Abstract
Magnetic resonance (MR) methods enable noninvasive, regional tumor therapy response assessment, but associations between MR parameters, underlying biology, and therapeutic effects must be investigated. The aim of this study was to investigate response assessment efficacy and biological associations of MR parameters in a neuroendocrine tumor (NET) model subjected to radionuclide treatment. Twenty-one mice with NETs received 177Lu-octreotate at day 0. MR experiments (day −1, 1, 3, 8, and 13) included T2-weighted, dynamic contrast-enhanced (DCE) and diffusion-weighted imaging (DWI) and relaxation measurements (T1/T2*). Tumor tissue was analyzed using proteomics. MR-derived parameters were evaluated for each examination day and for different radial distances from the tumor center. Response assessment efficacy and biological associations were evaluated using feature selection and protein expression correlations, respectively. Reduced tumor growth rate or shrinkage was observed until day 8, followed by reestablished growth in most tumors. The most important MR parameter for response prediction was DCE-MRI–derived pretreatment signal enhancement ratio (SER) at 40% to 60% radial distance, where it correlated significantly also with centrally sampled protein CCD89 (association: DNA damage and repair, proliferation, cell cycle arrest). The second most important was changed diffusion (D) between day −1 and day 3, at 60% to 80% radial distance, where it correlated significantly also with peripherally sampled protein CATA (association: oxidative stress, proliferation, cell cycle arrest, apoptotic cell death). Important information regarding tumor biology in response to radionuclide therapy is reflected in several MR parameters, SER and D in particular. The spatial and temporal information provided by MR methods increases the sensitivity for tumor therapy response.
Introduction & Aim
Ionizing radiation is known to induce DNA damage, but it can also affect intra- and intercellular signaling pathways [1]. It is therefore likely that radiotherapy, where tumor cell death is the desired end point, influences other mechanisms that affect the potential of cure, such as angiogenesis, protein integrity, invasiveness, and metastatic potential [1], [2]. Conventional radiotherapy response assessment relies on the reduction of tumor size. However, increased understanding of response mechanisms, individual treatment optimization, and early response assessment require methods that are sensitive to cellular and microenvironmental changes that precede gross morphological changes [3], [4].
Several noninvasive methods are available for tumor response evaluation after radiation therapy, such as positron emission tomography (PET), single photon emission computed tomography, computed tomography (CT), and ultrasound [5]. The spatial heterogeneity of tumor tissue combined with the fact that many modern therapeutics target specific features in the tumor microenvironment makes high spatial resolution a favorable feature of assessment methods, which limits the use of PET, single photon emission computed tomography, and ultrasound. CT offers excellent spatial resolution, but the physiological information and soft tissue contrast currently offered by CT are limited, and the subject is exposed to radiation that may confound studies on radiation therapy.
Magnetic resonance (MR) methods are noninvasive and provide excellent spatial resolution and soft tissue contrast. It is also possible to extract spatially resolved functional parameters with the potential as biomarkers for radiotherapy response [6], [7].
Diffusion weighted imaging (DWI) can be used to estimate the apparent diffusion coefficient (ADC) of tissue water molecules, which has been correlated with cellular density [10], [9]. The intravoxel incoherent motion (IVIM) model is related to the ADC model, but in addition to estimating the water diffusion, it can also estimate the voxel volume fraction of actively perfused capillaries [10]. IVIM-DWI is a promising alternative to perfusion assessments without the use of exogenous contrast agents, and it has been associated with both blood perfusion and interstitial fluid pressure [11], [15]. Dynamic contrast-enhanced (DCE) MRI can be used to probe tissue and vascular architecture, such as vascular permeability, vascular surface area, and extracellular extravascular space. DCE parameters also reflect blood flow and volume, and tracer distribution reveals accessibility of tumor regions to oxygen, nutrients, and therapeutic agents [13], [14]. Furthermore, magnetic relaxation times (T1, T2, and T2*) are sensitive to tissue molecular organization, macromolecules, and paramagnetic deoxyhemoglobin caused by, e.g., restructuring of microvasculature and hemorrhage. T1 has been used to assess response to antiangiogenic therapy in glioblastoma patients, and pretreatment T2* was associated with overall survival in patients with colorectal liver metastases [18], [16].
Although promising, diverging or contradictory results have been observed in studies where MR methods were used for tumor tissue characterization, such as both positive and negative correlations between ADC and cell density in different tumors of the same tumor type [17]. The biological complexity of tumor tissue, with seemingly chaotic biological and physiological structures, will inevitably influence MR parameters and hence the reproducibility of MR methods. A multivariable regression approach that combines information from multiparametric MRI experiments may yield models that account for confounding variables and thereby increase reproducibility. However, the risk for overfitting will increase if inefficacious variables with little or redundant information regarding treatment response are included, and the biological interpretability will be low [18]. Proper choice of MR techniques and MR-derived tumor parameters for efficacious response assessment is thus important, especially for evaluation of novel treatments. To the best of our knowledge, such systematic investigations are scarce.
Neuroendocrine tumors (NETs) are the most frequent malignancies of the small intestine, with increasing incidence during the last decades [19]. Patients with NETs are often diagnosed at a late stage, when curative surgery cannot be performed. However, NETs overexpress somatostatin receptors, which makes radionuclide therapy with 177Lu-labeled somatostatin analogues, such as 177Lu-[DOTA0, Tyr3]-octreotate (177Lu-octreotate), a therapeutic option. 177Lu-octreotate has been successfully used in patients with inoperable disease and is associated with increased overall survival and improved quality of life [20]. However, cure after 177Lu-octreotate therapy is rare, and there are few biomarkers that accurately predict disease progression and therapeutic response in individual patients. The nature of NETs, including slow growth and high vascularization, makes methods that are sensitive to functional tissue parameters interesting. Since several of the available MR methods are sensitive to perfusion related effects, they may provide useful biomarkers for therapy response assessment of NETs.
The aim of this study was to identify and evaluate potential MR-derived biomarkers for tumor response to 177Lu-octreotate therapy in an animal model of small intestine NET.
Materials and Methods
Animals, Tumor Model, and Radiopharmaceutical
Four-week-old female BALB/c nude mice (n = 21; Charles River, Japan and Germany) were subcutaneously transplanted into the neck region with samples from the human small intestine NET cell line GOT1 [21] under anesthesia (Ketaminol vet., Intervet AB, Sweden, 50 mg/ml and Domitor vet., Orion Pharma Animal Health, Sweden, 1 mg/ml; antidote: Antisedan, Orion Pharma Animal Health, Sweden, 5 mg/ml). The animals received standard diet and water ad libitum, and at the start of the experiment, the tumor diameters were between 10 and 20 mm.
The Gothenburg Ethical Committee on Animal Research approved this study.
177Lu-octreotate with a specific activity of 26 MBq/μg octreotate was prepared according to manufacturer's instruction (IDB Holland, the Netherlands). The fraction of peptide-bound 177Lu was >98%, as determined by instant thin layer chromatography (ITLC SG, PALL Corporation, USA), with 0.1 M sodium citrate (pH 5; VWR International AB, Sweden) as mobile phase. 177Lu activity in syringes was measured before and after injection using a well-type ionization chamber (CRC-15R; Capintec, IA, Florham Park, NJ).
MRI Examinations
All MR experiments were performed on a 7-T MR system with 400-mT/m gradients (Bruker BioSpin MRI GmbH, Germany; software: ParaVision 5.1), a four-channel array rat brain receiver coil, and a 72-mm volume transmit coil or a 50-mm quadrature transmit/receive volume coil (RAPID Biomedical GmbH, Germany). To establish similar tumor position on repeated experiments, a plastic animal bed supported the animal in supine position, and the tumor was immobilized by fitting it into a hole cut out from the bed. Field homogeneity within the tumor was improved by applying gel (Lectro Derm, Handelsvaruhuset Viroderm, Sweden) around the tumor in the animal bed cutout, resulting in a tissue-gel-air interface. A plastic film confined the gel, and the receiver coil was mounted immediately under the plastic film, with coil elements within a few millimeters from the tumor. For this setup, the 72-mm transmit volume coil was used, and no respiratory triggering was necessary. Three animals were imaged using the 50-mm transmit/receive coil, in prone position, without gel but with respiratory triggering.
