Abstract
Currently, it is believed that osteoclasts positive for tartrate-resistant acid phosphatase (TRAP+) are the exclusive bone-resorbing cells responsible for focal bone destruction in inflammatory arthritis. Recently, a mouse model of cherubism (Sh3bp2KI/KI) with a homozygous gain-of-function mutation in the SH3-domain binding protein 2 (SH3BP2) was shown to develop auto-inflammatory joint destruction. Here, we demonstrate that Sh3bp2KI/KI mice also deficient in the FBJ osteosarcoma oncogene (c-Fos) still exhibit noticeable bone erosion at the distal tibia even in the absence of osteoclasts at 12 weeks old. Levels of serum ICTP, a marker of bone resorption generated by matrix metalloproteinases (MMPs), were elevated, while levels of serum CTX, another resorption marker produced by cathepsin K, were not increased. Collagenolytic MMP levels were increased in the inflamed joints of the Sh3bp2KI/KI mice deficient in c-Fos. Resorption pits contained a large number of F4/80+ macrophages and genetic depletion of macrophages rescued these erosive changes. Importantly, administration of NSC405020, an MMP14 inhibitor targeted to the hemopexin (PEX) domain, suppressed bone erosion in c-Fos-deficient Sh3bp2KI/KI mice. After activation of the NF-κB pathway, M-CSF-dependent macrophages from c-Fos-deficient Sh3bp2KI/KI mice expressed increased amounts of MMP14 compared to wild-type macrophages. Interestingly, RANKL-deficient Sh3bp2KI/KI mice failed to show notable bone erosion, while c-Fos deletion did restore bone erosion to the RANKL-deficient Sh3bp2KI/KI mice, suggesting that osteolytic transformation of macrophages requires both loss-of-function of c-Fos and gain-of-function of SH3BP2 in this model. These data provide the first genetic evidence that cells other than osteoclasts can cause focal bone destruction in inflammatory bone disease and suggest that MMP14 is a key mediator conferring pathological bone-resorbing capacity on c-Fos-deficient Sh3bp2KI/KI macrophages. In summary, the paradigm that osteoclasts are the exclusive cells executing inflammatory bone destruction may need to be re-evaluated based on our findings with c-Fos-deficient cherubism mice lacking osteoclasts.
Keywords: Cherubism, SH3BP2, c-Fos, MMP14, Non-canonical osteoclasts/Osteolytic macrophages, Osteoclast-independent inflammatory bone destruction
Introduction
Osteoclasts are multinucleated cells of monocyte-macrophage origin that resorb mineralized bone by localized secretion of acids and proteinases such as cathepsins and matrix metalloproteinases (MMPs). Osteoclasts characteristically stain positive (+) for tartrate-resistant acid phosphatase (TRAP) on bone surfaces.(1) The major cytokine that regulates osteoclast differentiation is the receptor activator of nuclear factor-κB ligand (RANKL) and activating signals from its receptor RANK are required for the differentiation.(2,3) Signaling mechanisms underlying osteoclastogenesis have been extensively studied and NFATc1 (nuclear factor of activated T-cells, cytoplasmic 1) was identified as the master transcription factor for osteoclast differentiation and function.(2,3) FBJ osteosarcoma oncogene (c-Fos), a member of the AP-1 family of transcription factors, plays an essential role in the robust induction of NFATc1 in differentiating osteoclasts.(4,5) Therefore, mice lacking c-Fos or RANK/RANKL exhibit osteopetrosis due to the lack of TRAP+ osteoclasts.(6–12) In view of compelling genetic evidence that c-Fos- or RANKL-deficient mice with arthritis are completely protected from bone resorption (13,14), there has been a strong consensus in bone biology that osteoclasts are the only bone-resorbing cells that mediate bone erosion in inflammatory arthritis. (3,15,16)
Cherubism (OMIM#118400) is an autoinflammatory disorder of the craniofacial skeleton characterized by expansile maxillary and mandibular bone destruction due to the development of fibrous/inflammatory lesions. Mutations in SH3-domain binding protein 2 (SH3BP2) that uncouple SH3BP2 from poly(ADP-ribose)polymerase tankyrase 1 and 2 are responsible for this rare genetic condition.(17,18) Mutant SH3BP2 protein escapes from ubiquitin-mediated proteasome degradation due to the lack of ADP-ribosylation, resulting in stabilization of the SH3BP2 protein. Thus, increased levels of SH3BP2 activate downstream signaling pathways in osteoclasts and macrophages in a gain-of-function manner leading to tumor necrosis factor (TNF)-α-dependent bone destruction in the knock-in (KI) mouse model of cherubism (Sh3bp2KI/KI).(19–21)
Here, we show that Sh3bp2KI/KI mice that also lack c-Fos still exhibit substantial bone erosion at the distal tibia even though the mice have no TRAP+ osteoclasts. This result demonstrates a novel mechanism of inflammatory bone resorption by cells other than osteoclasts. Indeed, previous studies have suggested that cells other than osteoclasts can mediate bone resorption via different mechanisms.(22–25) Specifically, bisphosphonates failed to provide significant protection against focal bone erosion in rheumatoid arthritis (RA) patients, suggesting that a part of inflammatory bone destruction could occur independently of osteoclasts.(26–29)
Our study provides the first genetic evidence for inflammatory bone destruction that occurs independently of TRAP+ osteoclasts and demonstrates that macrophages can be converted to osteolytic cells under inflammatory conditions when SH3BP2- and c-Fos-mediated pathways are both dysregulated. The long-standing paradigm that osteoclasts are the exclusive bone-resorbing cells in inflammatory bone diseases may need to be re-evaluated based on our findings with c-Fos-deficient cherubism mice.
Materials and Methods
Mice
All animal experiments in this study were performed under an animal experimental protocol approved by the UMKC IACUC. Sh3bp2KI/KI mice were reported previously.(19) c-Fos-deficient (c-Fos−/−, #002293), Ranklfl/fl (#018978), EIIa-Cre (#003724), Mx1-Cre (#003556), and Csf1rfl/fl (#021212) mice were obtained from the Jackson laboratory. RANKL-deficient (Rankl−/−) mice were created by crossing Ranklfl/fl mice with EIIa-Cre mice. All mice were crossed and created on the mix background of C57BL/6 and 129X1/SvJ under specific pathogen free conditions. Csf1rfl/fl mice with Mx1-Cre were injected with poly(I:C) (250 mg/body, GE Healthcare) intraperitoneally three times with 2-day intervals at 1 week old to induce Cre expression. Csf1rfl/fl mice without Mx1-Cre were also injected with poly(I:C) as controls. There were no significant gender differences in phenotypes, thus samples from both males and females were pooled for statistical analysis.
Ex-vivo microCT (μCT) imaging
Left hind limbs were subjected for bone erosion analysis with μCT. Tissues were fixed with 4% paraformaldehyde (PFA) for 24 hours and soaked in 70% ethanol for a scan with the Skyscan1174 (Bruker) with following conditions: 80 kV X-ray energy, 6.67 μm pixel size, and 0.4 degree rotation step with 3000 ms of exposure time. Scanned data were reconstructed with NRecon software (Bruker) with 0 to 0.15 of dynamic range. Three-dimensional (3D) images were created using CTVox software (Bruker).
Quantification of bone erosion
Reconstructed data with NRecon were segmented by CTAn software (Bruker) to define the 1 mm from distal end of the tibia as a region of interest. 3D images of periosteal surface from 4 directions (anterior, posterior, lateral, and medial views) were used to measure eroded bone surface area (%) using ImageJ (NIH). Measurements were performed by personnel blinded to genotypes and inhibitor administration. Average value of 4 directions was used as eroded bone area % for each mouse.
In vivo μCT analysis
Right hind limbs were scanned with the vivaCT 40 (Scanco) with 10.5 μm of resolution under anesthesia with ketamine (80 mg/kg) and xylazine (10 mg/ml). In vivo μCT scanning was performed sequentially with 4-week intervals from 8 to 16 weeks old. A threshold of 270 was used to distinguish mineralized tissues from non-mineralized tissues.
Histology
Hind limbs were fixed with 4% PFA for 24 hours, decalcified with EDTA (0.5M, pH 7.2) for 3 weeks, and embedded in paraffin. Liver tissues were fixed with 4% PFA for 24 hours and paraffin-embedded. Six μm sections were subjected to hematoxylin and eosin (H&E) and TRAP staining. For immunohistochemical staining, tissues were decalcified with Immunocal (Decal Chemical Corp.) for 24 hours.
Serum ELISA
Mice were fasted for 6 hours before blood collection. Mouse TNF-α Duo-Set ELISA kit (R&D), mouse TRAP5b assay kit (Immunodiagnostic Systems), mouse ICTP ELISA kit (Biotang Inc.), and RatLaps™ CTX-I EIA kit (Immunodiagnostic Systems) were used. Serum was separated from blood with Vacutainer collection tube (BD) and stored at −80 °C until use.
RNA and protein extraction from ankle joints or macrophages
Ankle joints without skin were dissected out, immediately frozen by liquid nitrogen, and crushed in powder using a tissue pulverizer (Cellcrusher Limited) for RNA or protein isolation. Cultured macrophages were washed with ice-cold PBS twice, and lysed for RNA or protein extraction. RNA was extracted by Tripure reagent (Roche). Protein was extracted by cell lysis buffer (1% Triton X-100, 25 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 10% glycerol, 2.5 mM sodium pyrophosphate, 0.7 mM β-glycerophosphate) with protease and phosphatase inhibitor cocktails (Sigma-Aldrich).
Reverse transcription-quantitative PCR (RT-qPCR) analysis
Five hundred nanograms of total RNA was transcribed to cDNA using the High Capacity cDNA Reverse Transcription Kit (Life Technologies). qPCR reactions were performed with the StepOne Plus system (Life Technologies) using Maxima SYBR green mix (Thermo Scientific) or Taqman Gene Expression master mix (Life Technologies). qPCR primers and probes used in this study are listed in Supplemental Table 1. Relative gene expression levels were calculated using a relative-standard curb method. Each gene expression levels were normalized by expression levels of Hprt.
Histomorphometry of liver inflammation
Inflammatory infiltrates in liver tissue were quantified as previously described.(21) Briefly, total inflammatory area in the liver stained with H&E was measured as pixels with ImageJ (NIH), which was divided by the pixels of the entire tissue excluding areas of vasculature to calculate the proportion of inflammatory area (%). Liver images were taken with a 4X objective lens (Nikon E800). Results from four images from different areas of each tissue section were averaged.
