Abstract
Rationale
Primary Ciliary Dyskinesia (PCD) is a genetically heterogeneous condition characterized by progressive lung disease arising from abnormal cilia function. Approximately half of patients have situs inversus. The estimated prevalence of PCD in the UK South Asian population is 1:2,265. Early, accurate diagnosis is key to implementing appropriate management but clinical diagnostic tests can be equivocal.
Objectives
To determine the importance of genetic screening for PCD in a UK South Asian population with a typical clinical phenotype, where standard testing is inconclusive, and assess the relationship between gene mutations and diagnostic phenotype.
Methods
Next-generation sequencing was used to screen 86 South Asian patients who had a clinical history consistent with PCD. The effect of a CCDC103 p.His154Pro missense variant compared to other dynein arm-associated gene mutations on diagnostic/phenotypic variability was tested. CCDC103 p.His154Pro variant pathogenicity was assessed by oligomerisation assay.
Results
Sixteen of 86 (19%) carried a homozygous CCDC103 p.His154Pro mutation found to disrupt protein oligomerisation. Variable diagnostic test results were obtained including normal nasal nitric oxide levels, normal ciliary beat pattern and frequency and a spectrum of partial and normal dynein arm retention. Fifteen (94%) patients or their sibling(s) had situs inversus suggesting CCDC103 p.His154Pro patients without situs inversus are missed.
Conclusions
The CCDC103 p.His154Pro mutation is commoner than previously thought in the South Asian community and causes PCD that can be difficult to diagnose using pathology-based clinical tests. Genetic testing is important where there is a strong clinical phenotype with inconclusive standard diagnostic tests.
Keywords: primary ciliary dyskinesia, respiratory tract, cilia, diagnosis, CCDC103, mutation, genetic testing
INTRODUCTION
Primary ciliary dyskinesia (PCD; OMIM: 244400) is an inherited disorder affecting motile cilia. Patients usually present with a history of neonatal respiratory distress and suffer from lifelong symptoms of chronic wet cough and rhinitis. Recurrent chest infections ultimately lead to bronchiectasis and a progressive decline in lung function.1 Approximately half of patients have situs inversus and other situs abnormalities, due to ciliary dysmotility in the embryonic node.2 Fertility can also be affected by defective cilia in the fallopian tubes and non-motile sperm tail flagella.
The estimated prevalence of PCD in the UK is 1:15,000, but as high as 1:2,265 in the UK South Asian population.1,3 Early diagnosis is important to maintain lung function, and appropriately treat symptoms to reduce morbidity and mortality.4 Diagnosis can be complex and requires a combination of tests for cilia functional and ultrastructural defects.5,6 PCD is caused by mutations in genes encoding proteins conferring structural stability to the cilia and governing ciliogenesis. It is genetically heterogeneous with >200 individual mutations in more than 30 genes known to cause PCD, accounting for approximately 65% of cases.1,7–11 UKGTN-approved tests are offered in two centres (http://www.labs.gosh.nhs.uk/media/764464/ciliopathies_v8.pdf).12
CCDC103 mutations were first reported, in 2012, in patients with dynein arm loss and a typical clinical phenotype13. CCDC103 is an oligomeric protein of the cilia thought to participate in microtubule stabilization that facilitates dynein arm attachment and ciliary motility.13 A missense variant, p.His154Pro (rs145457535), was previously described as a hypomorphic mutation since mutant p.His154Pro CCDC103 induced intermediate partially rescued disease phenotypes when expressed in a zebrafish CCDC103-null model, suggesting some retained protein function.13 In cilia from p.His154Pro patients, including from a South Asian family, some cilia showed a partial dynein arm defect with the outer dynein arms (ODA) at least partially assembled, compared to loss-of-function CCDC103 mutations causing complete ODA loss. In agreement with this, loss-of-function CCDC103 mutation patients had largely static cilia, whilst patients carrying a homozygous p.His154Pro mutation exhibit a mixed (static and motile) ciliary beat pattern.13
We conducted genetic screening of 86 PCD patients of South Asian (mostly Pakistani) origin, detecting significant numbers (19%) were homozygous for CCDC103 p.His154Pro variant. Amongst these, many were at high risk of being mis-diagnosed without genetic testing, due to normal diagnostic results obtained in PCD investigations. Using electron microscopy and protein biochemistry we have sought to further determine the pathogenic nature of the p.His154Pro mutation.
METHODS
Patient selection
Eighty-six patients of South Asian (primarily Pakistan) descent with clinical signs and symptoms of PCD were identified from the UK National PCD Diagnostic and Management Services at Leeds and Bradford, The Royal Brompton Hospital, London, University Hospital Southampton, Birmingham Children’s Hospital and Leicester Royal Infirmary.1,14
Genotyping
All participants gave written informed consent to take part in this study. The protocol was approved by the London Bloomsbury Research Ethics Committee (08/H0713/82). High throughput screening used next-generation sequencing, either whole exome sequencing (WES) or targeted gene panel sequencing. Sequencing and variant identification methods are published for WES,12,15 or used custom gene panels (Illumina TruSeq Custom Amplicon or Agilent SureSelect Focused Exome (proprietary product) and SureSelectXT custom panel design systems) and a standardised variant calling pipeline.16 Sanger sequencing was used for variant confirmation, familial segregations and the replication study.
Comparator group
The comparator patient group consisted of 16 of the 86 individuals tested. This group was closely age and gender matched to the CCDC103 p.His154Pro group, all had a dynein arm defect on electron microscopy, but all were proven negative for CCDC103 p.His154Pro mutation.
FEV1 measurements
Spirometry was performed according to American Thoracic Society/European Respiratory Society recommendations.17 Forced expiratory volume in 1 second (FEV1) z-scores were calculated using the Global Lungs Initiative parameters.18
Diagnostic tests
Screening and diagnostic testing was performed according to the PCD National Service protocols. Investigations included nNO, nasal brushing analysed by HSVM for ciliary beat frequency and pattern19 and quantitative electron microscopy for ciliary ultrastructure.23 (Full details in OLS). When results were inconclusive or inconsistent, patients were offered repeat testing.