Field homogeneity was optimized using a field map–based shimming technique (Bruker MAPSHIM macro) until the full-width-at-half-maximum of the water spectral peak of a voxel within the tumor could not be further reduced or until it reached ~40 Hz (0.13 ppm).
MRI examinations were performed on day −1, 1, 3, 8, and 13. On day 0, 15 MBq 177Lu-octreotate was administered intravenously in a tail vein, resulting in an extrapolated absorbed tumor dose of up to 4.0 Gy calculated according to the MIRD formalism [22]. The amount of 177Lu-octreotate was chosen to give only partial remission to enable further analyses.
The animals underwent IVIM-DWI, T1 and T2* mapping, DCE-MRI, and T2 weighted MRI. The MRI pulse sequence parameters used are stated in Table 1. Total experiment time was ca. 1.5 h.
Table 1.
Pulse Sequence Parameters for the MR Measurements
| MR Technique & Pulse Sequence | Pulse Sequence Parameters |
|---|---|
|
DWI 2D SE-EPI |
Three orthogonal gradient directions, gradient separation/duration: 9/4 ms, 9 b values: 0, 5, 10, 20, 35, 50, 75, 100, 200, 400, 600, and 800 s/mm2, TR: 1500 ms, TE: 21 ms, number of averages/segments: 3/1 Effective bandwidth ≈ 300 kHz Partial Fourier acceleration: 1.5 Pixel size: 3202 μm2, slice thickness: 1000 μm, Slice gap: 500 μm Fat suppression: frequency selective Scan time < 6 minutes |
|
T2*-mapping Multiple echo gradient echo (MGE) |
10 echoes (TE): 5, 10, 15, … 50 ms TR: 2000 ms, number of averages: 1, flip angle = 30° Slice positions imported from DWI experiment Pixel size: 1602 μm2, slice thickness: 1000 μm, Slice gap: 500 μm No fat suppression Scan time < 4 minutes |
|
T1-mapping 2D RARE (RAREVTR) |
Seven TR: 13,000, 9000, 4500, 2500, 1500, 750, 300 ms TE: 24 ms, number of averages: 1, RARE factor: 4 Refocusing flip angle: 180°, Pixel size = 2802 μm2, slice thickness: 1000 μm, single slice Slice positions imported from DWI experiment (central slice) No fat suppression Scan time < 10 minutes |
|
DCE-MRI 2D RARE |
Number of repetitions (dynamics): 100, temporal resolution: 4.2 s (varied slightly with FOV), contrast injection during sixth dynamic TR: 300 ms, TE: 24 ms, number of averages: 1, RARE factor: 4 Partial Fourier acceleration: 1.5 Pixel size = 2802 μm2, slice thickness: 1000 μm, single slice Slice positions imported from DWI experiment (central slice) Fat suppression: frequency selective Scan time < 8 minutes |
|
T2-weighted MRI 2D RARE |
TR: 4190 ms, TE: 45 ms, number of averages: 2, RARE factor: 6 Pixel size: (160 ± 50)2 μm2 (varied with FOV) Slice thickness: 700 μm, no slice gap Fat suppression: frequency selective Scan time < 4 min |
SE-EPI, spin-echo echo planar imaging; RARE, rapid acquisition with relaxation enhancement; RAREVTR, RARE with variable repetition time; TR, repetition time; TE, echo time; FOV, field of view.
During imaging, anesthesia was maintained using air and isoflurane (2 %-3 %, Isoba vet., Schering-Plow Animal-health, Denmark), body temperature was maintained using a heating pad and circulating warm water, and respiration was monitored with a pressure sensitive pad (SA Instruments, Inc., NY). During the DCE experiment Gd-DTPA contrast agent (0.1 M, 0.3 mmol/kg bodyweight, Dotarem, Gothia Medical, Sweden) was administered in a tail vein via an infusion line reaching outside the magnet bore.
After the final MRI session, animals were given a lethal i.p. injection of sodium pentobarbitone (Pentobarbitalnatrium vet., Apotek Produktion & Laboratorier AB, Sweden, 60 mg/ml) followed by a heart incision. The tumors were removed under sterile conditions, and central and peripheral tumor samples were collected and snap frozen in liquid nitrogen for quantitative proteomics.
MRI Analysis Software
Model fitting and analyses were performed in MATLAB (R2015b, The MathWorks, Inc., MA) using built-in functions, in-house developed scripts, functions, and graphical user interfaces, unless otherwise stated. A 2×2 median filter was applied to all image data before parameter calculations. Table 2 gives a description of all derived MR parameters and their definition.
Table 2.
Definition of the MR Parameters
| Parameter | Description | |
|---|---|---|
| DWI | ||
|
IVIM S(b) = S0(1 − f ) ∙ e−bD + S0f ∙ e−b(D + D∗) S0: signal without diffusion weighting Model parameters were determined using a voxelwise Bayesian method with uniform prior distributions and mode as central tendency measure [52] Parameter limits were D: [0,5] μm2/ms, f: [0,1], D*: [0,1000] μm2/ms, S0: [0, 2xSmax], where Smax is the maximum measured signal |
||
| D | Diffusion coefficient | The tissue water diffusion coefficient |
| D* | Pseudodiffusion coefficient | Perfusion-related pseudodiffusion coefficient of incoherently flowing blood in the tissue |
| f | Perfusion fraction | Signal fraction from incoherently flowing tissue water (blood) |
|
Apparent diffusion S(b) = S0e−b ∙ ADC S0: signal without diffusion weighting The model was fitted using least squares with two b values (0 and 800 s/mm2.) |
||
| ADC | Apparent diffusion coefficient | The apparent diffusion coefficient affected by both diffusion and perfusion |
| Relaxation time mapping | ||
|
T1 relaxation S(TR) = signal intensity for repetition time TR A = signal bias C = signal intensity after complete T1 relaxation with no bias The parameters were estimated by a least squares fit |
||
| T1 | T1 time | Longitudinal relaxation time of tissue |
|
T2* relaxation S(TE) is the signal intensity for echo time TE A = signal bias C = signal intensity without T2* relaxation and no bias T2* was determined by least squares fit. The goodness-of-fit parameter R2 was used to exclude voxels with R2< 0.4 |
||
| T2* | T2* time | Transversal relaxation time of tissue including effects from magnetic field inhomogeneity |
| DCE-MRI | ||
|
Semiquantitative characteristics S(t), a continuous representation of signal intensity as a function of time, was determined by fitting a smoothing spline (smoothing parameter = 0.01) S0, the baseline signal intensity, was defined as the mean of the precontrast dynamics σvoxel, the voxel noise, was defined as the standard deviation of the residuals of S(t) for each voxel S(t) > S0 + 5 ∙ σvoxel defined significant contrast enhancement σimage, the image noise, was defined as the mean of σvoxel S0 > 5 ∙ σimage defined the threshold for inclusion of voxels in the analysis Smax was defined as the maximum value of S(t) for each voxel |
||
| AT | Arrival time | Time after injection required for signal intensity to reach significant enhancement |
| TOP | Time of peak intensity | Time after injection required to reach maximum signal intensity |
| TTP | Time to peak | Time between AT and TOP |
| SEmax | Relative, maximum signal enhancement | Maximum signal intensity relative to baseline signal intensity: (Smax– S0)/S0 |
| SE60 | Relative signal enhancement at 60 s | Signal intensity at 60 s after injection relative to signal baseline intensity: [S(60 s) – S0]/S0 |
| SER | Signal enhancement ratio | Ratio of early (55 s) and late (300 s) relative signal enhancements |
| CER | Contrast enhancement ratio | Ratio of maximum signal intensity and baseline signal intensity: Smax/S0 Missing value if no significant enhancement was obtained |
| AUCn | Normalized area under the curve | Area under S(t) between 0 and 5 minutes, normalized to S0 |
| WI | Wash in | Maximum time derivative of S(t) between AT and TOP |
| WO | Wash out | Maximum negative time derivative of S(t) between TOP and the last dynamic. Missing value if <5 dynamics left after TOP |
| BE | Brevity of enhancement | Time between the time points of WI and WO |
| IS | Initial slope | Average rate of signal enhancement between TOP and contrast injection: (Smax − S0)/TOP |
| NS | Negative slope | Average rate of signal decrease between TOP and the last dynamic. Missing value if <5 dynamics left after TOP |
Tumor Response and Data Collection
The response variable was defined as the tumor volume change after treatment, as determined by the linear regression coefficient of the relative tumor volumes on day −1, 1, 3, and 8.