Transmission electron microscope (TEM)
Distal tibiae were dissected, fixed with 2.5% glutaraldehyde, and decalcified with 0.5M EDTA (pH 7.2). Samples were embedded in epoxy resin. One hundred nanometer sections stained with uranyle acetate and lead citrate were observed using the Philips CM12 TEM at the UMKC School of Dentistry.
Fetal liver-derived macrophage colony-stimulating factor (M-CSF)-dependent macrophage culture and inhibitor treatment
Fetal livers obtained from E15.5 to 18.5 embryos were used as a source of M-CSF-dependent macrophages (30,31), because adult c-Fos-deficient mice have little bone marrow cells due to severe osteopetrosis.(6,7). After lysis of red blood cells, fetal liver cells were incubated with α-MEM supplemented with 10% FBS and penicillin/streptomycin for 3 hours on petri dishes in order to let stromal cells adhere on the surface of petri dishes. Non-adherent cells were collected and cultured on tissue culture-treated dishes in the presence of M-CSF (25 ng/ml, Peprotech) to obtain M-CSF-dependent macrophages. Culture medium containing M-CSF was changed every other day. For inhibitor assays, non-adherent fetal liver cells were cultured in the presence of M-CSF. After 48 hours, inhibitors were added. RNA and protein were extracted at 48 to 96 hours after addition of inhibitors. BMS-345541(32) (IKK inhibitor) and FK506(33) (NFAT inhibitor) were obtained from Sigma-Aldrich. SP600125(34) (JNK inhibitor), SB203580(35) (p38 inhibitor) were purchased from Millipore. R406(36) (SYK inhibitor), U73122(37) (PLC inhibitor) were obtained from Selleck chemicals. U0126(38) (MEK inhibitor) was purchased from Calbiochem. For Western blotting analysis, non-adherent fetal liver cells were cultured in the presence of M-CSF for 72 hours, then adherent M-CSF-dependent macrophages were harvested and re-seeded (2.6 × 104 cells/cm2) on culture dishes with M-CSF. After 24 hours, macrophages were starved for serum and M-CSF for 6 hours, then re-stimulated with M-CSF (25 ng/ml).
Osteoclast differentiation assay
Non-adherent fetal liver cells were cultured on petri dishes with M-CSF for 3 days. Adherent M-CSF-dependent macrophages were harvested with Cellstripper (Corning) and seeded on 8-well chamber slides, 48-well culture plates, or 100 mm culture dishes at a density of 2.6 × 104 cells/cm2. After 24 hours, M-CSF-dependent macrophages were stimulated with mouse RANKL (50 ng/ml, Peprotech) or RANKL/TNF-α (50 ng/ml and 100 ng/ml, respectively, Peprotech) for up to 10 days to induce osteoclast differentiation. Culture medium containing the cytokines was replaced every other day. TRAP-positive cells on 48-well plates were visualized with a TRAP staining kit (Sigma-Aldrich) and TRAP positive cells that have more than 3 nuclei were counted as TRAP-positive multinucleated cells (TRAP+ MNCs). For visualization of actin ring and nuclei, cells were co-stained with Phalloidin conjugated with CruzFluorTM 488 (Santa Cruz) and DAPI on 8-well chamber slides, and observed with the Nikon TE800 fluorescent microscope.
Western blotting
Five μg of protein samples were resolved by SDS-PAGE with reducing condition and transferred to nitrocellulose membrane. After blocking with 5% skim milk in TBS supplemented 1% Tween-20 (TBST), membranes were incubated with primary antibodies in 5% BSA/TBST for overnight at 4°C followed by incubation with appropriate HRP-conjugated species-specific secondary antibodies (Cell Signaling Technology). Bands were detected using SuperSignal West Dura or Femto chemiluminescent substrate (Thermo Scientific) and visualized by the ImageQuant LAS-4000 (GE Healthcare). Antibodies against MMP2 (ab37150) and MMP14 (ab53712) were obtained from abcam. Anti-SH3BP2 monoclonal antibody (clone 1E9) was purchased from Abnova. Antibody against NFATc1 (7A6) was obtained from Santa Cruz Biotechnology. Antibodies against phospho-IKKα/β (#2697), IKKβ (#2678), phospho-JNK (#4668), JNK (#9258), phospho-ERK (#4370), ERK (#4695), phospho-p38 (#4511), and p38 (#9212) were obtained from Cell Signaling Technology.
Immunohistochemical staining
Tissue sections were deparaffinized and rehydrated. For antigen retrieval, sections were treated with 20 μg/ml of proteinase K for 3 min and endogenous peroxidase was blocked with 3% H2O2/PBS solution. After blocking with 2% serum, which is from the same species as the secondary antibodies, sections were incubated overnight at 4°C with antibodies against F4/80 (Clone A3-1, AbD serotec), MMP2 (ab37150, abcam), or MMP14 (ab53712, abcam). After washing with PBS, sections were incubated with appropriate biotinylated secondary antibodies (Vector Labratories) for 60 min at room temperature and treated with Vectastain elite ABC kit (Vector Laboratories). Sections were colored using ImmPACT DAB (Vector Laboratories) and counter stained with Hematoxylin.
MMP14 inhibitor treatment
For inhibition of MMP14 activity in vivo, c-Fos−/− Sh3bp2KI/KI mice were administrated with NSC405020(39) (50 mg/kg/day, AdooQ Bioscience) or DX-2400(40) (40 and 200 mg/kg, every other day, Kadmon Corp.) subcutaneously from 4 to 10 weeks old. For the control of DX-2400 administration, human IgG (Equitech Bio.) was injected.
In vitro osteolytic activity assay
Collagenolytic activity of M-CSF-dependent macrophages was assessed by measuring fluorescence intensity of europium-labeled type I collagen fibers in culture supernatant using the OsteoLyse Assay Kit (Lonza). Mineral absorption capacity was assessed by measuring fluorescence intensity of fluoresceinamine-labeled calcium phosphate in culture supernatant using the Bone Resorption Assay Kit (Cosmo Bio). After culture of non-adherent fetal liver cells for 3 days with M-CSF on petri dishes, M-CSF-dependent macrophages were harvested and seeded on the plates from the kits described above at a density of 7.8 × 104 cells/cm2 and treated with 50 ng/ml of RANKL or 100 ng/ml of TNF-α in the presence of M-CSF for 7 days. Fluorescence intensity in culture supernatant was measured with the DTX880 Multimode Detector (Beckman Coulter).
Dentin resorption assay
Following the 3-day culture of non-adherent fetal liver cells with M-CSF on petri dishes, M-CSF-dependent macrophages were harvested and seeded on dentin slices in 96-well culture plates at a density of 7.8 × 104 cells/cm2. After 24 hours, M-CSF-dependent macrophages were stimulated with mouse RANKL (50 ng/ml) and TNF-α (100 ng/ml) for 28 days. Culture medium containing the cytokines was replaced every other day. Cells were removed mechanically by scrubbing with toothbrush and Kimwipes. Resorption pits on dentin slices were observed using the Philips XL30 scanning electron microscope (SEM) at the UMKC School of Dentistry.
Statistics
Statistical analysis was performed by the two-tailed unpaired Student’s t-test to compare two groups or by one-way ANOVA with Tukey-Kramer post-hoc test to compare three or more groups. GraphPad Prism 5 (GraphPad Software) and SPSS Statistics 20 (IBM) were used for all statistical analyses. p values less than 0.05 were considered to be significant.
Results
Cherubism mice also deficient in c-Fos exhibit bone erosion
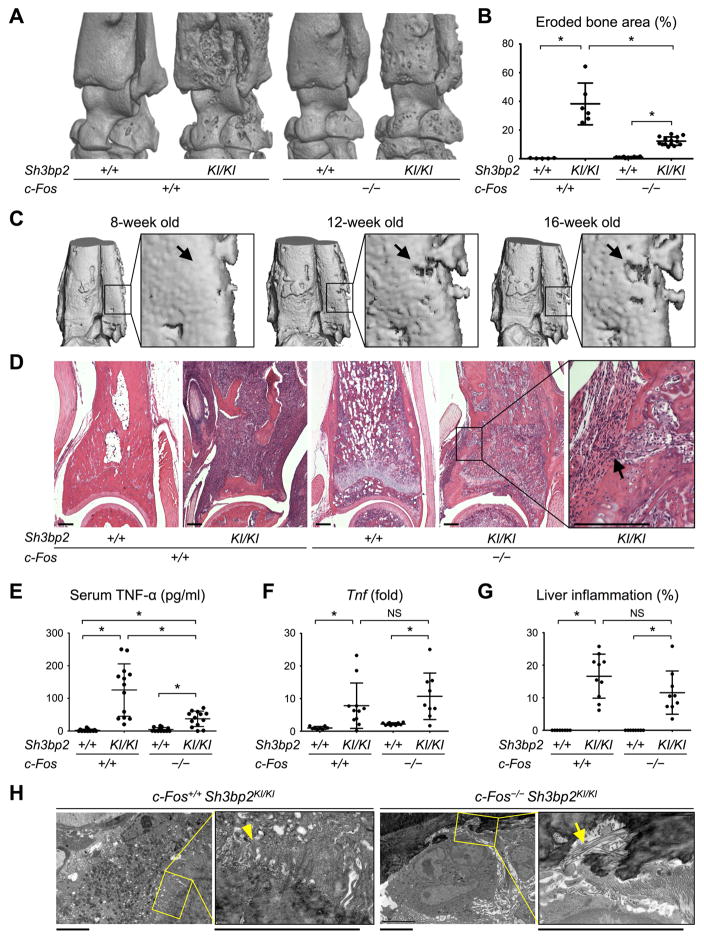
A knock-in (KI) mouse model of cherubism (Sh3bp2KI/KI) develops auto-inflammation responsible for joint destruction.(19) We initiated this study with a hypothesis that depletion of TRAP+ osteoclasts would rescue the Sh3bp2KI/KI mice from bone destruction. Because c-Fos is a transcription factor essential for osteoclastogenesis (6,7), Sh3bp2KI/KI mice were bred on a c-Fos-deficient background to generate Sh3bp2KI/KI mice lacking osteoclasts. At 12 weeks of age, μCT analysis was performed on the ankle joints of the double mutant mice. Unexpectedly, all c-Fos-deficient (c-Fos−/−) Sh3bp2KI/KI mice still developed profound eroded surfaces on the tibia, talus, and calcaneus (Fig. 1A), while eroded surface was approximately 60% less than c-Fos-sufficient (c-Fos+/+) Sh3bp2KI/KI mice (Fig. 1B). In vivo μCT analysis suggested that the erosion occurs postnatally (Fig. 1C). H&E staining revealed that c-Fos-deficient Sh3bp2KI/KI mice developed inflammatory infiltrates on their distal tibial surfaces that appeared to be responsible for the formation of erosion pits (Fig. 1D). Inflammation in the mice was associated with elevation of serum TNF-α levels compared to c-Fos-deficient Sh3bp2+/+ and wild-type (c-Fos+/+ Sh3bp2+/+) mice (Fig. 1E). Tnf expression levels in the ankle joints and areas of inflammation in the liver were comparable to c-Fos-sufficient Sh3bp2KI/KI mice (Fig. 1, F and G). Transmission electron microscopic analysis revealed exposed collagen fibers from mineralized bone matrix underneath inflammatory cells on bone surfaces of the c-Fos-deficient Sh3bp2KI/KI mice, but ruffled borders commonly associated with osteoclasts were absent (Fig. 1H). While almost all cells in inflammatory lesions were mononucleated cells, a small number of multinucleated cells were observed on the bone surface of the double mutant mice (Supplemental Fig. 1). In contrast to a previous c-Fos-deficient mouse model of inflammatory bone disease (13), our results show that Sh3bp2KI/KI mice develop erosive bone destruction in the absence of c-Fos.