Protein biochemistry on recombinant CCDC103 protein
Site-directed mutagenesis (QuikChange kit, Agilent Technologies UK Ltd) was used to generate the p.His154Pro mutation in an N-terminal His10 tagged H. sapiens CCDC103 cDNA synthesized using Escherichia coli codon bias which was subcloned into pET16b vector.20 Following transformation into E. coli BL21 (DE3), protein expression was induced by addition of 2 mM IPTG for two or more hours. Following sonication, His10-tagged proteins were dissolved in 8 M urea, very slowly diluted into 1 litre of 20 mM Tris.Cl pH8.0 150 mM NaCl then purified by Ni2+-affinity chromatography as described previously. Samples concentrated by ultrafiltration were subject to gel filtration in a calibrated Superose 6 10/300 column attached to an ÄktaPurifier-10 chromatography workstation.20
Statistical analysis
No power calculation was performed as the study size was opportunistic. The CCDC103 p.His154Pro group and the comparator group were closely matched for age and gender. Data was not normally distributed and therefore groups were compared using non-parametric statistical tests. P<0.05 was considered statistically significant.
RESULTS
A PCD-causing mutation CCDC103 p.His154Pro is prevalent in UK individuals of South Asian origin with a clinical phenotype of PCD
Genetic screening using next generation sequencing (either WES or gene panel) revealed 16 patients (19% of a total of 86 patients of South Asian descent) from 12 independent families. All were of consanguineous origin and homozygous for a previously published single base change (c.461A>C) mutation in CCDC103 that results in the amino acid substitution p.His154Pro (Table 1)
Table 1.
Clinical history of CCDC103 p.His154Pro cases summarising diagnostic investigations
| Demographics | Symptoms | Diagnostic test results | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Sex | Age (yrs) | Age diag (yrs) | Family History | FEV1 z-score | Main Symptoms | Situs | Nasal NO (nl/min) | CBF (Hz) | Ciliary beat pattern | Electron microscopy (1st brushing) | Electron microscopy (repeat brushing) | Genetics: sequencing protocol |
| 1 | M | 7 | 7 | No | −3.45 | Chronic cough, developmental delay | SI | ND | 0 | Immotile | IDA +ODA | IDA +ODA | CM Watson et al. 2014 |
| 2 | M | 4 | 1 | No | −2.25 | Nasal discharge | SI | 1 | 0.95 | Mostly immotile with occasional residual movement | IDA +ODA | IDA +ODA | CM Watson et al. 2014 |
| 3 | M | 12 | 10 | No | −2.75 | Neonatal respiratory distress, nasal discharge | SS | 11 | 2.9 | Mostly immotile with occasional residual movement | IDA +ODA | IDA +ODA | CM Watson et al. 2014 |
| 4 | M | 13 | <1 | No | ND | Neonatal respiratory distress, Gilbert syndrome, epilepsy | SI | 7 | 5.9 | Mostly immotile with occasional residual movement | IDA +ODA | IDA +ODA | CM Watson et al. 2014 |
| 5 | M | 13 | 6 | Sib 6 | −1.52 | Recurrent chest infections | SI | 87 | 5.79 | Normal ciliary beat pattern reduced frequency | IDA +ODA | IDA +ODA | WES |
| 6 | F | 15 | 4 | Sib 5 | −0.61 | Recurrent chest infections, Eustachian tube dysfunction, nasal discharge | SS | 43 | 7.66 | Normal ciliary beat pattern reduced frequency | IDA +ODA | IDA +ODA | WES |
| 7 | F | 18 | 9 | No | −1.58 | Recurrent chest infections, nasal discharge, bilateral glue ear | SI | 111 | 10.6 | Mixed Sample 1: normal ciliary beat pattern sample 2: Immotile | IDA +ODA | IDA +ODA | Illumina TruSeq custom gene panel |
| 8 | M | 9 | 6 | No | −0.08 | Wet cough, recurrent chest infections, conductive hearing loss | SI | 57 | 8.8 | Dyskinesia | IDA | IDA | Agilent SureSelect Focused Exome |
| 9 | F | 7 | 7 | CHD | 0.43 | Chronic cough | SI | 33 | 10.9 | Normal ciliary beat pattern | Normal | ND | CM Watson et al. 2014 |
| 10 | M | 10 | <1 | Sib 11 & 12 | −1.32 | Neonatal respiratory distress, nasal discharge | SI | 26 | 11.5 | Mixed: Normal areas, immotile areas, dyskinetic areas | Normal | Normal | Illumina TruSeq custom gene panel |
| 11 | F | 21 | 10 | Sib 10 & 12 | ND | Recurrent chest infections, asthma, persistent collapse of right lower lobe | SI | 293 | 8.5 | Mixed: Mostly reduced forward and recovery stroke | IDA | Normal | Illumina TruSeq custom gene panel |
| 12 | M | 13 | <1 | Sib 10 &11 | −1.24 | Neonatal respiratory distress, nasal discharge | SS | 214 | 16.3 | Normal ciliary beat pattern | IDA | Normal | Illumina TruSeq custom gene panel |
| 13 | F | 29 | 29 | No | ND | Bronchiectasis, infertility | SI | 239 | 4.31 | Mixed: Mostly reduced forward and recovery stroke. Static patches | Normal | Insufficient | Agilent SureSelectXT Ciliome_651 |
| 14 | F | 1 | 1 | No | ND | Neonatal respiratory distress, nasal discharge, wet cough | SI | ND | 16.76 | Dyskinesia | Normal | Inconclusive | Agilent SureSelectXT Ciliome_651 |
| 15 | F | 21 | 21 | Sib 16 | ND | Recurrent chest infections in childhood. No ear, chest or nasal symptoms in adulthood | SI | 151 | 10.9 | Normal ciliary beat pattern | IDA | Normal | Agilent SureSelectXT Ciliome_651 |
| 16 | M | 18 | 18 | Sib 15 | ND | Sensory neural hearing impairment, recurrent chest infections, chronic nasal congestion and rhinitis | SI | 276 | 9.9 | Mixed. 80% normal ciliary beat pattern. 20% immotile cilia on strips. | IDA | Normal | Agilent SureSelectXT Ciliome_651 |
Normal test results are highlighted in bold. Age diag, age diagnosis confirmed; SI, situs inversus; SS, situs solitus; NO, nitric oxide; CBF, ciliary beat frequency; IDA, inner dynein arm; ODA, outer dynein arm; ND, not done.