Tumor volumes were calculated from T2-weighted images by manually delineating tumor borders on all slices and multiplying the total number of tumor voxels by the voxel volume [23]. Partial volume effects were minimized by tracing the region of interest (ROI) border on the tumor side where the intensity gradient from background hypointensity to tumor hyperintensity was no longer visible.
Day 1 measurements were included after an interim data evaluation suggesting early effects. Hence, data were collected from 6 animals on day −1, 3, 8, and 13 and from 11 animals on day −1, 1, 3, 8, and 13. One animal died before the T2-weighted anatomical imaging on day 1 (volume was calculated from the IVIM-DWIs), and three animals had reduced study time points due to technical reasons, which also necessitated the switch to a volume coil. For the latter four animals, data were only collected on days −1 and 1.
Definition of Tumor Regions for Local Response Assessment
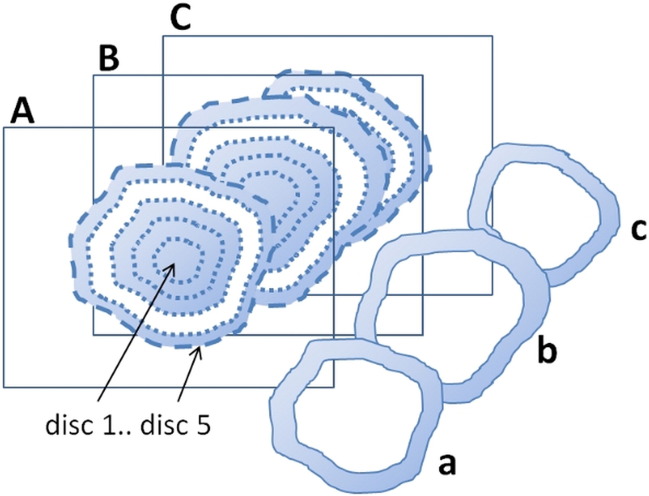
An ROI was manually delineated on the tumor borders on the precontrast DCE image of each animal and study time point. The corresponding slice on the IVIM-DWI (b = 800 s/mm2) and MGE (TE = 5 ms) series was located, and the tumor was delineated also on them. Each ROI was automatically replicated to form five concentric ROIs with stepwise decreased radius. The five ROIs thus defined one central region and four annular disc shaped regions, and parameter data were evaluated within single discs as well as the entire tumor region (all discs together). For improved statistics, discs were created also on the two most adjacent slices on the IVIM-DWI and MGE, and data from these discs were merged to data from the corresponding central slice disc. Figure 1 illustrates the disc definition.
Figure 1.
Illustration of the extraction of data from an annular disc-shaped region of the parametric tumor maps (disc 4 in this example). Tumor borders on the central (B) and adjacent (A and C) image slices were manually delineated (dashed outer lines). The ROIs were automatically replicated with stepwise decreased size (dotted lines, 20% of original distance between centroid and periphery removed in each step). Data from the region constituting an annular disc between two adjacent ROIs (b) were extracted and merged to the data from the corresponding discs in the adjacent slices (a and c). DCE-MRI was only acquired from one slice (B), and DCE parameters were thus only acquired from (b).
Partial volume voxels were avoided in the delineation process as described for the tumor volume calculation and by only evaluating data from central tumor slices.
Definition of MR Features
Henceforth, an MR feature is either 1) the disc median or the tumor median (all discs) prior to therapy (day −1) or 2) the change in the corresponding median from day A to B.
A change from day A to B (Δday A:B) was defined in two ways, both as the absolute change: value(B) − value(A), and as the relative change: [value(B) − value(A)]/value(A). Changes were calculated for the following combinations of days: A:B = −1:1, −1:3, −1:8, −1:13, 1:3, 3:8, and 8:13.
E.g., ADCtumor−1 is the median tumor ADC on day −1, whereas ADCdisc 2−1 : 3rel is the relative change of the median ADC in disc 2 from day −1 to day 3. Tumor volume before treatment is denoted Voltumor−1.
MR Data Analysis and Statistics
Prior to feature calculations, MR parameters were transformed to approximately normal distributions using the Box-Cox power transformation and standardized to have zero mean value and a unit standard deviation. The linear correlation between individual features and the response variable was calculated, and hierarchical clustering was performed in R (http://www.r-project.org, version 3.1.1).
In the subsequent analysis, features with more than 50% missing values were discarded. For other features, stochastic data imputation using random values from a normal distribution was used to replace remaining missing values. A sparse multiparametric model was fitted using the lasso method [24] with the regularization parameter chosen based on test mean squared error (mse) from N-fold cross-validation. To better handle the high number of features, the lasso was preconditioned using the supervised principal components method [25]. The threshold for the linear regression coefficient (β) and the number of principal components (up to five) used by the supervised principal components were chosen based on mse from N-fold cross-validation.
The imputation process and subsequent analysis were repeated 5000 times, with the β coefficients returned from the lasso cumulatively added for returning features, thus yielding feature specific β sums.
Quantitative Proteomics and Correlation with MR Features
Peripheral and central tumor tissue samples were collected from 3 tumors on day 1 and 12 tumors on day 13. Samples were prepared and analyzed as described elsewhere (Sci Rep, submitted). Briefly, tissue samples were homogenized in 50 mM triethylammonium bicarbonate (Fluka, Sigma Aldrich) and 2% sodium dodecyl sulfate, and 30 μg total protein per sample was taken for trypsin digestion (Pierce Trypsin Protease, MS Grade, Thermo Fisher Scientific) using the filter-aided sample preparation modified from Ref. [26]. Peptides were labeled with TMT 10plex (Thermo Scientific) for relative quantification, pooled, and fractionated using high-pH reversed-phase chromatography (Waters XBridge BEH C18 3.0 × 150 mm, 3.5 μm).
Proteomics data were acquired using an Easy nanoLC1000 coupled to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific). During a 60-minute acetonitrile gradient in 0.2% formic acid, positive ions with an m/z range of 380 to 1200 and a resolution of 120,000 were scanned. Mass spectrometry (MS) scans were selected for fragmentation (MS2) by collision-induced dissociation for identification in the ion trap, while MS3 fragments produced by high-energy collision dissociation were detected in the Orbitrap. MS ions were isolated in the quadrupole with a 1.6-m/z window and a dynamic exclusion of 30 seconds for already identified m/z values.
Protein identification and quantification were performed using Proteome Discoverer version 1.4 (Thermo Fisher Scientific, Waltham, MA) with the integrated Mascot search engine (Matrix Science, Boston, MA) using the Homo sapiens SwissProt database version March 2015 (Swiss Institute of Bioinformatics, Switzerland). The following settings were used in Mascot: Trypsin as cleaving enzyme (allowing zero miscleavages), 5 ppm MS peptide tolerance, fragment mass tolerance of 500 millimass units, methionine oxidation as dynamic modification, and TMT labeling (lysines or peptide N-termini) as well as cysteine alkylation as fixed modifications. Resulting proteins passing a false discovery rate of 1% and containing at least one unique peptide were further analyzed and associated with biological functions using the Gene Ontology database (http://www.geneontology.org) [27].
Quantification was performed by ratio calculation of the reporter ion intensities. The protein quantity that was analyzed was the protein amount extracted from an individual tissue sample normalized to the mean amount of the same protein in all tissue samples. The resulting distribution of the normalized protein quantity was transformed to be approximately normally distributed using the MATLAB boxcox function before the correlation with response was calculated. Only proteins with correlations yielding P values < .01 were analyzed further. Pairwise correlation analysis was performed between MR features with high β sums and proteins significantly correlated with response.
Reported correlations refer to Pearson correlation coefficients with corresponding P values derived from Student's t distribution under the assumption of no correlation at all. Where statistical significance is stated, multiple comparisons were corrected for using the Benjamini and Yekutieli method (q = 0.05).