Fig. 1. c-Fos-deficient Sh3bp2KI/KI mice develop inflammatory bone erosion.
(A) μCT images of the ankle joint at 12 weeks old. (B) Quantitative measurement of eroded bone surface area at the distal tibia. (C) In vivo μCT images of the distal tibia from the same c-Fos−/− Sh3bp2KI/KI mouse at different ages. Arrows indicate a location of bone erosion that appeared after 12 weeks old. (D) H&E staining of the distal tibia showing inflammatory lesions invading into marrow spaces (arrow). 12 weeks old. Bar = 200 μm. (E) Serum TNF-α levels quantitated by ELISA at 12 weeks old. (F) qPCR analysis of Tnf mRNA expression levels in ankle joint tissue at 12 weeks old. Average expression level in wild-type mice is set as 1. (G) Histomorphometric analysis of liver lesions in 12-week-old mice. (H) Transmitted electron microscope images on the surface of the distal tibia. 12 weeks old. Arrow and arrowhead indicate collagen fibers and ruffled border, respectively. Bar = 5 μm. Data are presented as mean ± SD. *p < 0.05. ANOVA with Tukey-Kramer post-hoc test. NS = not significant.
c-Fos-deficient Sh3bp2KI/KI mice have no TRAP-positive osteoclasts
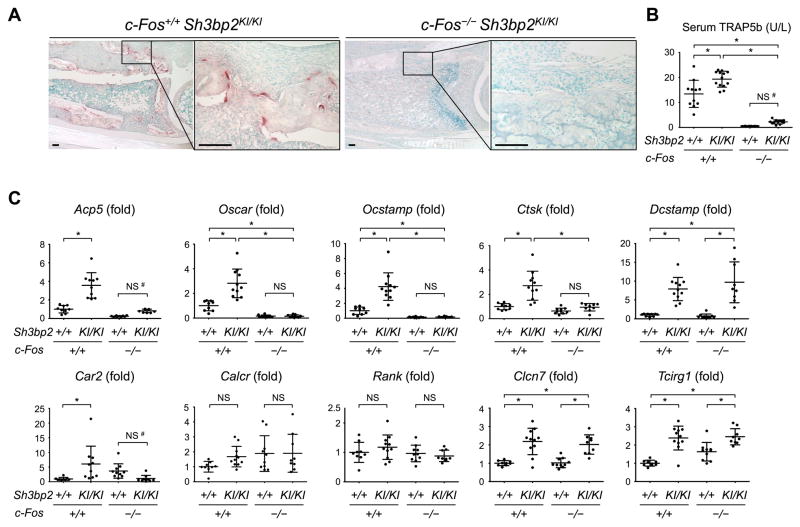
To confirm the lack of canonical osteoclasts in c-Fos-deficient Sh3bp2KI/KI mice, TRAP staining was performed on bone tissue adjacent to the inflammatory lesions. Consistent with previous results (8,13,41), lack of c-Fos completely blocked the formation of TRAP+ osteoclasts in Sh3bp2KI/KI mice (Fig. 2A). Also, serum levels of TRAP5b, a marker for active osteoclasts, were lower than in c-Fos-sufficient Sh3bp2KI/KI and wild-type mice (Fig. 2B). Activated macrophages are most likely to be the source of the TRAP5b slightly elevated in c-Fos-deficient Sh3bp2KI/KI mice compared to c-Fos-deficient Sh3bp2+/+ mice (Fig. 2B).(42,43) Absence of osteoclasts at the inflamed ankle joint of the c-Fos-deficient Sh3bp2KI/KI mice was further confirmed by Oscar and Ocstamp expression levels lower than wild-type mice (Fig. 2C). No significant increase in expression of Acp5, Oscar, Ocstamp, Ctsk, Car2, Calcr, and Rank compared to c-Fos-deficient Sh3bp2+/+ mice verified the lack of osteoclasts in c-Fos-deficient Sh3bp2KI/KI mice (Fig. 2C). Interestingly, Dcstamp, Clcn7, and Tcirg1 expression in c-Fos-deficient Sh3bp2KI/KI mice was higher than wild-type mice and comparable to c-Fos-sufficient Sh3bp2KI/KI mice (Fig. 2C). These data demonstrate that knocking out of the c-Fos gene effectively blocks the formation of osteoclasts in Sh3bp2KI/KI mice.
Fig. 2. c-Fos-deficient Sh3bp2KI/KI mice develop no TRAP+ osteoclasts.
(A) TRAP staining of inflammatory lesions at the distal tibia. 12 weeks old. Bar = 100 μm. (B) ELISA of serum TRAP5b at 12 weeks old. (C) qPCR analysis of osteoclast-associated genes in ankle joint tissue at 12 weeks old. Average expression levels in wild-type mice are set as 1. Data are presented as mean ± SD. *p < 0.05. ANOVA with Tukey-Kramer post-hoc test. NS = not significant. #: p < 0.05 with two-tailed t-test (B, C).
Osteoclast precursors from c-Fos-deficient Sh3bp2KI/KI mice fail to form TRAP-positive osteoclasts
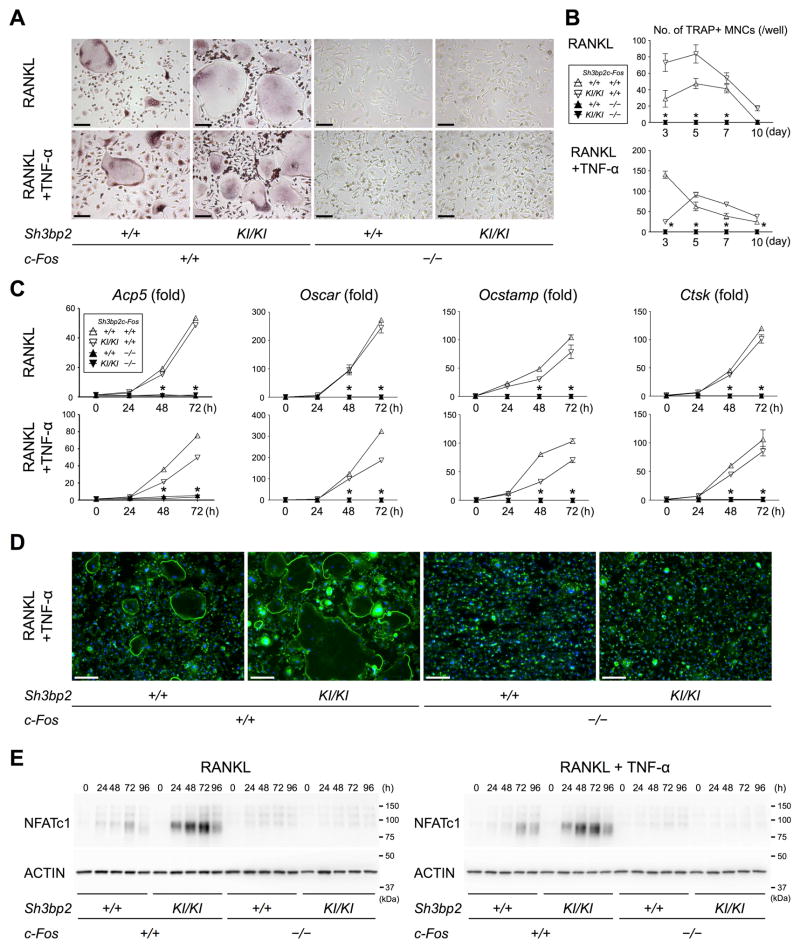
To confirm the lack of in vitro capacity of c-Fos-deficient Sh3bp2KI/KI myeloid cells to differentiate into osteoclasts, M-CSF-dependent macrophages from fetal liver of the c-Fos-deficient Sh3bp2KI/KI mice were stimulated either with RANKL or with RANKL/TNF-α. Formation of TRAP+ multinucleated cells was completely abrogated in the M-CSF-dependent macrophages from c-Fos-deficient Sh3bp2KI/KI mice under both culture conditions (Fig. 3, A and B). No increase in osteoclast marker gene expression was observed during the culture period compared to M-CSF-dependent macrophages from c-Fos-sufficient Sh3bp2KI/KI and wild-type mice (Fig. 3C). Actin ring formation typically seen in TRAP+ multinucleated osteoclasts was not observed in c-Fos-deficient Sh3bp2KI/KI macrophage cultures with RANKL/TNF-α (Fig. 3D).
Fig. 3. M-CSF-dependent macrophages from c-Fos-deficient Sh3bp2KI/KI mice fail to differentiate to TRAP+ osteoclasts.