In a small predictive study within this investigation, we specifically screened a subset of twelve ungenotyped South Asian PCD patients with reported ODA defects or normal ultrastructure for the CCDC103 p.His154Pro variant by Sanger sequencing. This detected one case (8%) carrying the mutation in homozygous form.
Whole exome sequence data obtained from 1,542 unaffected parents with similar ethnic backgrounds participating in the Born-in-Bradford cohort (UK South Asian, primarily Pakistani heritage)21 revealed six heterozygous carriers of the CCDC103 p.His154Pro variant (E. Sheridan, unpublished data). The ExAc database22 containing exome sequencing data from 60,706 unrelated non-PCD individuals records an allele frequency for p.His154Pro three times as high in 8,256 South Asian individuals (0.003) compared to 33,345 North Europeans (0.001). No p.His154Pro homozygote individuals were present in the entire Born-in-Bradford or ExAc cohorts.
Amongst the sixteen CCDC103 p.His154Pro homozygote PCD patients identified, normal diagnostic test results were apparent as highlighted in Table 1. Although PCD was strongly suspected in all cases, five of the sixteen patients (case 9 and 13-16 in Table 1) did not receive a definitive disease diagnosis until their genotype was confirmed. Strikingly in the case of patient 15, this individual had been deemed to not have PCD and was discharged from respiratory care. She was re-tested due to her situs inversus and the finding that her brother, who remained under clinical suspicion, had this CCDC103 mutation. Thirteen of the 16 CCDC103 p.His154Pro patients (81%) have situs inversus, two have a sibling with situs inversus and only one child did not have a family history of situs inversus. Situs inversus affects approximately 50% patients with PCD with dynein arm defects2 and affected 56% of the comparator group in this study (Table 2). 30% of the comparator group had no family history of situs inversus.
Table 2.
Comparison of mean values of clinical tests obtain in CCDC103 p.His154Pro cases versus a comparator group
| Clinical features | N | CCDC103 p.His154Pro | N | Comparator group |
|---|---|---|---|---|
| Age (mean SD) | 16 | 11.8 (4.8) | 16 | 11.9 (8.8) |
| Gender (% Male) | 16 | 67% | 16 | 63% |
| Nasal Nitric Oxide (nl/min) | 14 | 63 (63) | 9 | 12 (7)** |
| Ciliary beat frequency (Hz) | 16 | 7.5 (4.7) | 16 | 1.3 (3.6)** |
| Proportion of IDA absent on TEM | 15 | 46% | 16 | 63% |
| Proportion of ODA absent on TEM | 15 | 27% | 16 | 89%** |
| FEV1 z score (median IQR) | 10 | −1.4 (−0.8, −2.1) | 13 | −1.8 (−1.5, −2.1) |
| Situs Inversus | 16 | 81% | 16 | 56% |
The CCDC103 p.His154Pro cases are described in Table 1 and the comparator group are South Asian origin CCDC103 p.His154Pro-negative cases with a confirmed absent dynein arms defect. Data shown as the mean, with standard deviation shown in brackets (StDev).
p<0.005.
Despite these findings, FEV1 measurements showed that impact on lung function was similar in the group of 16 p.His154Pro patients compared to a comparator group (Table 1, Figure 1). The comparator group of 16 South Asian individuals with PCD all had dynein arm deficiency, but due to different genetic causes since this group comprised 3 DNAAF1, 2 DNAAF3, 2 DNAH5, 3 LRRC6 and 2 ZMYND10 cases whilst 4 were genetically undefined. The mean difference for clinical measures between the CCDC103 p.His154Pro and comparator groups is presented in Table 2, showing an equivalent age and gender composition.
Figure 1. Comparison of predicted FEV1 in CCDC103 p.His154Pro cases versus a comparator group.
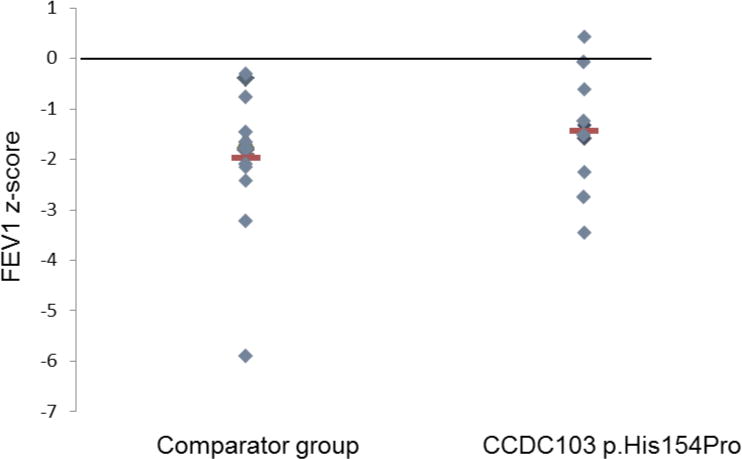
The CCDC103 p.His154Pro cases are described in Table 1 and the comparator group are South Asian origin CCDC103 p.His154Pro-negative cases with a confirmed absent dynein arms defect.