Results
Tumor Volume Response
The noncurative amount of 177Lu-octreotate resulted in a transient volume reduction in approximately 50% of the tumors, whereas the other tumors showed reduced growth rate. After 8 to 13 days, the growth rate of all tumors was comparable to untreated tumors (doubling time approximately 2 weeks, not shown) (Figure 2).
Figure 2.
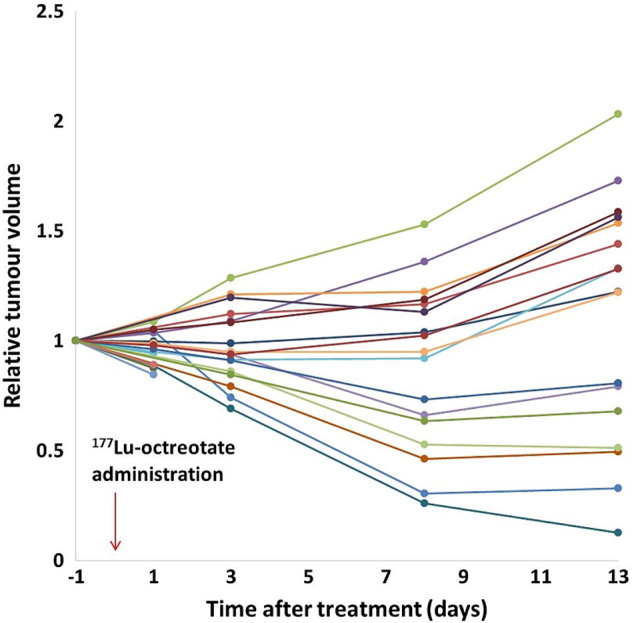
MR-derived, individual, relative tumor volumes over time after 177Lu-octreotate therapy (n = 21). In ~50% of the tumors, the volume is transiently reduced after treatment, whereas growth rate is reduced in the other half. After day 8, increasing volume is observed in most tumors again.
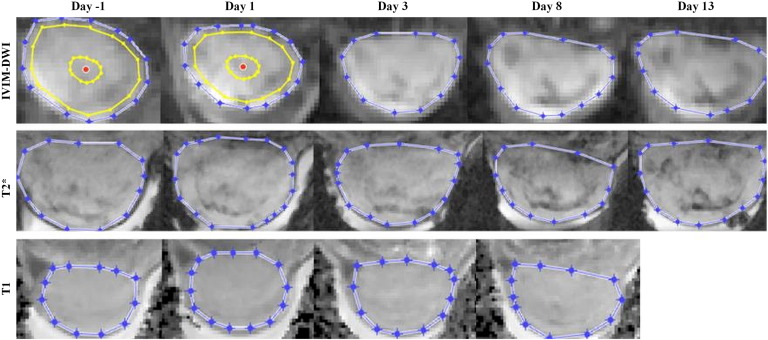
Tumor tissue was easily discernible from background on images from all involved MR methods and time points, facilitating delineation and data extraction (Figure 3).
Figure 3.
Representative images of a central tumor section from the IVIM-DWI (b = 600 s/mm2), T2*-mapping (TE = 5 ms), and T1-mapping (T1 value) experiments. Manual tumor delineations are shown in blue. Two examples of the automatically calculated tumor centroid (red dot) are shown with the first and fourth automatically reproduced delineations (yellow) that were used to define tumor discs.
Correlations Between Single Features and Tumor Response
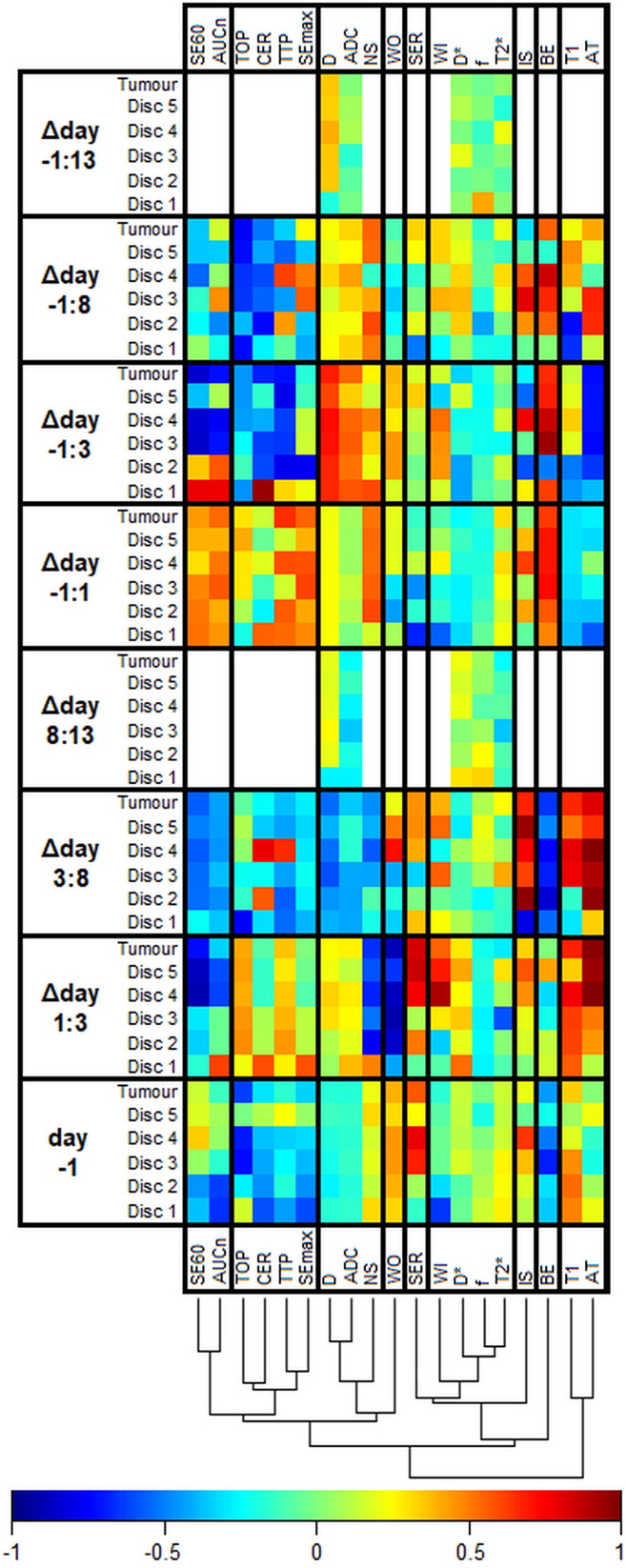
The heat map in Figure 4 shows the correlations between single features and response from simple regression analysis. The color representation of regression coefficients makes it possible to discern patterns, such as similarities between features and variations within tumors. One example is the gradually increasing correlation between arrival time and response from central (disc 1) to peripheral (disc 5) parts of the tumor in the Δday 1:3 combination. Effects on the correlations attributable to the definition of change (absolute or relative) were in general small (<0.1, data not shown). For some parameters, however, there was a change of arithmetical sign of the correlation and/or a substantial variation in magnitude, resulting in highly different correlations (Figure S1).
Figure 4.
The heat map shows the Pearson correlation coefficient between MR features and response in NETs after 177Lu-octreotate therapy (white = not measured). MR parameters are defined in Table 2, and Δday a:b indicates the relative change of an MR parameter from day a to b ([value day a – value day b]/value day a). The dendrogram shows the results of a cluster analysis of the correlations with the columns as input vectors. The cluster analysis was conducted to facilitate perception of patterns and similarities between MR parameters and how they were affected by therapy. The vertical help lines in the heat map were added to separate highly dissimilar clusters.