(A) TRAP staining of M-CSF-dependent macrophages from fetal liver cultured in the presence of RANKL or RANKL/TNF-α for 5 days. Bar = 100 μm. (B) Number of TRAP+ multinucleated cells per well of cultures in (A). n = 6. (C) qPCR analysis of osteoclast-associated genes in fetal liver-derived M-CSF-dependent macrophages stimulated with RANKL or RANKL/TNF-α. Average expression levels in wild-type mice at 0 hour are set as 1. n = 3. (D) Phalloidin staining of actin rings. Fetal liver-derived M-CSF-dependent macrophages were stimulated with RANKL/TNF-α for 5 days. Bar = 100 μm. (E) Western blotting analysis of NFATc1 with cell lysates from fetal liver-derived M-CSF-dependent macrophages cultured in the presence of RANKL or RANKL/TNF-α. Data are presented as mean ± SD. *p < 0.05 vs. c-Fos-sufficient mice. ANOVA with Tukey-Kramer post-hoc test.
Since robust induction of NFATc1 is a molecular hallmark of differentiating osteoclasts(5), we next examined whether NFATc1 increases in M-CSF-dependent macrophages from c-Fos-deficient Sh3bp2KI/KI mice in response to RANKL or RANKL/TNF-α. In agreement with a previous report (4), lack of c-Fos failed to induce NFATc1 auto-amplification even with the gain-of-function mutation of SH3BP2 (Fig. 3E), suggesting that c-Fos is a transcription factor downstream of SH3BP2 in osteoclastogenesis. Lack of c-Fos did not affect SH3BP2 protein levels in the macrophages (Supplemental Fig. 2). Taken together, our results show that osteoclast precursor cells from c-Fos-deficient Sh3bp2KI/KI mice are not able to differentiate into mature TRAP+ osteoclasts, suggesting that bone erosion in c-Fos-deficient Sh3bp2KI/KI mice was due to cells other than osteoclasts.
Increased MMP expression in joint tissue of c-Fos-deficient Sh3bp2KI/KI mice
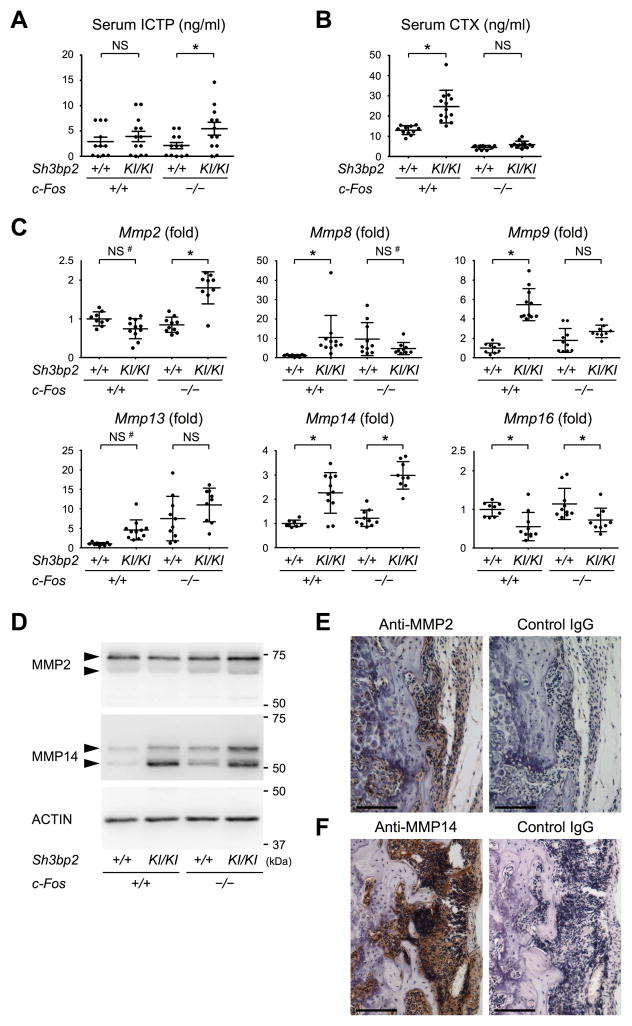
We found that serum levels of cross-linked carboxyterminal telopeptide of type I collagen (ICTP), a marker of bone resorption generated by MMPs (44), were elevated in c-Fos-deficient Sh3bp2KI/KI mice compared to c-Fos-deficient Sh3bp2+/+ mice (Fig. 4A). In contrast, serum C-terminal crosslinked telopeptide of type I collagen (CTX) levels, another bone resorption marker produced by cathepsin K from osteoclasts (44), were comparable between c-Fos-deficient Sh3bp2KI/KI and c-Fos-deficient Sh3bp2+/+ mice (Fig. 4B). These results suggest that MMPs participate in the degradation of bone matrices in c-Fos-deficient Sh3bp2KI/KI mice. Expression analysis of collagenolytic MMPs (45) in ankle joint tissue (without skin) revealed that Mmp2 and Mmp14 levels in c-Fos-deficient Sh3bp2KI/KI mice are higher than c-Fos-deficient Sh3bp2+/+ mice, which correlated with the appearance of bone erosion (Fig. 4C). Expression levels of non-collagenolytic MMPs were also elevated (Supplemental Fig. 3). Western blotting analysis confirmed the elevation of MMP2 and MMP14 proteins in the joint tissue of the c-Fos-deficient Sh3bp2KI/KI mice compared to c-Fos-deficient Sh3bp2+/+ mice that do not have bone erosion (Fig. 4D). Consistent with these results, lesions within pits and on the surface of bone were positive for MMP2 and MMP14 (Fig. 4, E and F). Collectively, our results suggest that MMP2 and/or MMP14 participate in an osteoclast-independent degradative mechanism of bone in c-Fos-deficient Sh3bp2KI/KI mice.
Fig. 4. MMP2 and MMP14 expression levels are increased in joint tissue of c-Fos-deficient Sh3bp2KI/KI mice.
(A) ELISA for serum ICTP at 12 weeks old. (B) ELISA for serum CTX at 12 weeks old. (C) qPCR analysis of collagenotylic MMPs in joint tissue at 12 weeks old. Mmp1 expression was not analyzable due to low expression levels in both SYBR green and Taqman assays (Supplemental Table 1). Average expression levels in wild-type mice are set as 1. (D) Western blotting analysis for MMP2 and MMP14 with cell lysates from the ankle joints. (E, F) Immunohistochemical staining of MMP2 and MMP14 with tissue sections of the distal tibia. Bar = 100 μm. Data are presented as mean ± SD. *p < 0.05 with two-tailed t-test (A, B) or ANOVA with Tukey-Kramer post-hoc test (C). NS = not significant. #: p < 0.05 with two-tailed t-test.
Macrophages are responsible for bone erosion of c-Fos-deficient Sh3bp2KI/KI mice
Erosion pits were filled with cells that stain strongly for F4/80, a marker for macrophages encoded by the Adgre1 gene (Fig. 5A). Expression levels of macrophage marker genes were elevated in the ankle joint of c-Fos-deficient Sh3bp2KI/KI mice compared to c-Fos-deficient Sh3bp2+/+ and wild-type mice (Fig. 5B). These results led us to hypothesize that macrophages are primarily responsible for the bone erosion in c-Fos-deficient Sh3bp2KI/KI mice.
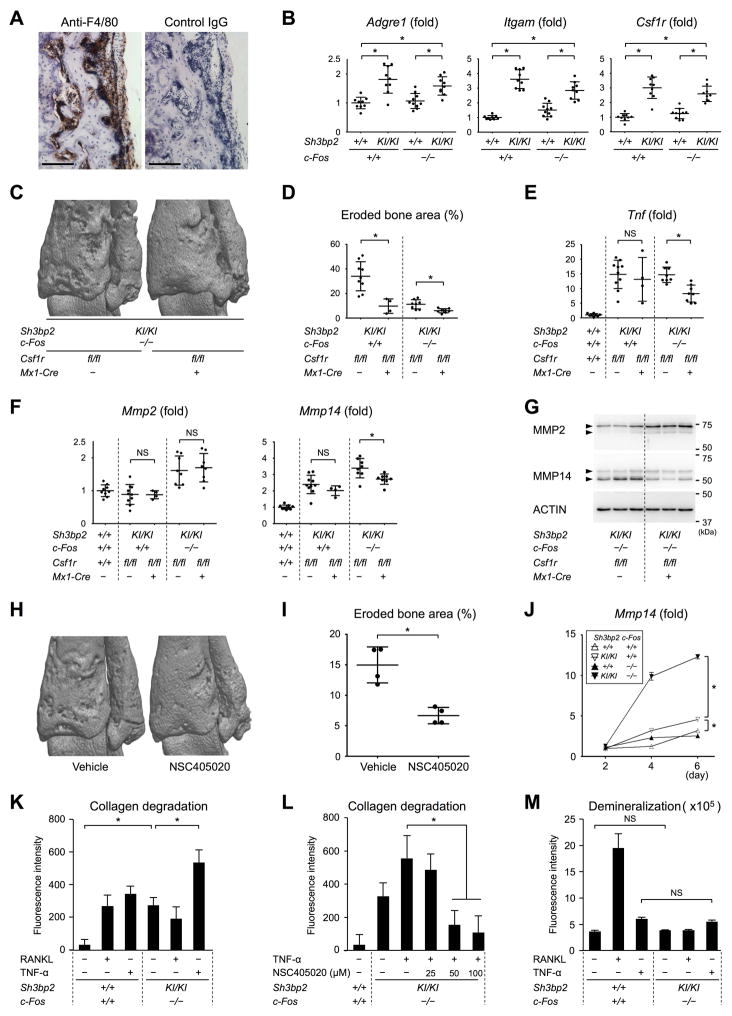
Fig. 5. Macrophages expressing MMP14 are responsible for bone erosion in c-Fos-deficient Sh3bp2KI/KI mice.