PCD caused by the CCDC103 p.His154Pro mutation can be associated with normal nasal nitric oxide (nNO) results
We proceeded to retrospectively analyse clinical phenotypes of the sixteen affected South Asian patients carrying the homozygous p.His154Pro missense mutation in more detail, as presented in Table 1. Of the 16 p.His154Pro patients tested, seven (43%) had normal nNO levels, above 77nl/min which is a recommended diagnostic cut off.23 Of these 7 patients, most (patients 5, 7, 11, 12 and 13) were subject to repeat measurements at subsequent clinic appointments. Patient 12 displayed persistent values within the normal range up to 5 years from the first measurement. Mean nNO was significantly higher than the low levels consistently detected in the comparator PCD patient group (Table 2).
PCD caused by the CCDC103 p.His154Pro mutation can be associated with areas of normal ciliary beat frequency on high speed video light microscopy
High-speed video microscopy revealed that amongst the 16 homozygous p.His154Pro individuals, nine (56%) had ciliary beat frequencies within the normal range of 8.5-16.8Hz (average 11.6Hz)19 (Table 1). The other seven p.His154Pro patients (patients 1-6 and 13) had a reduced beat frequency, on average 3.9Hz (range 0-7.7Hz), which is more in keeping with typical results found for PCD patients and more specifically found for CCDC103 patients expressing loss-of-function alleles.13,24 The comparator group of PCD patients displayed a reduced ciliary beat frequency, on average 1.3Hz (Table 2), which is similar to the reported reduced ciliary beating seen in published PCD cases associated with other causes of dynein arm loss. In half the p.His154Pro patients the beat pattern of cilia was completely or largely normal in some strips of epithelium (patients 5-7, 9, 10, 12, 15, 16, Table 1) whilst static, slow or dyskinetic in others. Some His154Pro patients (7, 10, 16) demonstrated a full beat pattern and almost immotile cilia together within the same sample (Supplementary videos 1-3). Therefore the motility in p.His154Pro individuals was variable, compared to the fully motile cilia of healthy controls (Supplementary video 4) and the completely static cilia seen in patients with LRCC6 and ZMYND10 defects from the comparator group (Supplementary video 5).
PCD caused by the CCDC103 p.His154Pro mutation can be associated with normal ultrastructural appearance by electron microscopy
Of the 16 CCDC103 p.His154Pro individuals, seven patients (44%) (cases 1-7 in Table 1) had a defect of the ciliary inner and outer dynein arms demonstrated by transmission electron microscopy (Table 1). The other nine p.His154Pro individuals (cases 8-16) had TEM that either showed an absence of the inner dynein arm, or that was considered normal or inconclusive despite extensive interrogation. The spectrum of ultrastructural defects found in p.His154Pro individuals is illustrated with representative examples in Figure 2. Overall, the inner dynein arm appeared to be the most affected structure in these cases, with significant retention of outer dynein arms. Quantification of these dynein arm defects is shown in Figure 3. Published normal range counts from >200 non-PCD respiratory controls are also shown for comparison (Figure 3, grey box). Notably, samples from four CCDC103 p.His154Pro patients were within this normal range. When a partial defect of the outer dynein arm was seen, an assessment of proximity to the epithelial cell surface - as judged by the presence of neighboring microvilli - showed that ODA loss typically occurred at the distal end towards the tip of the cilia in the sample.
Figure 2. Transmission electron microscopy of CCDC103 p.His154Pro patients.
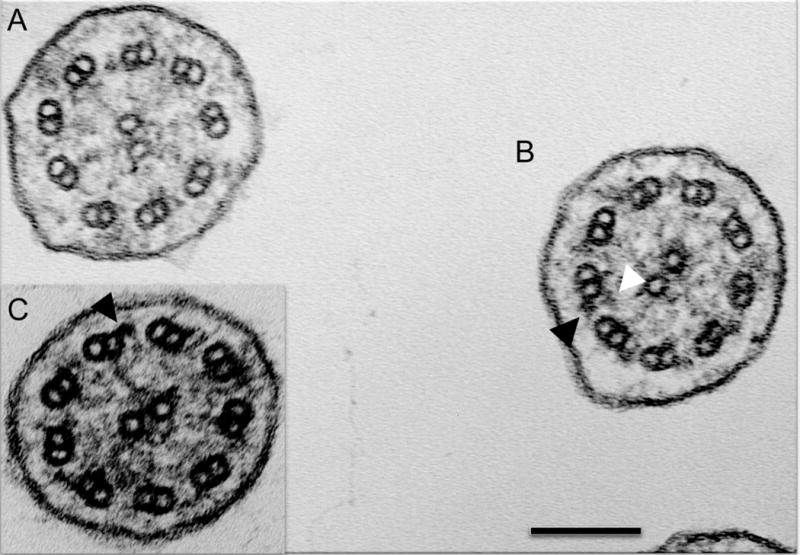
Representative cilia cross sections show (A) absent outer and partially absent inner dynein arms, (B) presence of outer dynein arms only and presence of both inner and outer dynein arms within the same micrograph. (C) Inset shows presence of outer but not inner dynein arms. Black arrows indicate example outer dynein arms. White arrows indicate inner dynein arms. Scale bar, 100 nm.
Figure 3. Quantitative transmission electron microscopy survey of inner and outer dynein arm loss in CCDC103 p.His154Pro patients versus a comparator group.
