IEEE; 2015. pp. 2940–2943. [DOI] [PubMed] [Google Scholar]
- 6.Long B, Koyfman A. Best clinical practice: Current controversies in pulmonary embolism imaging and treatment of subsegmental thromboembolic disease. The Journal of emergency medicine. 2017;52(2):184–193. doi: 10.1016/j.jemermed.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 7.Martinez JLA, Sánchez FJA, Echezarreta MAU, García IV, Álvaro JR. Central versus peripheral pulmonary embolism: Analysis of the impact on the physiological parameters and long-term survival. North American journal of medical sciences. 2016;8(3):134. doi: 10.4103/1947-2714.179128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman BE, Lee S, Kang HP, Chapman WW. Document-level classification of ct pulmonary angiography reports based on an extension of the context algorithm. Journal of biomedical informatics. 2011;44(5):728–737. doi: 10.1016/j.jbi.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. arXiv preprint arXiv:1301.3781. [Google Scholar]
- 10.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Advances in neural information processing systems. 2013:3111–3119. [Google Scholar]
- 11.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. Journal of the American Medical Informatics Association. 1994;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hripcsak G, Austin JH, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports 1. Radiology. 2002;224(1):157–163. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]
- 13.Dreyer KJ, Kalra MK, Maher MM, Hurier AM, Asfaw BA, Schultz T, Halpern EF, Thrall JH. Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: validation study 1. Radiology. 2005;234(2):323–329. doi: 10.1148/radiol.2341040049. [DOI] [PubMed] [Google Scholar]
- 14.Christensen LM, Haug PJ, Fiszman M. Proceedings of the ACL-02 workshop on Natural language processing in the biomedical domain. Vol. 3. Association for Computational Linguistics; 2002. Mplus: a probabilistic medical language understanding system; pp. 29–36. [Google Scholar]
- 15.Martinez D, Ananda-Rajah MR, Suominen H, Slavin MA, Thursky KA, Cavedon L. Automatic detection of patients with invasive fungal disease from free-text computed tomography (ct) scans. Journal of biomedical informatics. 2015;53:251–260. doi: 10.1016/j.jbi.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Sohn S, Ye Z, Liu H, Chute CG, Kullo IJ. Identifying abdominal aortic aneurysm cases and controls using natural language processing of radiology reports. AMIA Summits on Translational Science Proceedings. 2013;2013:249. [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen DH, Patrick JD. Supervised machine learning and active learning in classification of radiology reports. Journal of the American Medical Informatics Association. 2014;21(5):893–901. doi: 10.1136/amiajnl-2013-002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. [accessed: 2017-09-26];Clever terminology in github. https://github.com/stamang/CLEVER/blob/master/res/dicts/base/clever_base_terminology.txt.
- 19.Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, Jonquet C, Rubin DL, Storey M-A, Chute CG, et al. Bioportal: ontologies and integrated data resources at the click of a mouse. Nucleic acids research. 2009;37(suppl 2):W170–W173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maaten Lvd, Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research. 2008 Nov;9:2579–2605. [Google Scholar]
- 21.Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. Journal of computational and applied mathematics. 1987;20:53–65. [Google Scholar]
- 22.Agresti A. A survey of exact inference for contingency tables. Statistical science. 1992:131–153. [Google Scholar]
- 23.Chapman WW, Nadkarni PM, Hirschman L, D’avolio LW, Savova GK, Uzuner O. Overcoming barriers to nlp for clinical text: the role of shared tasks and the need for additional creative solutions. 2011 doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]