(A) Immunohistochemical staining of inflammatory lesions at the distal tibia from c-Fos-deficient Sh3bp2KI/KI mouse with anti-F4/80 antibody. 12 weeks old. Bar = 100 μm. (B) qPCR analysis of macrophage marker gene expression in the ankle joints. (C) μCT images of the distal tibia at 12 weeks old. (D) Quantitative measurement of eroded bone surface area at the distal tibia. (E) qPCR analysis of Tnf expression in the ankle joints. (F) qPCR analysis of Mmp2 and Mmp14 expression in the ankle joints. (G) Western blotting analysis of MMP2 and MMP14 with cell lysates from the ankle joints. (H) μCT images of the distal tibia from Sh3bp2KI/KI mice treated with or without an MMP14 inhibitor NSC405020 (50 mg/kg/day) from 4 to 10 weeks old. (I) Quantitative measurement of eroded bone surface area at the distal tibia of Sh3bp2KI/KI mice treated with or without NSC405020 from 4 to 10 weeks old. (J) Comparison of Mmp14 expression levels in fetal liver-derived M-CSF-dependent macrophages by qPCR. n = 3. (K) Measurement of collagenolytic activity of fetal liver-derived M-CSF-dependent macrophages. n = 6. (L) Measurement of collagenolytic activity of fetal liver-derived M-CSF-dependent macrophages in the presence or absence of NSC405020. n = 6. (M) Measurement of mineral resorption capacity of fetal liver-derived M-CSF-dependent macrophages. n = 6. Macrophages were cultured for 7 days on the plates coated with europium-labeled type I collagen fibers (K, L) or on the plates coated with fluoresceinamine-labeled calcium phosphate (M). Average expression levels in wild-type mice are set as 1 (B, E, F). Average expression level in wild-type mice at day 2 is set as 1 (J). Data are presented as mean ± SD. *p < 0.05 with two-tailed t-test (D, E, F, I) or ANOVA with Tukey-Kramer post-hoc test (B, J, K, L, M). NS = not significant.
Colony-stimulating factor 1 receptor (CSF1R) is a receptor for M-CSF that is required for the maintenance of tissue macrophage populations. Functional ablation of CSF1R or M-CSF causes severe deficiency of macrophages and osteoclasts.(46,47) To determine whether macrophage depletion suppresses bone resorption in c-Fos-deficient Sh3bp2KI/KI mice, Mx1-Cre Csf1rfl/fl mice were created on an osteoclast-free c-Fos−/− Sh3bp2KI/KI background and depletion of M-CSF-dependent macrophages was induced in the mice by administrating pI:pC three times at one week of age.(48) MicroCT analysis revealed that macrophage depletion effectively ameliorates bone erosion in the c-Fos-deficient Sh3bp2KI/KI mice at 12 weeks of age (Fig. 5, C and D). This suppression was accompanied by a reduction in Tnf expression in the ankle joints (Fig. 5E). Mmp14 expression was also suppressed in the joints of macrophage-depleted c-Fos-deficient Sh3bp2KI/KI mice compared to non-depleted control c-Fos-deficient Sh3bp2KI/KI mice, while Mmp2 expression was comparable (Fig. 5F). Consistent with the results, MMP14 protein levels were also decreased, although MMP2 was elevated (Fig. 5G). These data indicate that macrophages that express MMP14 are responsible for bone erosion in c-Fos-deficient Sh3bp2KI/KI mice.
Next, we examined whether pharmacological inhibition of MMP14 rescues the bone erosion. Administration of NSC405020, an MMP14 inhibitor that suppresses the collagenolytic activity of MMP14 by interacting with its hemopexin (PEX) domain (39), reduced bone erosion in c-Fos-deficient Sh3bp2KI/KI mice (Fig. 5, H and I). Also, we discovered that Mmp14 expression in M-CSF-dependent macrophages derived from fetal liver of c-Fos-sufficient Sh3bp2KI/KI mice is increased compared to wild-type macrophages, and this increase was further augmented by the lack of c-Fos by 3.8-fold (Fig. 5J). These findings suggest that the SH3BP2- and c-Fos-mediated signaling mechanisms regulating MMP14 expression in macrophages merits further study.
To confirm whether c-Fos-deficient Sh3bp2KI/KI macrophages have the capacity to degrade bone collagen and demineralize bone, M-CSF-dependent macrophages from c-Fos-deficient Sh3bp2KI/KI mice were cultured on plates coated either with europium-labeled type I collagen fibers or with fluorescein-labeled calcium phosphate. The fluorescent intensity of culture supernatants revealed that c-Fos-deficient Sh3bp2KI/KI macrophages have an increased capacity for collagen degradation compared to wild-type macrophages and that this capacity is enhanced in the presence of TNF-α (Fig. 5K). This increase was abrogated by treatment with NSC405020 (Fig. 5L). In contrast, the capacity for mineral dissolution was comparable between c-Fos-deficient Sh3bp2KI/KI and wild-type macrophages in the presence or absence of TNF-α (Fig. 5M). Finally, we confirmed that c-Fos-deficient Sh3bp2KI/KI macrophages do create resorption pits on dentine slices when stimulated with RANKL and TNF-α, although resorption area and depth of pits were smaller than those in wild-type macrophage cultures (Supplemental Fig. 4). In summary, the data indicate that macrophages in c-Fos-deficient Sh3bp2KI/KI mice have an increased capacity to degrade collagen and that MMP14 plays a critical role in macrophage-mediated bone resorption in c-Fos-deficient Sh3bp2KI/KI mice.
NF-κB signaling mediates increased Mmp14 expression in c-Fos-deficient Sh3bp2KI/KI macrophages
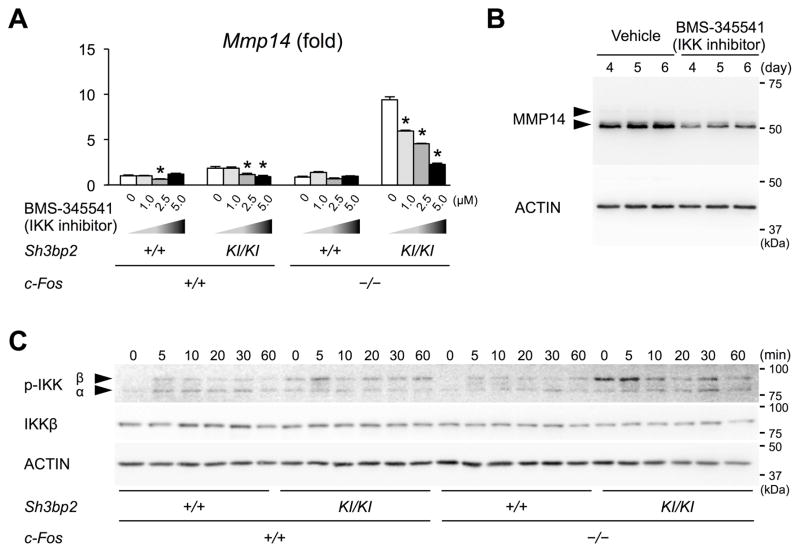
We next sought to determine the molecular mechanism by which c-Fos-deficient Sh3bp2KI/KI macrophages increase Mmp14 expression. Fetal liver cells were cultured in the presence of M-CSF with or without inhibitors that block a series of signaling pathways. Among all inhibitors tested, BMS-345541, an IκB kinase (IKK) inhibitor that blocks NF-κB-dependent transcriptional activation, was most effective in suppressing the expression of Mmp14 in c-Fos-deficient Sh3bp2KI/KI macrophages in a dose-dependent manner (Fig. 6A, Supplemental Fig. 5). MMP14 protein levels were also decreased in c-Fos-deficient Sh3bp2KI/KI macrophages when cultured with BMS-345541 (Fig. 6B). We further found that the basal phosphorylation level of IKKβ is elevated in the M-CSF-dependent c-Fos-deficient Sh3bp2KI/KI macrophages, and which was further elevated after re-stimulation with M-CSF (Fig. 6C). Activation of ERK and p38 pathways downstream of CSF1R was not significantly changed in c-Fos-deficient Sh3bp2KI/KI macrophages compared to wild-type macrophages (Supplemental Fig. 6). Finally, phosphorylation of JNK was sustained longer in c-Fos-deficient Sh3bp2KI/KI macrophages, but a JNK inhibitor SP600125 failed to suppress the elevated Mmp14 expression in the double mutant macrophages (Supplemental Fig. 5). All the above inhibitors are commercially available and their selectivities have been widely accepted. These data suggest that increased MMP14 expression in c-Fos-deficient Sh3bp2KI/KI macrophages is mediated primarily by activation of the NF-κB pathway.
Fig. 6. NF-κB pathway is responsible for increased MMP14 expression in c-Fos-deficient Sh3bp2KI/KI macrophages.
(A) Comparison of Mmp14 expression levels in fetal liver-derived M-CSF-dependent macrophages by qPCR. Non-adherent fetal liver cells were cultured with M-CSF for 2 days, then further cultured with M-CSF in the presence or absence of BMS-345541 for 2 days. Average expression level in wild-type mice with no inhibitor is set as 1. n = 3. Data are representative of three independent experiments. (B) Western blotting analysis of MMP14 in M-CSF-dependent macrophages treated with or without BMS-345541. Non-adherent fetal liver cells were cultured with M-CSF for 2 days, then further cultured with M-CSF in the presence or absence of BMS-345541 for 4 days (day 3–6). (C) Western blotting analysis of phosphorylated IKKα/β. Non-adherent fetal liver cells were cultured with M-CSF for 4 days, then M-CSF-dependent macrophages were starved for serum and M-CSF for 6 hours followed by re-stimulation with M-CSF (25 ng/ml). *p < 0.05 decreased vs. 0 μM in each group. ANOVA with Tukey-Kramer post-hoc test.