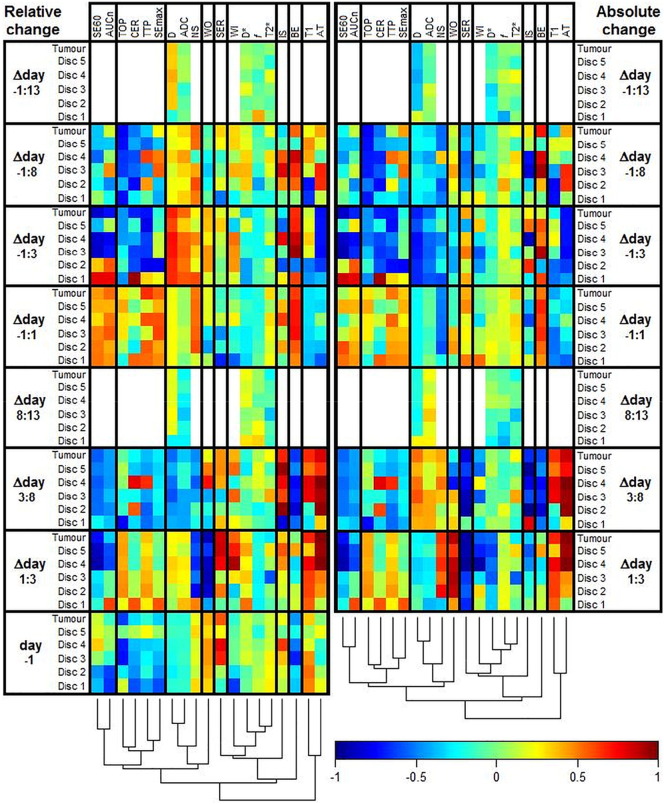
Figure S1.
The heat map shows the Pearson correlation coefficient between MR features and response in NETs after 177Lu-octreotate therapy (white = not measured). The dendrogram shows the results of a cluster analysis of the correlations with the columns as input vectors. The cluster analysis was conducted to facilitate perception of patterns and similarities between MR parameters and how they were affected by therapy. The vertical help lines in the heat map were added to separate highly dissimilar clusters. MR parameters are defined in Table 2, and Δday A:B indicates the change of an MR parameter from day A to day B. The left part of the figure is identical to Figure 4 and thus constitutes features defined as the relative change ([value(dayB) − value(dayA)]/value(dayA)). The right part shows the same evaluation but constitutes features defined as the absolute change (value(dayB) − value(dayA)). Note the additional bottom rows that show correlations between the pretreatment feature values and response.
The correlation coefficient between pretreatment tumor volume (Voltumor−1) and response was −0.60, suggesting that treatment effect improved with larger pretreatment tumor volume.
Redundancy, Predictive Power, and Relative Importance of MR-Derived Tumor Characteristics
The dendrogram in Figure 4 shows that there were similarities between some of the MR parameters regarding how they were affected by treatment, which indicate that they reflect related biological information. As expected, ADC and D were first grouped by the clustering algorithm. The IVIM-DWI perfusion fraction (f) and T2* were also grouped early, whereas, e.g., T1 and brevity of enhancement (BE) appear to be differently associated with response.
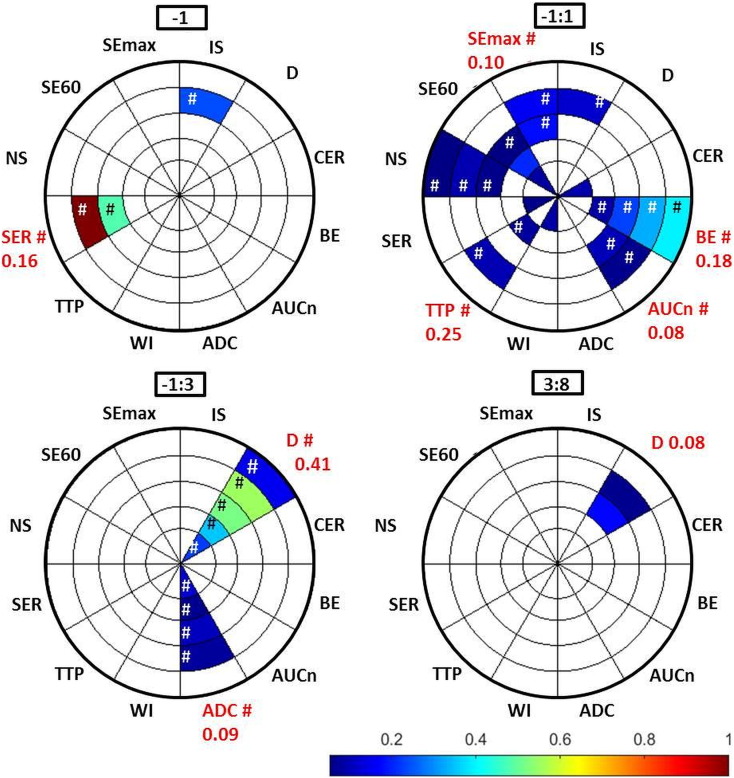
The feature selection method regarded SERdisc 4−1 the most efficacious MR feature (relative β sum = 1), followed by SERdisc 3−1. The second most important MR parameter was related to diffusion, where Ddisc 4−1 : 3rel reached a relative β sum of approximately 0.5. Figure 5 graphically displays MR features reaching relative β sums > 0.05. The displayed features were derived from 12 different MR parameters, of which 5 were included only after separate disc evaluation, whereas tumor median sufficed for the remaining 7 MR parameters. These results indicate that the sensitivity of several MR methods could be improved by regional evaluation of therapy response.
Figure 5.
The relative efficacy of MR features in assessment of NET response to 177Lu-octreotate therapy, as indicated by the relative β sums (color bar), from feature selection. Disc number and MR parameter are indicated by the sections of the pie chart. Note that several of the evaluated combinations of days are left out since they did not contain features that reached β sums high enough for display (limit set to exclude features with β sum < 0.05 for clarity of display), and that most features are concentrated close to treatment (−1:3 and −1:1). Hashtag (#) indicates that the feature was based on relative change (cf.Figure 4 and S1 for arithmetical sign of correlation).
The combination of day −1 and day 1 (Δday −1:1) rendered most MR features with β sums > 0.05 (24 MR features from 10 MR parameters), indicating that a considerable amount of biological information related to response may be available already very early after treatment (Figure 5). Note that evaluation of Δdays −1:8, −1:13, 1:3, 3:8, and 8:13 did not yield β sums > 0.05. These combinations of days are therefore not shown in Figure 5.
Affected Protein Expression Levels
In total, expression ratios for 5381 different proteins were determined in the liquid chromatography–mass spectrometry analysis. Of these, 104 proteins yielded correlations with the response variable with P values < .01, whereof 68, 28, and 8 were found in central only, peripheral only, and both central and peripheral tumor samples, respectively. Of these proteins, 66 can be found in different categories of biological processes that are related to response to radiation.
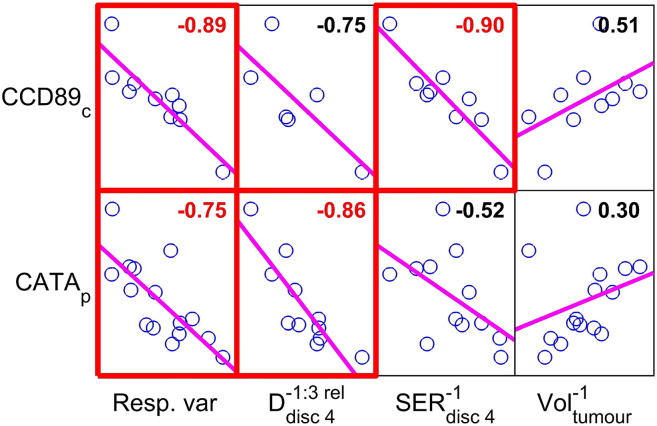
The evaluation of associations between MR features reaching high β sums in feature selection and the 66 proteins associated with radiation response yielded two proteins of particular interest, CATA (catalase, encoded by CAT) and CCD89 (coiled-coil domain containing 89, encoded by CCDC89). CATA is associated with oxidative stress, proliferation, cell cycle arrest, and apoptotic cell death, and CCD89 is associated with DNA damage and repair, proliferation, and cell cycle arrest [34]. In addition to their strong and statistically significant correlations with response, these proteins were strongly correlated with the MR features regarded highly efficacious by the feature selection, SERdisc 4−1 and Ddisc 4−1 : 3rel. SERdisc 4−1 correlated with centrally sampled CCD89 (r = −0.9, P = .03), and Ddisc 4−1 : 3rel correlated with peripherally sampled CATA (r = −0.86, P = .03) (Figure 6).