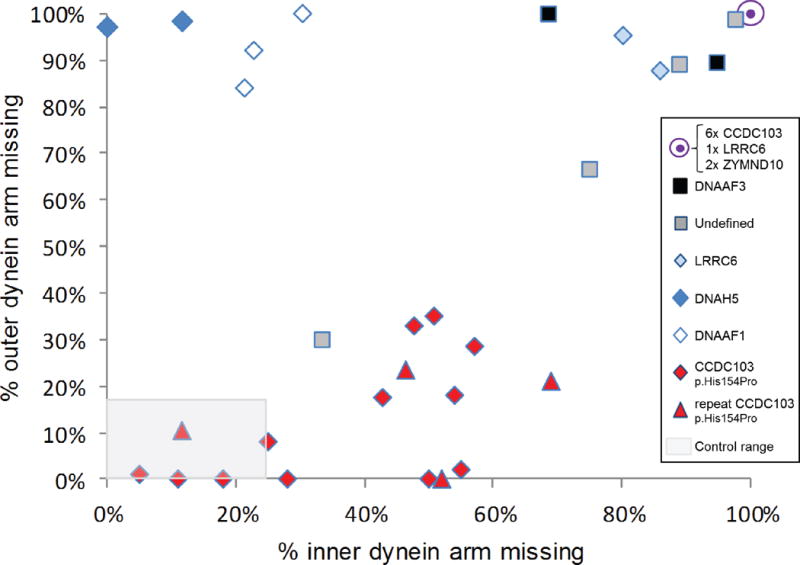
By surveying >100 cross sections in each patient sample we performed quantitative electron microscopy to determine the percentage of arm defects in cilia from individuals homozygous for the hypomorphic p.His154Pro CCDC103 mutation. Red diamonds and triangles indicate results from 16 CCDC103 p.His154Pro homozygote PCD patients, where triangles indicates the result of 4 repeat nasal brushings performed on patients marked by a diamond. The comparator group of 16 individuals with PCD, indicated by other symbols as shown, consists of 3 cases with DNAAF1 mutations (open diamonds), 2 DNAAF3 (black squares), 2 DNAH5 (dark blue diamonds), 3 LRRC6 (2 as light blue diamonds, 1 contained within the filled purple circle), 2 ZMYND10 (contained within the filled purple circle) and 4 cases in whom no mutations in known genes could be identified (grey squares). 6 CCDC103 p.His154Pro samples (27%) of p.His154Pro samples showed complete lack of both outer and inner dynein arms comparable to other gene mutations in the graph (purple circle). The grey shaded area represents normal range counts from >200 non PCD respiratory controls.30 Four CCDC103 p.His154Pro patients had counts within this normal range (one is a repeat sample (triangle) which showed similar data). Individuals with CCDC103 p.His154Pro mutation have a trend towards a distinctive pattern of partial loss of dynein arms that diverges from total dynein arm loss in the comparator group.
A summary shown in Table 2 highlights that the majority of CCDC103 p.His154Pro cases clearly differed from the non-CCDC103 comparator group, which have a near complete absence of both dynein arms. This is highly significant in terms of outer dynein arm loss (p<0.05), with only 27% ODA absence in the CCDC103 p.His154Pro cilia compared to 89% in the comparator group. Inner dynein arms were also clearly more retained in CCDC103 p.His154Pro cilia, but this difference did not reach statistical significance. The overall relative lack of disturbance to dynein arm structures means that CCDC103 p.His154Pro TEM overlaps with that of non PCD controls.
Biochemical analysis of CCDC103 p.His154Pro mutation reveals an abrogation of its oligomerisation capacity
We conducted gel filtration of purified CCDC103 p.His154Pro protein compared to the normal protein using previously established methods,20 to assess the functional viability of the mutant form of the protein. The p.His154Pro variant appears to be a highly disruptive mutation that reduces protein folding ability and almost completely abolishes the ability of CCDC103 to oligomerize beyond a dimer, as shown in the chromatogram (Figure 4).
Figure 4. CCDC103 p.His154Pro oligomerisation capacity.
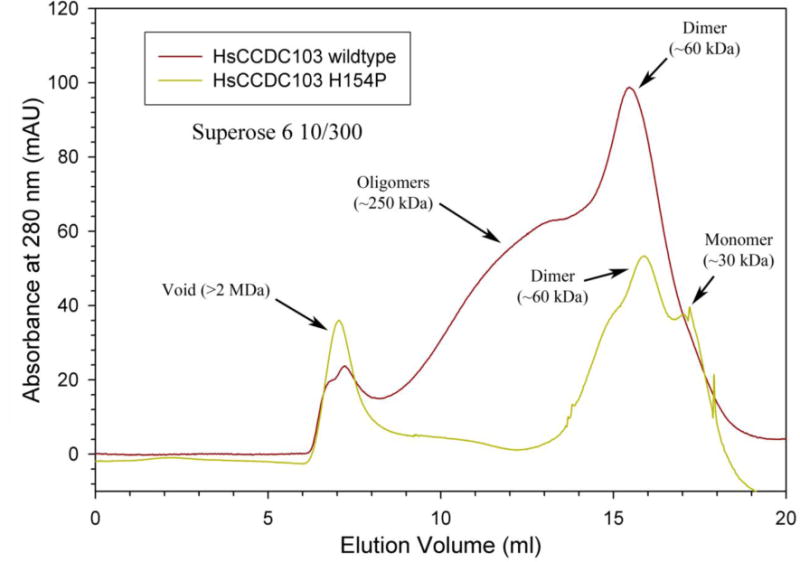
Chromatograms of wildtype (red trace) and His154Pro (green trace) CCDC103 native proteins separated in a calibrated Superose 6 10/300 gel filtration column. The data is plotted as absorbance at 280 nm (in mAU) against elution volume (ml). Both proteins show strong dimer peaks at ~60 kDa. However, only the wildtype form generates a series of higher-order oligomers with an approximate mass of ~250 kDa. Aggregated material (>2 MDa) eluted in the void volume.
DISCUSSION
We report from genetic screening of a PCD cohort ascertained through UK National PCD Services that the CCDC103 p.His154Pro mutation accounts for approximately one fifth of cases in the South Asian PCD community. Therefore a significant risk of disease arises from the presence of this important common mutation, to the extent that a predictive screen for p.His154Pro in the highly consanguineous South Asian community picked up one case out of 12 where the patient selection was based purely on ethnic origin and a TEM result.