RANKL deficiency protects against bone erosion in Sh3bp2KI/KI mice and c-Fos deficiency restores bone erosion to the RANKL-deficient Sh3bp2KI/KI mice
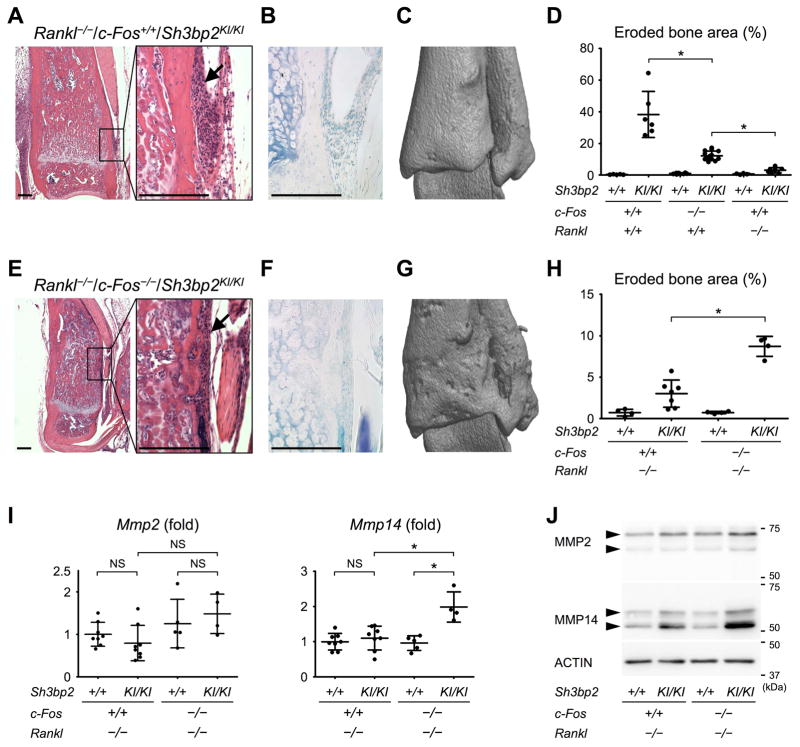
To examine whether other Sh3bp2KI/KI mice lacking osteoclasts also develop bone erosion, Sh3bp2KI/KI mice also deficient in RANKL were created. Similar to the c-Fos-deficient Sh3bp2KI/KI mice, RANKL-deficient Sh3bp2KI/KI mice developed inflammatory lesions at the distal tibia (Fig. 7A) with no TRAP+ multinucleated osteoclasts on the bone surfaces (Fig. 7B). Interestingly, a few of TRAP+ cells were detected in a small number of RANKL-deficient Sh3bp2KI/KI mice (Supplemental Fig. 7).(20) We found that RANKL-deficient Sh3bp2KI/KI mice exhibit significantly less bone erosion compared to the c-Fos-deficient Sh3bp2KI/KI mice (Fig. 7, C and D). We have further deleted the c-Fos gene on the RANKL-deficient Sh3bp2KI/KI background. We discovered that c-Fos/RANKL-deficient Sh3bp2KI/KI mice develop inflammatory lesions comparable to the c-Fos-deficient Sh3bp2KI/KI mice with no TRAP+ osteoclasts (Fig. 7, E and F) and that c-Fos deletion restores bone erosion to the RANKL-deficient Sh3bp2KI/KI mice (Fig. 7, G and H). These results suggest that both loss-of-function of c-Fos and gain-of-function of SH3BP2 are required for the macrophage-mediated bone erosion in c-Fos-deficient Sh3bp2KI/KI mice. Comparison of Mmp14 expression levels in ankle joint tissue between RANKL-deficient Sh3bp2KI/KI mice without noticeable bone erosion and c-Fos/RANKL-deficient Sh3bp2KI/KI mice with a considerable amount of bone erosion revealed that elevated Mmp14 mRNA expression correlates well with the appearance of bone erosion (Fig. 7I). In agreement with this result, MMP14 protein level was elevated in the joint of c-Fos/RANKL-deficient Sh3bp2KI/KI mice compared to that for RANKL-deficient Sh3bp2KI/KI mice (Fig. 7J). In contrast, MMP2 levels were not significantly changed (Fig. 7, I and J). These data support MMP14’s role as a key mediator of bone resorption in c-Fos-deficient Sh3bp2KI/KI mice.
Fig. 7. Both c-Fos deficiency and SH3BP2 gain-of-function are required for osteolytic transformation of macrophages.
(A, E) H&E staining of the distal tibia showing inflammatory lesions (arrows). 12 weeks old. Bar = 200 μm. (B, F) TRAP staining images of inflammatory lesions at the distal tibia at 12 weeks old. Bar = 200 μm. (C, G) μCT images of the distal tibia at 12 weeks old. (D, H) Quantitative measurement of eroded bone surface area at the distal tibia. (I) Mmp2 and Mmp14 expression levels in the ankle joints. Average expression levels in wild-type mice are set as 1. (J) Western blotting analysis of MMP2 and MMP14 with cell lysates from ankle joint tissue. Data are presented as mean ± SD. *p < 0.05. ANOVA with Tukey-Kramer post-hoc test. NS = not significant.
Discussion
Previous studies have shown that osteoclasts play an indispensable role in inflammatory bone destruction and established a definitive consensus that osteoclasts are the exclusive cells responsible for bone resorption in pathological conditions.(3,15,49) This dogma is based on convincing genetic evidence that bone erosion is absent in osteoclast-free mouse models of inflammatory bone disease such as c-Fos-deficient mice that overexpress TNF-α (13) and RANKL-deficient mice that develop autoimmune arthritis.(14) However, this paradigm should now be re-evaluated as a result of our studies with osteoclast-free Sh3bp2KI/KI mice.
We have shown that Sh3bp2KI/KI mice also deficient in c-Fos display a considerable amount of inflammatory focal bone erosion, although the mice have no TRAP+ osteoclasts. We have further shown that myeloid cells from the mice are not able to differentiate into TRAP+ osteoclasts under RANKL or RANKL/TNF-α stimulation. These data demonstrate that c-Fos-deficient Sh3bp2KI/KI mice are the first compelling animal model of inflammatory arthritis that develops bone erosion independently of osteoclasts. Mechanistically, we have shown that macrophages expressing MMP14 are primarily responsible for this osteoclast-independent bone resorption. Taken together, our results provide the evidence that dysregulated macrophages can substitute for osteoclasts in arthritis to cause bone destruction. Given the bone-resorbing capacity of c-Fos-deficient Sh3bp2KI/KI macrophages, we suggest classifying these cells as a subtype of osteoclasts (e.g. non-canonical osteoclasts or osteolytic macrophages).
Indeed, the possibility of osteoclast-independent pathological bone destruction has been implied previously. Nakamura et al. reported that administration of bisphosphonates to collagen-induced arthritis mice failed to provide full protection against focal bone erosion.(24) One explanation for this unsuccessful protection is that bone resorption could be caused by cells other than osteoclasts such as macrophages and/or granulocytes under inflammatory conditions. Ralston et al., also discussed this possibility in human RA.(26) Supporting this assumption, Kadono et al. reported a rare case of osteopetrosis associated with joint inflammation due to RA.(50) In this patient, the protection against bone destruction was not complete, and bone erosion was still observed in the patient’s carpal bones. These clinical findings demonstrate that cells other than fully functional canonical osteoclasts can cause significant inflammatory bone destruction in human arthritis.
On the other hand, we have previously shown that bone structural properties are impaired in Sh3bp2KI/KI mice.(51) In addition, our preliminary study showed that c-Fos-deficient mice exhibit reduced bone mineral density compared to wild-type mice and that Sh3bp2KI/KI alleles decrease the density further (data not shown). These data suggest that impaired matrix composition and reduced mineral deposition of bone may increase the susceptibility to bone degradation by activated macrophages in c-Fos-deficient Sh3bp2KI/KI mice. In this context, we presume that bone fragility due to chronic exposure to inflammatory cytokines or gene mutations/polymorphisms that affect the function of osteoblasts and/or osteocytes is a prerequisite for bone destruction mediated by non-canonical bone resorbing cells.
Interestingly, we found that expression levels of the chloride channel Clcn7 and V-ATPase subunit Tcirg1, both of which are required in osteoclasts for reducing the local pH to resorb bone inorganic minerals, are elevated in the joins of c-Fos-deficient Sh3bp2KI/KI mice compared to c-Fos-deficient Sh3bp2+/+ mice (Fig. 2C). This suggests that activated macrophages in inflammatory lesions of the c-Fos-deficient Sh3bp2KI/KI mice can create an acidic extracellular environment suitable for removing bone minerals.
We showed that depletion of RANKL results in protection against bone destruction in Sh3bp2KI/KI mice. Specifically, denosumab, an inhibitory antibody against RANKL, is effective for the treatment of giant cell tumors that exhibit indistinguishable histological characteristics from cherubism.(52) These findings suggest that RANKL is a therapeutic target for prevention and treatment of excessive jawbone resorption in cherubism. Our study may also have implications for the treatment of osteopetrosis due to lack of canonical osteoclasts (53) through the induction of osteolytic macrophages.
Both RANKL and c-Fos are essential for TRAP+ osteoclast formation.(8–10) However, while the absence of RANKL rescues bone erosion in Sh3bp2KI/KI mice, an absence of c-Fos failed to protect against bone erosion. The phenotypic difference in bone erosion between the two distinct osteoclast-depleted Sh3bp2KI/KI models may be due to the greater activity of inflammatory macrophages in c-Fos-deficient Sh3bp2KI/KI mice than in RANKL-deficient Sh3bp2KI/KI mice. In fact, the average Tnf level in joint tissue of c-Fos-deficient Sh3bp2KI/KI mice is 2.1-fold higher than RANKL-deficient Sh3bp2KI/KI mice (data not shown). Previous reports that c-Fos limits inflammatory responses and acts as an anti-inflammatory factor in macrophages and dendritic cells by suppressing NF-κB pathway and inflammatory cytokine production (54–56) support this notion.
Collagenase MMP14 is the major MMP expressed by synoviocytes in RA.(57,58) This fact identified MMP14 as a promising drug target for the protection against joint damage in inflammatory bone diseases. For example, DX-2400 is a human MMP14 inhibitory antibody that targets the catalytic domain of MMP14. It represses proteolytic activity of MMP14 leading to a blockade of proMMP2 processing required for collagen degradation by active MMP2.(40) A recent study showed that treatment of the mice with collagen-induced arthritis with this antibody had minimal protective effect on bone erosion, while it reduced the progression of the joint inflammation and prevented cartilage degradation.(59) When we administered DX-2400 to c-Fos-deficient Sh3bp2KI/KI mice from 4 to 10 weeks of age, bone erosion was not suppressed compared to the human IgG control group (data not shown). In contrast, we have shown that administration of NSC405020 successfully suppresses the bone erosion of c-Fos-deficient Sh3bp2KI/KI mice.(39) NSC405020 inhibits collagen degradation differently by interacting with the hemopexin (PEX) domain of MMP14. The PEX domain of MMP14 is known to be critical for the homodimerization of MMP14 that is essential for type I collagen degradation, but the domain is not required for the activation of proMMP2 on cell surface.(60,61) It is also reported that NSC405020 suppresses the collagenolytic activity of MMP14 without affecting the catalytic function of MMP14 necessary for proMMP2 processing.(39) Therefore, comparison of the effect of these two MMP14 inhibitors that target the different domains of MMP14 suggests that inhibition against the PEX domain suppresses MMP14-mediated bone resorption more efficiently than against the catalytic domain.