Figure 6.
The left column of the scatter plot matrix shows the correlation between the two proteins CCD89 and CATA and tumor response to 177Lu-octreotate therapy (Resp. var). The day −1 tumor volume (Voltumor−1) was a good predictor of response per se, but it was not correlated with CCD89 or CATA on a statistically significant level (right column). The two MR features that were regarded most important by the feature selection method, i.e., early increased diffusion D in peripheral tumor (day −1 to day 3 in disc 4) and pretreatment signal enhancement ratio SER in peripheral tumor (day −1, disc 4) did, however, show strong and statistically significant correlation with CATA and CCD89, respectively (middle columns). Red fonts and boxes indicate statistical significance (P < .05) after correction for multiple comparisons. Protein suffixes p and c indicate peripheral and central tumor tissue sample region, respectively. Least squares reference lines of each scatter plot have slopes equal to the displayed correlation coefficients. It should be noted that the arithmetical sign of a correlation can only be unambiguously determined by conferring Figure 4 and S1.
Interestingly, although pretreatment tumor volume Voltumor−1 per se was a good predictor of response, it did not correlate significantly with significant proteins or with any high β sum MR features (Figure 6).
Discussion
This study was set up to identify potential MR-derived biomarkers for early NET tissue response after 177Lu-octreotate therapy. We used a model with subcutaneous GOT1 xenografts on nude mice. This model has shown retained characteristic properties of NETs, such as expression of somatostatin receptors and slow growth rate [21]. It has also been shown that 177Lu-octreotate induces cell cycle arrest, apoptosis, and dose-dependent tumor volume reduction and cure in GOT1 tumor-bearing mice [28]. Furthermore, molecular response of noncurative treatment of GOT1 tumors using 177Lu-octreotate has been observed at longer time points after administration [22], [33].
The noncurative 177Lu-octretotate treatment resulted in a range of responses, from tumors showing an initially decreasing volume to tumors showing only slightly reduced growth rate (Figure 2). Almost all tumors did, however, show regrowth between day 8 and 13. The noncurative treatment schedule was chosen to enable response evaluation with MRI and quantitative proteomics during a longer time period and to reflect a wide range of biological responses. We related tumor volume to the volume before treatment (day −1) and defined the mean change in relative volume up to day 8 as our response variable, excluding day 13 volume due to the regrowth. This definition made it possible to use the same definition for all subjects regardless of missing values due to, e.g., some tumors not being examined on day 1.
The feature selection results showed that two MR features were more important than others regarding monitoring response to therapy: the SER prior to therapy and changes in D after therapy. The biophysical interpretation of SER, defined as the early to late relative signal enhancement after contrast injection, is not straightforward. However, Li et al. [30] demonstrated a close relationship between SER and the more physiologically understood pharmacokinetic redistribution rate constant (kep) using both computer simulations and in vivo data from a DCE study of a mouse model of breast cancer. Low kep has been correlated with lower histologic and nuclear grade and better prognosis in breast cancer [31], [32]. In locally advanced breast cancer, SER significantly discriminated patients showing pathologic complete response to neoadjuvant chemotherapy from patients with residual disease, and SER correlated positively with changes in glucose transport rate from blood to tumor tissue, as estimated by dynamic 18F-FDG [33]. In the present study, pretreatment measurement of SER in peripheral parts of the tumor was regarded particularly important for prediction of tumor response (Figure 5), with lower SER indicating better response. There was also a strong and statistically significant correlation between SERdisc 4−1 and centrally sampled protein CCD89 (Figure 6), previously associated with mitotic activity and increased H2AX phosphorylation (indicating DNA damage). Although the specificity of the evaluated MR techniques for specific protein levels would be low, the obtained correlation suggests that it might be possible to monitor levels of clinically interesting proteins by indirectly monitoring their biological manifestation to which the MR method is sensitive. The specific function of CCD89 is yet not well known, which makes its association with MR features or response presently difficult to interpret. It should also be noted that some correlations, such as the one between SER and CCD89, were between MR features and proteins from different parts of the tumor.
The diffusion features Ddisc 3−1 : 3 rel and Ddisc 4−1 : 3rel measured by DWI were also regarded important by feature selection (relative β sum ~0.5), and they showed strong and positive correlation with response. DWI has gained acceptance in both clinical and preclinical settings, and there is a relatively high consensus regarding its potential in tumor therapy assessments [35]. The most common parameter derived from DWI is ADC, but IVIM-DWI measurements are gaining interest. Both ADC and D are reduced in regions of restricted molecular movement due to, e.g., cell and organelle membranes [35]. Measurements of diffusion using D are less biased by perfusion-related pseudodiffusion compared with ADC, and the sensitivity to cell density should thus be higher than for ADC. Since radiotherapy is highly capable of reducing proliferation and activating cell death mechanisms such as apoptosis, with subsequent disintegration of membrane structures, ADC and D are both potent candidates for radiotherapy response assessment [2], [36]. Several studies have shown that ADC is related to both cellular density and tumor aggressiveness [10], [8], [37], [45], [39], [40], [41], [42]. Rijswijk et al. [43] compared diffusion estimated by D with ADC in soft tissue tumors in patients with tumors of different type and location. As expected, they found that ADC was significantly higher than D in all tumors and that D could separate malignant from nonmalignant tumors, whereas ADC could not. In the present study, it thus seems like the molecular diffusion in relatively peripheral tumor parts increased to a greater extent in more responding tumors (shrinking) and that D may be used to assess response at a very early stage. Figure 4 shows a close similarity between ADC and D since they are grouped first by clustering. D, however, outperformed ADC in feature selection, probably due to less perfusion sensitivity. Figure 5 shows that several diffusion features defined from D on Δday −1:3 and 3:8 were regarded highly important, whereas fewer features from ADC reached β sums > 0.05 (display limit) on Δday −1:3 and with lower β sums than D features for corresponding Δdays and discs. It was also found that Ddisc 4−1 : 3rel correlated strongly with peripheral expression of CATA (r = −0.86). Ionizing radiation induces production of reactive oxygen species, such as hydrogen peroxide, which can cause DNA damage with subsequent cell cycle arrest or cell death [44]. CATA acts by catalyzing the conversion of hydrogen peroxide into oxygen and water, and has been shown to protect irradiated tumor cells from apoptosis [45]. A previous study showed that CATA is increased in liver tissue of mice exposed to 137Cs [46].The peripherally increased levels of CATA in the tumors responding well to the treatment could thus be due to the oxidative stress imposed by irradiation. The association between CATA and D thus seems reasonable and supports the motivation to use perfusion-corrected diffusion measures to evaluate therapy response after radiotherapy.
An interesting observation is that pretreatment tumor volume (Voltumor−1) was correlated with response such that better therapeutic results were seen for larger Voltumor−1. A possible explanation for this is that the greater volume allows for higher absorbed dose to the tumor cells (since the range of the most abundantly emitted beta-particles is comparable to tumor radii). However, Voltumor−1 acquired a very low β sum in the feature selection (outperformed by hundreds of features), and it was not significantly correlated with relevant proteins. It is thus likely that the biological features that are important for tumor response also affect Voltumor−1 but are better reflected by other MR features such as features sensitive to blood supply. The Voltumor−1 feature would thus provide redundant information and be regarded less important by the lasso, as was observed.
Our results indicate that tumor cells in the peripheral regions responded better to treatment, with subsequent cell death followed by disintegration of membranes and increased diffusion some days later. We did not see the same effects on CATA or D in central tumor parts, which may indicate lower therapeutic effect by the 177Lu-octreotate, e.g., due to impaired delivery to central tumor or reduced radiosensitivity due to hypoxic conditions [47]. The latter explanation is in agreement with a previous study of the GOT1 tumor model, where higher proliferation was found in peripheral tumor parts, although there was a higher uptake of 177Lu-octreotate in central tumor parts [48].The proliferation would facilitate apoptosis induced by DNA damage, and the central uptake of 177Lu-octreotate indicates adequate delivery and that the lower response must be caused by something else, such as hypoxia.