Despite the relatively high prevalence of CCDC103 p.His154Pro, this mutation is not the sole cause of the increased incidence of PCD reported in the British South Asian community.3 Our screen also revealed patients in the group of 86 screened that carry causal mutations in an assortment of other genes including CCDC40, DNAAF1, DNAAF3, HEATR2, LRRC6, ZMYND10 and RSPH4A. Conversely, the CCDC103 p.His154Pro defect is not exclusive to the UK Asian community and has also been detected in a patient from an Irish travelling family25 as well as in two North European origin families with PCD from our studies (unpublished data).
This study highlights extensive variability in the diagnostic results from CCDC103 p.His154Pro mutations. Normal standard diagnostic test results have led to CCDC103 p.His154Pro patients being discharged from the PCD clinic, and in some cases the diagnosis of PCD in a CCDC103 p.His154Pro patient was only finally confirmed following the genetic results. Fifteen (94%) patients or their sibling(s) had situs inversus suggesting CCDC103 p.His154Pro patients without situs inversus are missed either due to lack of referral or due to normal diagnostic tests.
These patients represent an expanded phenotype for PCD, since without genetic results many in this group may not have been considered to meet the current UK clinical diagnostic criteria for PCD, resulting in uncertainty for the patient and their parents and unnecessary further investigation into the cause of their symptoms. One South Asian patient in the comparator group had a TEM phenotype similar to that of the CCDC103 p.His154Pro group. This individual also had a nNO level of >77nl/min, but a screen for the CCDC103 mutation was negative (Figure 3). This case, along with other patients in our clinic, suggests that there will be other mutations which cause PCD with non-diagnostic results.
Interestingly, functional preservation in CCDC103 p.His154Pro-mutant cilia is not apparently accompanied by significantly preserved lung function, since the FEV1 range in p.His154Pro patients is equivalent to the comparator group and respiratory capacity is equally reduced in these individuals. However, larger numbers of patients and controls should be analysed to verify this observation and genotype – phenotype relationships could be further investigated with more sensitive tests for staging lung disease such as Lung Clearance Index or CT scan. Patients with this mutation could represent an interesting cohort for targeted gene therapies due to their prevalence, but also because all the components required for normal ciliary beating appear to be present, albeit at variable levels.
The nNO levels within the normal range in CCDC103 p.His154Pro patients highlight the importance of considering the full clinical history in conjunction with nNO testing in patients with suspected PCD. Clinicians should proceed to further testing in cases with a high index of clinical suspicion, using the NO test as part of a multidisciplinary diagnostic protocol rather than a stand-alone screening test. This is not the first report of normal nNO results in patients with PCD. Some patients with RSPH1 mutations are also reported to have levels of NO close to normal and we expect further such cases to be reported as the complex genetic landscape of PCD diagnosis is unraveled further.26
The apparently normal/mixed beat pattern found in half of p.His154Pro cases was surprisingly high, but is supported by previous work indicating that this represents a hypomorphic allele in cilia function tests.13 Awareness of these cases is critical when assessing diagnostic samples in the laboratory. Due to the eye being naturally drawn to movement it is possible that the observer may inadvertently select beating strips over those that are static when scanning the sample at 5-20 times magnification before full analysis with high speed video, therefore inadvertently missing this defect when a significant portion of the sample has a normal co-ordinated beat. This places an emphasis on diagnostic centres to increase awareness and expertise of the operators when assessing nasal biopsies by high-speed video microscopy. HSVM does not measure the power of the ciliary beating, but in cases with a normal stroke and frequency there could be weakness in the strength of the beating that might not be detected by any of the current diagnostic tests.
In this study, TEM diagnosis often revealed a pattern of intermittent IDA and ODA loss in CCDC103 p.His154Pro patients which was distinct from cases of dynein arm absence due to mutations in other genes such as LRRC6, DNAAF1 and ZYMND10. The distinction was most significant for ODA retention with CCDC103 p.His154Pro. Despite analysis of >100 cilia cross sections, usually sufficient for diagnosis, the loss of dynein was not always detected by electron microscopy. However, it has long been accepted that normal ultrastructure by electron microscopy cannot exclude PCD.27,28 Our study appears to confirm in CCDC103 p.His154Pro patients the previous evidence that CCDC103 mutations confer a loss of distal ODAs containing DNAH9, not ODAs in the proximal half of the cilia closer to the epithelial surface.13
This study contributes strong evidence that CCDC103 p.His154Pro is pathogenic rather than a benign polymorphism. It is present at very low frequency in the non-PCD population and no CCDC103 p.His154Pro homozygote individuals were detected in large scale screening of 9,798 South Asian and 33,345 North European controls. Also, segregation analysis in the 16 families studied here showed an inheritance pattern consistent with recessive disease (data not shown). The CCDC103 protein remains poorly characterised but is considered important in stabilising microtubules by helping to generate a high-affinity site on the doublets for outer arm assembly, either through direct interactions or indirectly, by modifying the underlying microtubule lattice.20 Previous studies have shown that CCDC103 usually forms dimers and higher order oligomers, and that the microtubule-binding capacity is a property of the protein’s central region which contains a highly conserved RPAP3_C domain that functions in protein-protein interactions. The His154 amino acid is contained within the conserved RPAP3_C functional domain which spans residues 96-189 of the protein, therefore altering His154 might be expected to disrupt function. Our current results reveal that the His154Pro form retains the ability to dimerize but shows little oligomer formation, suggesting that this property is disrupted by the mutation and that CCDC103 may have two distinct self-interaction domains. We hypothesise that gene mutations causing instability or depletion of CCDC103 protein may make the attachment of the dynein arms more susceptible to physical, infective or inflammatory insult. We cannot determine if the presence of normal cilia in our patients is temporal or spatial however, as results appear to vary from one biopsy to the next e.g. in case 11, 12, 15 and 16 in Table 1; cell culture and repeating investigations may be useful in these cases.