In addition to its effectiveness in the treatment of giant cell tumors (52), denosumab also suppresses the progression of bone erosion in RA patients.(62) This result presents clear evidence that osteoclasts play a definitive role in bone destruction in RA in humans and suggests that the potential contribution of inflammatory macrophages to bone resorption in common forms of inflammatory bone disease is minimal. Nonetheless, insights into the mechanism of bone resorption in c-Fos-deficient Sh3bp2KI/KI mice should lead us to rethink current therapeutic approaches towards inflammatory bone diseases. In particular, new attention should be focused on osteolytic macrophages and collagenolytic activity of MMP14 as therapeutic targets not only for cherubism but also for RA, which showed an inadequate response to current anti-inflammation and anti-bone resorption therapies.(63,64)
Moreover, our discovery of osteolytic transformation of c-Fos-deficient Sh3bp2KI/KI macrophages may contribute to a new understanding of the pathology of diseases where an osteoclastic mechanism of bone resorption cannot be fully rationalized.(65,66) Signaling molecules involved in the macrophage-mediated bone resorption and remodeling might also be involved in the mechanisms of bone metastasis of tumors. (67)
In summary, inflammatory bone resorption in c-Fos-deficient Sh3bp2KI/KI mice is mediated by MMP14-expressing macrophages even in the absence of osteoclasts. This non-canonical bone resorption in mice provides new insights into the mechanism and treatment of bone loss in inflammatory bone diseases and offers an opportunity to re-evaluate what type of cells can resorb bone and whether activated macrophages are responsible for pathological bone resorption in humans.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases under Award Number R21AR070953 to YU as well as by the National Institute of Dental and Craniofacial Research under Award Number R01DE020835 and R01DE025870 to YU of the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. MK is a recipient of the fellowship from brain circulation program to develop new leaders for international dental education course through international collaborative dental research, Japan. We thank all members of the Bone Biology Research Program at the University of Missouri-Kansas City, School of Dentistry for critical suggestions. We appreciate Vladimir Dusevich and Donggao Zhao for technical assistance of TEM and SEM procedures. We also thank Joshua Prather for μCT analysis.
Footnotes
Disclosure
The authors have declared that no conflict of interest exists.
Authors’ roles: YU had the project idea. MK and YU initiated the project. MK, JPG, and YU designed the experiments. MK, KM, TM, TY, TY performed experiments. All authors contributed to the data analysis and interpretation of the results. MK, JPG, and YU wrote the manuscript and all authors approved the manuscript. MK and YU are responsible for the data integrity and analysis.
References
- 1.Teitelbaum SL, Tondravi MM, Ross FP. Osteoclasts, macrophages, and the molecular mechanisms of bone resorption. J Leukoc Biol. 1997;61(4):381–8. doi: 10.1002/jlb.61.4.381. [DOI] [PubMed] [Google Scholar]
- 2.Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7(4):292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- 3.Takayanagi H. Osteoimmunology and the effects of the immune system on bone. Nat Rev Rheumatol. 2009;5(12):667–76. doi: 10.1038/nrrheum.2009.217. [DOI] [PubMed] [Google Scholar]
- 4.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3(6):889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 5.Asagiri M, Sato K, Usami T, Ochi S, Nishina H, Yoshida H, et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med. 2005;202(9):1261–9. doi: 10.1084/jem.20051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang ZQ, Ovitt C, Grigoriadis AE, Mohle-Steinlein U, Ruther U, Wagner EF. Bone and haematopoietic defects in mice lacking c-fos. Nature. 1992;360(6406):741–5. doi: 10.1038/360741a0. [DOI] [PubMed] [Google Scholar]
- 7.Johnson RS, Spiegelman BM, Papaioannou V. Pleiotropic effects of a null mutation in the c-fos proto-oncogene. Cell. 1992;71(4):577–86. doi: 10.1016/0092-8674(92)90592-z. [DOI] [PubMed] [Google Scholar]
- 8.Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA, et al. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;266(5184):443–8. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- 9.Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397(6717):315–23. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 10.Kim N, Odgren PR, Kim DK, Marks SC, Jr, Choi Y. Diverse roles of the tumor necrosis factor family member TRANCE in skeletal physiology revealed by TRANCE deficiency and partial rescue by a lymphocyte-expressed TRANCE transgene. Proc Natl Acad Sci U S A. 2000;97(20):10905–10. doi: 10.1073/pnas.200294797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dougall WC, Glaccum M, Charrier K, Rohrbach K, Brasel K, De Smedt T, et al. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999;13(18):2412–24. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Sarosi I, Yan XQ, Morony S, Capparelli C, Tan HL, et al. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc Natl Acad Sci U S A. 2000;97(4):1566–71. doi: 10.1073/pnas.97.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redlich K, Hayer S, Ricci R, David JP, Tohidast-Akrad M, Kollias G, et al. Osteoclasts are essential for TNF-alpha-mediated joint destruction. J Clin Invest. 2002;110(10):1419–27. doi: 10.1172/JCI15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettit AR, Ji H, von Stechow D, Muller R, Goldring SR, Choi Y, et al. TRANCE/RANKL knockout mice are protected from bone erosion in a serum transfer model of arthritis. Am J Pathol. 2001;159(5):1689–99. doi: 10.1016/S0002-9440(10)63016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh NC, Gravallese EM. Bone loss in inflammatory arthritis: mechanisms and treatment strategies. Curr Opin Rheumatol. 2004;16(4):419–27. doi: 10.1097/01.bor.0000127824.42507.68. [DOI] [PubMed] [Google Scholar]
- 16.Schett G, Gravallese E. Bone erosion in rheumatoid arthritis: mechanisms, diagnosis and treatment. Nat Rev Rheumatol. 2012;8(11):656–64. doi: 10.1038/nrrheum.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueki Y, Tiziani V, Santanna C, Fukai N, Maulik C, Garfinkle J, et al. Mutations in the gene encoding c-Abl-binding protein SH3BP2 cause cherubism. Nat Genet. 2001;28(2):125–6. doi: 10.1038/88832. [DOI] [PubMed] [Google Scholar]
- 18.Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, et al. Loss of Tankyrase-Mediated Destruction of 3BP2 Is the Underlying Pathogenic Mechanism of Cherubism. Cell. 2011;147(6):1324–39. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueki Y, Lin CY, Senoo M, Ebihara T, Agata N, Onji M, et al. Increased myeloid cell responses to M-CSF and RANKL cause bone loss and inflammation in SH3BP2 “cherubism” mice. Cell. 2007;128(1):71–83. doi: 10.1016/j.cell.2006.10.047. [DOI] [PubMed] [Google Scholar]
- 20.Mukai T, Ishida S, Ishikawa R, Yoshitaka T, Kittaka M, Gallant R, et al. SH3BP2 Cherubism Mutation Potentiates TNF-alpha-Induced Osteoclastogenesis Via NFATc1 and TNF-alpha-Mediated Inflammatory Bone Loss. J Bone Miner Res. 2014;29(12):2618–2635. doi: 10.1002/jbmr.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshitaka T, Mukai T, Kittaka M, Alford LM, Masrani S, Ishida S, et al. Enhanced TLR-MYD88 signaling stimulates autoinflammation in SH3BP2 cherubism mice and defines the etiology of cherubism. Cell Rep. 2014;8(6):1752–66. doi: 10.1016/j.celrep.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mundy CR, Altman AJ, Gondek MD, Bandelin JG. Direct resorption of bone by human monocytes. Science. 1977;196(4294):1109–11. doi: 10.1126/science.16343. [DOI] [PubMed] [Google Scholar]
- 23.Teitelbaum SL, Stewart CC, Kahn AJ. Rodent peritoneal macrophages as bone resorbing cells. Calcif Tissue Int. 1979;27(3):255–61. doi: 10.1007/BF02441194. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura M, Ando T, Abe M, Kumagai K, Endo Y. Contrast between effects of aminobisphosphonates and non-aminobisphosphonates on collagen-induced arthritis in mice. Br J Pharmacol. 1996;119(2):205–12. doi: 10.1111/j.1476-5381.1996.tb15972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qing H, Ardeshirpour L, Pajevic PD, Dusevich V, Jahn K, Kato S, et al. Demonstration of osteocytic perilacunar/canalicular remodeling in mice during lactation. J Bone Miner Res. 2012;27(5):1018–29. doi: 10.1002/jbmr.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ralston SH, Hacking L, Willocks L, Bruce F, Pitkeathly DA. Clinical, biochemical, and radiographic effects of aminohydroxypropylidene bisphosphonate treatment in rheumatoid arthritis. Ann Rheum Dis. 1989;48(5):396–9. doi: 10.1136/ard.48.5.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maccagno A, Di Giorgio E, Roldan EJ, Caballero LE, Perez Lloret A. Double blind radiological assessment of continuous oral pamidronic acid in patients with rheumatoid arthritis. Scand J Rheumatol. 1994;23(4):211–4. doi: 10.3109/03009749409103063. [DOI] [PubMed] [Google Scholar]
- 28.Valleala H, Laasonen L, Koivula MK, Mandelin J, Friman C, Risteli J, et al. Two year randomized controlled trial of etidronate in rheumatoid arthritis: changes in serum aminoterminal telopeptides correlate with radiographic progression of disease. J Rheumatol. 2003;30(3):468–73. [PubMed] [Google Scholar]