To our knowledge, the spatial heterogeneity and temporal changes in tumor tissue after treatment are not well known for any tumor types or treatment modalities. Therefore, it is important to include repeated measurement in the analysis, as well as a regional evaluation of the resulting parameters. However, longitudinal assessments based on pixel-wise analysis require sophisticated image registration algorithms due to tissue alterations from treatment as well as regrowth. ROI analysis of large tumor regions does not require registration, but important information on regional response might be averaged out. In this study, we used a tradeoff between image detail and assessment time interval by analyzing intensity variations over time in contiguous disc-shaped tumor regions (Figure 1). This approach is sensitive to radial intensity variations, which is plausible in tumor tissue since solid tumors often present hypoxic cores and proliferating periphery [47], [49]. It does, however, not account for angular variations. The feature selection results in Figure 5 show several MR parameters with a radial variation, where disc separation was required to reach display limit, such as IS and SE60. Interestingly, some parameters (e.g., SE60 and normalized area under the curve (AUCn) for Δday −1:3, Figure 4) even show opposite correlation with response in central and peripheral tumor. Such features are most likely averaged out as nonsignificant on evaluation of ROI median values.
In this study, temporal variations were observed, e.g., in D, which was regarded as one of the most important features for Δday −1:3 (Figure 5), the only feature displayed for Δday 3:8, but not displayed at all for the other time intervals. SER and BE were also regarded very important, and they were only visible on day −1 and Δday −1:1, respectively (Figure 5). This indicates the importance of including a sufficient number of posttreatment measurements, at least in explorative studies such as this one. An important aspect on longitudinal parameter evaluation is the choice of absolute or relative change in parameter values between successive time points since it may significantly impact the sensitivity of the method and even alter the arithmetic sign of a correlation (cf. Figures 4 and S1).
Principal component analysis and ridge regression are techniques that are commonly used when analyzing multiple features based on relatively few observations in order to make predictions regarding, e.g., therapy response [18]. However, the resulting models are based on linear combinations of multiple features, which reduce the biological interpretability. By employing the lasso feature selection algorithm, we chose another strategy to address this problem. The lasso is designed to find sparse sets of highly efficacious features and thereby produce models with maintained interpretability. The lasso does, however, tend to arbitrarily choose one of two features if they are highly correlated, and similar to principal component and ridge regression analysis, it cannot handle missing values [50]. We used stochastic data imputation to handle missing values, which we also believe enabled nonbiased evaluation of contiguous tumor discs. Data from contiguous discs are probably often highly correlated (for the same MR parameter and day or Δday), and the lasso would therefore probably reject one of them. However, the 5000 repetitions of the stochastic data imputation should result in disc inclusion in proportion to their true relative efficacy.
Few studies have used feature selection techniques for the purpose of unraveling the most efficacious subset of high-dimensional MR feature sets. However, in a recent study on prostate cancer diagnostic performance, Haq et al. applied the lasso method to find the optimal set of principal components from T1 intensities and semiquantitative DCE-MRI features, and found that the set produced by the lasso method outperformed traditional pharmacokinetic parameters in receiver operating characteristic tests of diagnostic performance [51]. Based on our literature review, there seems to be no previous studies that combine the lasso method with data imputation in the repetitive manner for statistical evaluation, as done by us. By this approach, we were able to evaluate features that should be highly correlated with each other, such as features from adjacent disc within a tumor. The great number of repetitions does, however, make the method computationally demanding.
It should be noted that the results of this study may probably not be generalizable to other tumor types and treatment modalities. The differences in genetics and biology between tumor types and the complexity of the constitution of tumor tissues make it likely that other MR features are important in other studies. Due to the vast number of proteins associated with each tumor type, correlations between MR features, response, and protein expression levels will most likely be specific to the particular tumor treatment model system. Nevertheless, the method proposed by us should provide a means for evaluation, regardless of model system.
Another limitation of the current study is the choice of response variable. We defined response as the change in tumor volume after treatment. This definition implicitly states that reduced tumor volume is equivalent to successful treatment, although it may not reflect improved long-term survival of the patient. Furthermore, even if reduced volume does represent successful treatment, the volume is still just a manifestation of the biological processes that we correlate with the MR features. Hence, volume will not be a truly independent measure of response.
In this study, we did not correct the tumor volume for, e.g., necrotic regions, and the results may have been slightly different if such corrections were made. This would, however, require unambiguous knowledge of the tissues constituting the tumor, which would require ex vivo assessments. A possible means to measure viable tumor volume would be to use, e.g., PET methods that assess the metabolically active tumor volume. It should also be noted that the absorbed dose to tumor tissue may vary within a tumor and between tumors due to factors such as different tumor volume and impaired delivery to certain tumor regions.
We observed very early effects on an interim evaluation of data and decided to harvest tumor tissue for quantitative proteomics on day 1 for three animals. All analyses in this study, however, include data from all 15 animals, which may introduce bias or disguise findings that would appear in separate evaluations of early (day 1) effects and effects during tumor regrowth (days 8-13). Such a study would, however, require additional subjects for the early time points.
To our knowledge, studies that include both spatial and temporal assessments of multiple MR features of generically different MR techniques have not previously been published, which makes this study unique in the information it provides for future studies on multiparametric MR for tumor tissue response assessments.
Conclusions
Several of the studied MR parameters show great potential for studies on NET characteristics in the context of 177Lu-octreotate therapy, but the importance of evaluating solid tumors on a regional level and longitudinally by noninvasive means, such as multiparametric MR methods, must be emphasized.
Two of the MR parameters, the DCE-MRI–derived SER and the IVIM-DWI–derived D, seem highly efficacious for response assessment in NETs after 177Lu-octreotate therapy but only when they are evaluated at a particular point in time relative to treatment and in a particular tumor region. The perfusion corrected tissue diffusion measure D seems to be more sensitive and specific to effects of 177Lu-octreotate on NETs and should be preferred over the commonly used ADC measure.
Although pretreatment tumor volume was a good predictor of NET response to 177Lu-octreotate therapy, the MR features selected by feature selection seem to reflect more biologically specific response mechanisms, as demonstrated by the strong correlations with certain proteins.
The following are the supplementary data related to this article.
Acknowledgments
Acknowledgements
The authors are grateful to Lilian Karlsson and Ann Wikström for their skillful technical assistance.
We are also grateful to the Proteomics Core Facility at Sahlgrenska Academy, Gothenburg University, who performed the analysis for protein quantification and Inga-Britt and Arne Lundbergs Research Foundations for the donation of the Orbitrap Fusion Tribrid MS instrument used in this analysis.
Funding
This work was supported by the Swedish Research Council (grant number 21073); the Swedish Cancer Society (grant numbers 21073, 2016/692); BioCARE—a National Strategic Research Program at the University of Gothenburg; the King Gustav V Jubilee Clinic Cancer Research Foundation; the Sahlgrenska University Hospital Research Funds; the Assar Gabrielsson Cancer Research Foundation; the Adlerbertska Research Fund; the Wilhelm and Martina Lundgren science trust fund; and the Royal Society of Arts and Sciences in Gothenburg (K.V.V.S.).
Contributor Information
Mikael Montelius, Email: mikael.montelius@radfys.gu.se.
Johan Spetz, Email: spetz@hsph.harvard.edu.
Oscar Jalnefjord, Email: oscar.jalnefjord@vgregion.se.
Evelin Berger, Email: evelin.berger@gu.se.
Ola Nilsson, Email: ola.nilsson@vgregion.se.
Maria Ljungberg, Email: maria.ljungberg@vgregion.se.
Eva Forssell-Aronsson, Email: eva.forssell_aronsson@radfys.gu.se.