In conclusion, the CCDC103 p.His154Pro variant is prevalent in the UK South Asian community and likely to be found in South Asian patients worldwide. Therefore this patient group should undergo genetic testing for p.His154Pro, especially if (partial) dynein arm absence is suspected. These patients are frequently a diagnostic dilemma, due to inconclusive results of multiple clinical diagnostic tests. This study expands the diagnostic phenotype which we consider to be PCD, since in some cases pathology-based tests can be equivocal as the cilia can beat at least partially in a co-ordinated manner, at the correct speed and may appear structurally normal. PCD is widely understood to be an underdiagnosed condition and this appears to be the case for CCDC103 p.His154Pro patients, who demonstrate a high level of situs inversus probably indicative of a lack of recognition making their diagnosis liable to be missed.29 We anticipate that studies such as this, in combination with easier access to high throughput and economically achievable genetic screening, should greatly increase disease recognition and understanding. We have highlighted the importance of multidisciplinary testing, repeat testing and genotyping in patients with a highly suggestive history for PCD.
Supplementary Material
Supplementary video 1 HSVM of p.His154Pro homozygote patient 10 captures fully beating cilia
Supplementary video 2 HSVM of p.His154Pro homozygote patient 10 captures mixed dyskinetic and faintly moving/immotile cilia
Supplementary video 3 HSVM of p.His154Pro homozygote patient 16 captures mixed beat pattern (immotile and beating)
Supplementary video 4 HSVM of patient carrying ZMYND10 mutations (static) from comparator group
Supplementary video 5 HSVM of health control (normal beat)
What is the key question?
Can gene sequencing improve diagnosis of the inherited respiratory condition primary ciliary dyskinesia in patients with unclear clinical diagnostic investigations?
What is the bottom line?
CCDC103 p.His154Pro missense mutations cause up to 20% PCD cases in UK South Asian populations but their diagnosis can be difficult in this group using standard clinical diagnostic tests where results are often normal, we therefore propose that genetic analysis is an essential part of the diagnostic algorithm to complement standard clinical tests to improve diagnostic accuracy.
Why read on?
Patients with primary ciliary dyskinesia may be missed using current pathology-based diagnostic protocols that do not include funded genetic screening.
For Twitter: 140 character conclusion
Diagnosis can be difficult in PCD with CCDC103 p.His154Pro mutations and genetic testing is essential in the high-risk UK South Asian community.
Acknowledgments
We are very grateful to the families with PCD who have participated in this study and to the UK PCD Family Support Group for their support. The research is supported by the BEAT-PCD: Better Evidence to Advance Therapeutic options for PCD network (COST Action 1407). We thank Louise Ocaka and Chela James (UCL GOSgene), Emily Frost (Royal Brompton Hospital) and Bruna Rubbo (University of Southampton) for experimental support and data analysis. A.B. was supported by the NIHR Respiratory Disease Biomedical Research Unit at the Royal Brompton and Harefield NHS Foundation Trust and Imperial College London. Work by A.S. is independent research funded by a postdoctoral research fellowship from the National Institute of Health Research and Health Education England, and the views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute of Health Research or the Department of Health. R.S.P-K. and S.M.K. are supported by NIH grant GM051293. M.S. is supported by a Radboudumc Hypatia Tenure Track fellowship, a Radboud University Excellence fellowship and received funding from the German research Foundation (DFG), collaborative Research Center (CRC) 1140 KIDGEM. This research and the Centre for Translational Omics (GOSgene) is supported by the National Institute for Health Research Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London. H.M.M. was supported by grants from Action Medical Research (GN2101), Newlife Foundation (10-11/15) and the Great Ormond Street Hospital Children’s Charity. Work in Southampton is supported by NIHR Respiratory Biomedical Research Unit and NIHR Wellcome Trust Clinical Research Facility.
References
- 1.Lucas JS, Burgess A, Mitchison HM, et al. Diagnosis and management of primary ciliary dyskinesia. Arch Dis Child. 2014;99(9):850–6. doi: 10.1136/archdischild-2013-304831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shapiro AJ, Davis SD, Ferkol T, et al. Laterality defects other than situs inversus totalis in primary ciliary dyskinesia: insights into situs ambiguus and heterotaxy. Chest. 2014;146(5):1176–86. doi: 10.1378/chest.13-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Callaghan C, Chetcuti P, Moya E. High prevalence of primary ciliary dyskinesia in a British Asian population. Arch Dis Child. 2010;95(1):51–2. doi: 10.1136/adc.2009.158493. [DOI] [PubMed] [Google Scholar]
- 4.Shah A, Shoemark A, MacNeill SJ, et al. A longitudinal study characterising a large adult primary ciliary dyskinesia population. Eur Respir J. 2016;48(2):441–50. doi: 10.1183/13993003.00209-2016. [DOI] [PubMed] [Google Scholar]
- 5.Lucas JS, Barbato A, Collins SA, Goutaki M, Behan L, Caudri D, Dell S, Eber E, Escudier E, Hirst RA, Hogg C, Jorissen M, Latzin P, Legendre M, Leigh MW, Midulla F, Nielsen KG, Omran H, Papon JF, Pohunek P, Redfern B, Rigau D, Rindlisbacher BF, Shoemark A, Snijders D, Tonia T, Titieni A, Walker WT, Werner C, Bush A, Kuehni CE. ERS Task Force guideline for the diagnosis of primary ciliary dyskinesia. Eur Resp J. 2016 in press. [Google Scholar]