- 29.Breuil V, Euller-Ziegler L. Bisphosphonate therapy in rheumatoid arthritis. Joint Bone Spine. 2006;73(4):349–54. doi: 10.1016/j.jbspin.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 30.Naito M, Takahashi K, Nishikawa S. Development, differentiation, and maturation of macrophages in the fetal mouse liver. J Leukoc Biol. 1990;48(1):27–37. doi: 10.1002/jlb.48.1.27. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Keogh RJ, Hunter MG, Mitchell CA, Frey RS, Javaid K, et al. SHIP2 is recruited to the cell membrane upon macrophage colony-stimulating factor (M-CSF) stimulation and regulates M-CSF-induced signaling. J Immunol. 2004;173(11):6820–30. doi: 10.4049/jimmunol.173.11.6820. [DOI] [PubMed] [Google Scholar]
- 32.Burke JR, Pattoli MA, Gregor KR, Brassil PJ, MacMaster JF, McIntyre KW, et al. BMS-345541 is a highly selective inhibitor of I kappa B kinase that binds at an allosteric site of the enzyme and blocks NF-kappa B-dependent transcription in mice. J Biol Chem. 2003;278(3):1450–6. doi: 10.1074/jbc.M209677200. [DOI] [PubMed] [Google Scholar]
- 33.Flanagan WM, Corthesy B, Bram RJ, Crabtree GR. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 1991;352(6338):803–7. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- 34.Bennett BL, Sasaki DT, Murray BW, O’Leary EC, Sakata ST, Xu W, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci U S A. 2001;98(24):13681–6. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuenda A, Rouse J, Doza YN, Meier R, Cohen P, Gallagher TF, et al. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364(2):229–33. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 36.Braselmann S, Taylor V, Zhao H, Wang S, Sylvain C, Baluom M, et al. R406, an orally available spleen tyrosine kinase inhibitor blocks fc receptor signaling and reduces immune complex-mediated inflammation. J Pharmacol Exp Ther. 2006;319(3):998–1008. doi: 10.1124/jpet.106.109058. [DOI] [PubMed] [Google Scholar]
- 37.Bleasdale JE, Thakur NR, Gremban RS, Bundy GL, Fitzpatrick FA, Smith RJ, et al. Selective inhibition of receptor-coupled phospholipase C-dependent processes in human platelets and polymorphonuclear neutrophils. J Pharmacol Exp Ther. 1990;255(2):756–68. [PubMed] [Google Scholar]
- 38.Duncia JV, Santella JB, 3rd, Higley CA, Pitts WJ, Wityak J, Frietze WE, et al. MEK inhibitors: the chemistry and biological activity of U0126, its analogs, and cyclization products. Bioorg Med Chem Lett. 1998;8(20):2839–44. doi: 10.1016/s0960-894x(98)00522-8. [DOI] [PubMed] [Google Scholar]
- 39.Remacle AG, Golubkov VS, Shiryaev SA, Dahl R, Stebbins JL, Chernov AV, et al. Novel MT1-MMP small-molecule inhibitors based on insights into hemopexin domain function in tumor growth. Cancer Res. 2012;72(9):2339–49. doi: 10.1158/0008-5472.CAN-11-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devy L, Huang L, Naa L, Yanamandra N, Pieters H, Frans N, et al. Selective inhibition of matrix metalloproteinase-14 blocks tumor growth, invasion, and angiogenesis. Cancer Res. 2009;69(4):1517–26. doi: 10.1158/0008-5472.CAN-08-3255. [DOI] [PubMed] [Google Scholar]
- 41.Matsuo K, Owens JM, Tonko M, Elliott C, Chambers TJ, Wagner EF. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat Genet. 2000;24(2):184–7. doi: 10.1038/72855. [DOI] [PubMed] [Google Scholar]
- 42.Modderman WE, Tuinenburg-Bol Raap AC, Nijweide PJ. Tartrate-resistant acid phosphatase is not an exclusive marker for mouse osteoclasts in cell culture. Bone. 1991;12(2):81–7. doi: 10.1016/8756-3282(91)90004-3. [DOI] [PubMed] [Google Scholar]
- 43.Janckila AJ, Parthasarathy RN, Parthasarathy LK, Seelan RS, Hsueh YC, Rissanen J, et al. Properties and expression of human tartrate-resistant acid phosphatase isoform 5a by monocyte-derived cells. J Leukoc Biol. 2005;77(2):209–18. doi: 10.1189/jlb.0504287. [DOI] [PubMed] [Google Scholar]
- 44.Garnero P, Ferreras M, Karsdal MA, Nicamhlaoibh R, Risteli J, Borel O, et al. The type I collagen fragments ICTP and CTX reveal distinct enzymatic pathways of bone collagen degradation. J Bone Miner Res. 2003;18(5):859–67. doi: 10.1359/jbmr.2003.18.5.859. [DOI] [PubMed] [Google Scholar]
- 45.Song F, Wisithphrom K, Zhou J, Windsor LJ. Matrix metalloproteinase dependent and independent collagen degradation. Front Biosci. 2006;11:3100–20. doi: 10.2741/2036. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, et al. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345(6274):442–4. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 47.Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99(1):111–20. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- 48.Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM, et al. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J Clin Invest. 2008;118(11):3775–89. doi: 10.1172/JCI35711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walsh NC, Gravallese EM. Bone remodeling in rheumatic disease: a question of balance. Immunol Rev. 2010;233(1):301–12. doi: 10.1111/j.0105-2896.2009.00857.x. [DOI] [PubMed] [Google Scholar]
- 50.Kadono Y, Tanaka S, Nishino J, Nishimura K, Nakamura I, Miyazaki T, et al. Rheumatoid arthritis associated with osteopetrosis. Mod Rheumatol. 2009;19(6):687–90. doi: 10.1007/s10165-009-0208-7. [DOI] [PubMed] [Google Scholar]
- 51.Wang CJ, Chen IP, Koczon-Jaremko B, Boskey AL, Ueki Y, Kuhn L, et al. Pro416Arg cherubism mutation in Sh3bp2 knock-in mice affects osteoblasts and alters bone mineral and matrix properties. Bone. 2010;46(5):1306–15. doi: 10.1016/j.bone.2010.01.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez-Pousa A, Martin Broto J, Garrido T, Vazquez J. Giant cell tumour of bone: new treatments in development. Clin Transl Oncol. 2015;17(6):419–30. doi: 10.1007/s12094-014-1268-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sobacchi C, Schulz A, Coxon FP, Villa A, Helfrich MH. Osteopetrosis: genetics, treatment and new insights into osteoclast function. Nat Rev Endocrinol. 2013;9(9):522–36. doi: 10.1038/nrendo.2013.137. [DOI] [PubMed] [Google Scholar]
- 54.Ray N, Kuwahara M, Takada Y, Maruyama K, Kawaguchi T, Tsubone H, et al. c-Fos suppresses systemic inflammatory response to endotoxin. Int Immunol. 2006;18(5):671–7. doi: 10.1093/intimm/dxl004. [DOI] [PubMed] [Google Scholar]
- 55.Maruyama K, Sano G, Ray N, Takada Y, Matsuo K. c-Fos-deficient mice are susceptible to Salmonella enterica serovar Typhimurium infection. Infect Immun. 2007;75(3):1520–3. doi: 10.1128/IAI.01316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida R, Suzuki M, Sakaguchi R, Hasegawa E, Kimura A, Shichita T, et al. Forced expression of stabilized c-Fos in dendritic cells reduces cytokine production and immune responses in vivo. Biochem Biophys Res Commun. 2012;423(2):247–52. doi: 10.1016/j.bbrc.2012.05.097. [DOI] [PubMed] [Google Scholar]
- 57.Sabeh F, Fox D, Weiss SJ. Membrane-type I matrix metalloproteinase-dependent regulation of rheumatoid arthritis synoviocyte function. J Immunol. 2010;184(11):6396–406. doi: 10.4049/jimmunol.0904068. [DOI] [PubMed] [Google Scholar]
- 58.Miller MC, Manning HB, Jain A, Troeberg L, Dudhia J, Essex D, et al. Membrane type 1 matrix metalloproteinase is a crucial promoter of synovial invasion in human rheumatoid arthritis. Arthritis Rheum. 2009;60(3):686–97. doi: 10.1002/art.24331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaneko K, Williams RO, Dransfield DT, Nixon AE, Sandison A, Itoh Y. Selective Inhibition of Membrane Type 1 Matrix Metalloproteinase Abrogates Progression of Experimental Inflammatory Arthritis: Synergy With Tumor Necrosis Factor Blockade. Arthritis Rheumatol. 2016;68(2):521–31. doi: 10.1002/art.39414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang P, Nie J, Pei D. The hemopexin domain of membrane-type matrix metalloproteinase-1 (MT1-MMP) Is not required for its activation of proMMP2 on cell surface but is essential for MT1-MMP-mediated invasion in three-dimensional type I collagen. J Biol Chem. 2004;279(49):51148–55. doi: 10.1074/jbc.M409074200. [DOI] [PubMed] [Google Scholar]
- 61.Itoh Y, Ito N, Nagase H, Evans RD, Bird SA, Seiki M. Cell surface collagenolysis requires homodimerization of the membrane-bound collagenase MT1-MMP. Mol Biol Cell. 2006;17(12):5390–9. doi: 10.1091/mbc.E06-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takeuchi T, Tanaka Y, Ishiguro N, Yamanaka H, Yoneda T, Ohira T, et al. Effect of denosumab on Japanese patients with rheumatoid arthritis: a dose-response study of AMG 162 (Denosumab) in patients with RheumatoId arthritis on methotrexate to Validate inhibitory effect on bone Erosion (DRIVE)-a 12-month, multicentre, randomised, double-blind, placebo-controlled, phase II clinical trial. Ann Rheum Dis. 2016;75(6):983–90. doi: 10.1136/annrheumdis-2015-208052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rubbert-Roth A, Finckh A. Treatment options in patients with rheumatoid arthritis failing initial TNF inhibitor therapy: a critical review. Arthritis Res Ther. 2009;11(Suppl 1):S1. doi: 10.1186/ar2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hero M, Suomalainen A, Hagstrom J, Stoor P, Kontio R, Alapulli H, et al. Anti-tumor necrosis factor treatment in cherubism - Clinical, radiological and histological findings in two children. Bone. 2013;52(1):347–53. doi: 10.1016/j.bone.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Silvestris F, Ciavarella S, De Matteo M, Tucci M, Dammacco F. Bone-resorbing cells in multiple myeloma: osteoclasts, myeloma cell polykaryons, or both? Oncologist. 2009;14(3):264–75. doi: 10.1634/theoncologist.2008-0087. [DOI] [PubMed] [Google Scholar]
- 66.Dellinger MT, Garg N, Olsen BR. Viewpoints on vessels and vanishing bones in Gorham-Stout disease. Bone. 2014;63:47–52. doi: 10.1016/j.bone.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 67.Sousa S, Maatta J. The role of tumour-associated macrophages in bone metastasis. J Bone Oncol. 2016;5(3):135–138. doi: 10.1016/j.jbo.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.