- 6.Jackson CL, Behan L, Collins SA, et al. Accuracy of diagnostic testing in primary ciliary dyskinesia. Eur Respir J. 2016;47(3):837–48. doi: 10.1183/13993003.00749-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Khouri E, Thomas L, Jeanson L, et al. Mutations in DNAJB13, Encoding an HSP40 Family Member, Cause Primary Ciliary Dyskinesia and Male Infertility. Am J Hum Genet. 2016;99(2):489–500. doi: 10.1016/j.ajhg.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeanson L, Copin B, Papon JF, et al. RSPH3 Mutations Cause Primary Ciliary Dyskinesia with Central-Complex Defects and a Near Absence of Radial Spokes. Am J Hum Genet. 2015;97(1):153–62. doi: 10.1016/j.ajhg.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallmeier J, Al-Mutairi DA, Chen CT, et al. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Genet. 2014;46(6):646–51. doi: 10.1038/ng.2961. [DOI] [PubMed] [Google Scholar]
- 10.Wallmeier J, Shiratori H, Dougherty GW, et al. TTC25 Deficiency Results in Defects of the Outer Dynein Arm Docking Machinery and Primary Ciliary Dyskinesia with Left-Right Body Asymmetry Randomization. Am J Hum Genet. 2016;99(2):460–9. doi: 10.1016/j.ajhg.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olbrich H, Cremers C, Loges NT, et al. Loss-of-Function GAS8 Mutations Cause Primary Ciliary Dyskinesia and Disrupt the Nexin-Dynein Regulatory Complex. Am J Hum Genet. 2015;97(4):546–54. doi: 10.1016/j.ajhg.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watson CM, Crinnion LA, Morgan JE, et al. Robust diagnostic genetic testing using solution capture enrichment and a novel variant-filtering interface. Hum Mutat. 2014;35(4):434–41. doi: 10.1002/humu.22490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panizzi JR, Becker-Heck A, Castleman VH, et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat Genet. 2012;44(6):714–9. doi: 10.1038/ng.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucas JS, Chetcuti P, Copeland F, et al. Overcoming challenges in the management of primary ciliary dyskinesia: the UK model. Paediatr Respir Rev. 2014;15(2):142–5. doi: 10.1016/j.prrv.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Le Quesne Stabej P, Williams HJ, James C, et al. STAG3 truncating variant as the cause of primary ovarian insufficiency. Eur J Hum Genet. 2016;24(1):135–8. doi: 10.1038/ejhg.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trump N, McTague A, Brittain H, et al. Improving diagnosis and broadening the phenotypes in early-onset seizure and severe developmental delay disorders through gene panel analysis. J Med Genet. 2016;53(5):310–7. doi: 10.1136/jmedgenet-2015-103263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beydon N, Davis SD, Lombardi E, et al. An official American Thoracic Society/European Respiratory Society statement: pulmonary function testing in preschool children. Am J Respir Crit Care Med. 2007;175(12):1304–45. doi: 10.1164/rccm.200605-642ST. [DOI] [PubMed] [Google Scholar]
- 18.Quanjer PH, Stanojevic S, Cole TJ, et al. Multi-ethnic reference values for spirometry for the 3-95-yr age range: the global lung function 2012 equations. Eur Respir J. 2012;40(6):1324–43. doi: 10.1183/09031936.00080312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chilvers MA, Rutman A, O’Callaghan C. Ciliary beat pattern is associated with specific ultrastructural defects in primary ciliary dyskinesia. J Allergy Clin Immunol. 2003;112(3):518–24. doi: 10.1016/S0091-6749(03)01799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King SM, Patel-King RS. The oligomeric outer dynein arm assembly factor CCDC103 is tightly integrated within the ciliary axoneme and exhibits periodic binding to microtubules. J Biol Chem. 2015;290(12):7388–401. doi: 10.1074/jbc.M114.616425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheridan E, Wright J, Small N, et al. Risk factors for congenital anomaly in a multiethnic birth cohort: an analysis of the Born in Bradford study. Lancet. 2013;382(9901):1350–9. doi: 10.1016/S0140-6736(13)61132-0. [DOI] [PubMed] [Google Scholar]
- 22.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leigh MW, Hazucha MJ, Chawla KK, et al. Standardizing nasal nitric oxide measurement as a test for primary ciliary dyskinesia. Ann Am Thorac Soc. 2013;10(6):574–81. doi: 10.1513/AnnalsATS.201305-110OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raidt J, Wallmeier J, Hjeij R, et al. Ciliary beat pattern and frequency in genetic variants of primary ciliary dyskinesia. Eur Respir J. 2014;44(6):1579–88. doi: 10.1183/09031936.00052014. [DOI] [PubMed] [Google Scholar]
- 25.Casey JP, Goggin P, McDaid J, et al. A case report of primary ciliary dyskinesia, laterality defects and developmental delay caused by the co-existence of a single gene and chromosome disorder. BMC Med Genet. 2015;16:45. doi: 10.1186/s12881-015-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knowles MR, Ostrowski LE, Leigh MW, et al. Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am J Respir Crit Care Med. 2014;189(6):707–17. doi: 10.1164/rccm.201311-2047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olbrich H, Schmidts M, Werner C, et al. Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am J Hum Genet. 2012;91(4):672–84. doi: 10.1016/j.ajhg.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwabe GC, Hoffmann K, Loges NT, et al. Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations. Hum Mutat. 2008;29(2):289–98. doi: 10.1002/humu.20656. [DOI] [PubMed] [Google Scholar]
- 29.Kuehni CE, Frischer T, Strippoli MP, et al. Factors influencing age at diagnosis of primary ciliary dyskinesia in European children. Eur Respir J. 2010;36(6):1248–58. doi: 10.1183/09031936.00001010. [DOI] [PubMed] [Google Scholar]
- 30.Shoemark A, Dixon M, Corrin B, et al. Twenty-year review of quantitative transmission electron microscopy for the diagnosis of primary ciliary dyskinesia. J Clin Pathol. 2012;65(3):267–71. doi: 10.1136/jclinpath-2011-200415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary video 1 HSVM of p.His154Pro homozygote patient 10 captures fully beating cilia
Supplementary video 2 HSVM of p.His154Pro homozygote patient 10 captures mixed dyskinetic and faintly moving/immotile cilia
Supplementary video 3 HSVM of p.His154Pro homozygote patient 16 captures mixed beat pattern (immotile and beating)
Supplementary video 4 HSVM of patient carrying ZMYND10 mutations (static) from comparator group
Supplementary video 5 HSVM of health control (normal beat)