References
- 1.Pouget J-P, Lozza C, Deshayes E, Boudousq V, Navarro-Teulon I. Introduction to radiobiology of targeted radionuclide therapy. Front Med. 2015;2(12):1–11. doi: 10.3389/fmed.2015.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orth M, Lauber K, Niyazi M, Friedl AA, Li M, Maihöfer C, Schüttrumpf L, Ernst A, Niemöller OM, Belka C. Current concepts in clinical radiation oncology. Radiat Environ Biophys. 2014;53(1):1–29. doi: 10.1007/s00411-013-0497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Marcus CD, Ladam-Marcus V, Cucu C, Bouché O, Lucas L, Hoeffel C. Imaging techniques to evaluate the response to treatment in oncology: current standards and perspectives. Crit Rev Oncol Hematol. 2009;72(3):217–238. doi: 10.1016/j.critrevonc.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Weber WA. Assessing tumor response to therapy. J Nucl Med. 2009;50(Suppl 1):1S–10S. doi: 10.2967/jnumed.108.057174. [DOI] [PubMed] [Google Scholar]
- 6.Jentsch C, Beuthien-Baumann B, Troost EGC, Shakirin G. Validation of functional imaging as a biomarker for radiation treatment response. Br J Radiol. 2015;88(1051) doi: 10.1259/bjr.20150014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rafat M, Ali R, Graves EE. Imaging radiation response in tumor and normal tissue. Am J Nucl Med Mol Imaging. 2015;5(4):317–332. [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Liu Y, Yu T, Ye N, Wang Q. Evaluation of apparent diffusion coefficient associated with pathological grade of lung carcinoma, before therapy. J Magn Reson Imaging. 2014;24(10) doi: 10.1002/jmri.24823. [DOI] [PubMed] [Google Scholar]
- 9.Verma S, Rajesh A, Morales H, Lemen L, Bills G, Delworth M, Gaitonde K, Ying J, Samartunga R, Lamba M. Assessment of aggressiveness of prostate cancer: correlation of apparent diffusion coefficient with histologic grade after radical prostatectomy. Am J Roentgenol. 2011;296(2):374–381. doi: 10.2214/AJR.10.4441. [DOI] [PubMed] [Google Scholar]
- 10.Le Bihan D, Breton E, Lallemand D, Aubin ML, Vignaud J, Laval-Jeantet M. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology. 1988;168(2):497–505. doi: 10.1148/radiology.168.2.3393671. [DOI] [PubMed] [Google Scholar]
- 11.Lee HJ, Rha SY, Chung YE, Shim HS, Kim YJ, Hur J, Hong YJ, Choi BW. Tumor perfusion-related parameter of diffusion-weighted magnetic resonance imaging: correlation with histological microvessel density. Magn Reson Med. 2014;71(4):1554–1558. doi: 10.1002/mrm.24810. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Decarlo L, Cho GY, Jensen JH, Sodickson DK, Moy L, Formenti S, Schneider RJ, Goldberg JD, Sigmund EE. Interstitial fluid pressure correlates with intravoxel incoherent motion imaging metrics in a mouse mammary carcinoma model. NMR Biomed. 2012;25(5):787–794. doi: 10.1002/nbm.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huuse EM, Moestue SA, Lindholm EM, Bathen TF, Nalwoga H, Krüger K, Bofin A, Mælandsmo GM, Akslen LA, Engebraaten O. In vivo MRI and histopathological assessment of tumor microenvironment in luminal-like and basal-like breast cancer xenografts. J Magn Reson Imaging. 2012;35(5):1098–1107. doi: 10.1002/jmri.23507. [DOI] [PubMed] [Google Scholar]
- 14.Jackson A, Li K-L, Zhu X. Semi-quantitative parameter analysis of DCE-MRI revisited: Monte-Carlo simulation, clinical comparisons, and clinical validation of measurement errors in patients with type 2 neurofibromatosis. PLoS One. 2014;9(3):e90300. doi: 10.1371/journal.pone.0090300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lescher S, Jurcoane A, Veit A, Bähr O, Deichmann R, Hattingen E. Quantitative T1 and T2 mapping in recurrent glioblastomas under bevacizumab: earlier detection of tumor progression compared to conventional MRI. Neuroradiology. 2015;57(1):11–20. doi: 10.1007/s00234-014-1445-9. [DOI] [PubMed] [Google Scholar]
- 16.Heijmen L, Ter Voert EEGW, Oyen WJG, Punt CJA, VanSpronsen DJ, Heerschap A, De Geus-Oei LF, Van Laarhoven HWM. Multimodality imaging to predict response to systemic treatment in patients with advanced colorectal cancer. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0120823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borren A, Moman MR, Groenendaal G, Boeken Kruger AE, van Diest PJ, van der Groep P, van der Heide UA, van Vulpen M, Philippens MEP. Why prostate tumour delineation based on apparent diffusion coefficient is challenging: an exploration of the tissue microanatomy. Acta Oncol. 2013;52(8):1629–1636. doi: 10.3109/0284186X.2013.787164. https://doi.org/10.3109/0284186x.0282013.0787164. [DOI] [PubMed] [Google Scholar]
- 18.Hastie T, Tibshirani R, Friedman J. Springer-Verlag; 2009. The Elements of Statistical Learning, 2nd edition. [Google Scholar]
- 19.Yao JC, Hassan M, Phan A, Dagohoy C, Leary C, Mares JE, Abdalla EK, Fleming JB, Vauthey JN, Rashid A. One hundred years after "carcinoid": epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol. 2008;26(18):3063–3072. doi: 10.1200/JCO.2007.15.4377. [DOI] [PubMed] [Google Scholar]
- 20.Swärd C, Bernhardt P, Ahlman H, Wängberg B, Forssell-Aronsson E, Larsson M, Svensson J, Rossi-Norrlund R, Kölby L. [177Lu-DOTA 0-Tyr 3]-octreotate treatment in patients with disseminated gastroenteropancreatic neuroendocrine tumors: the value of measuring absorbed dose to the kidney. World J Surg. 2010;34(6):1368–1372. doi: 10.1007/s00268-009-0387-6. [DOI] [PubMed] [Google Scholar]
- 21.Kölby L, Bernhardt P, Ahlman H, Wängberg B, Johanson V, Wigander A, Forssell-Aronsson E, Karlsson S, Ahrén B, Stenman G. A transplantable human carcinoid as model for somatostatin receptor-mediated and amine transporter-mediated radionuclide uptake. Am J Pathol. 2001;158(2):745–755. doi: 10.1016/S0002-9440(10)64017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalmo J, Spetz J, Montelius M, Langen B, Arvidsson Y, Johansson H, Parris TZ, Helou K, Wängberg B, Nilsson O. Priming increases the anti-tumor effect and therapeutic window of 177Luoctreotate in nude mice bearing human small intestine neuroendocrine tumor GOT1. EJNMMI Res. 2017;7(1):6. doi: 10.1186/s13550-016-0247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montelius M, Ljungberg M, Horn M, Forssell-Aronsson E. Tumour size measurement in a mouse model using high resolution MRI. BMC Med Imaging. 2012;12(1) doi: 10.1186/1471-2342-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc B Methodol. 1996:267–288. [Google Scholar]
- 25.Paul D, Bair E, Hastie T, Tibshirani R. "Preconditioning" for feature selection and regression in high-dimensional problems. Ann Stat. 2008:1595–1618. [Google Scholar]
- 26.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kölby L, Bernhardt P, Johanson V, Schmitt A, Ahlman H, Forssell-Aronsson E, Mäcke H, Nilsson O. Successful receptor-mediated radiation therapy of xenografted human midgut carcinoid tumour. Br J Cancer. 2005;93(10):1144–1151. doi: 10.1038/sj.bjc.6602845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spetz J, Langen B, Rudqvist N, Parris TZ, Helou K, Nilsson O, Forssell-Aronsson E. Hedgehog inhibitor sonidegib potentiates 177Lu-octreotate therapy of GOT1 human small intestine neuroendocrine tumors in nude mice. BMC Cancer. 2017;17(1):528. doi: 10.1186/s12885-017-3524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li K-L, Henry RG, Wilmes LJ, Gibbs J, Zhu X, Lu Y, Hylton NM. Kinetic assessment of breast tumors using high spatial resolution signal enhancement ratio (SER) imaging. Magn Reson Med. 2007;58(3):572–581. doi: 10.1002/mrm.21361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JY, Kim SH, Kim YJ, Kang BJ, An YY, Lee AW, Song BJ, Park YS. Enhancement parameters on dynamic contrast enhanced breast MRI: do they correlate with prognostic factors and subtypes of breast cancers? Magn Reson Imaging. 2015;33(1):72–80. doi: 10.1016/j.mri.2014.08.034. [DOI] [PubMed] [Google Scholar]
- 32.Koo HR, Cho N, Song IC, Kim H, Chang JM, Yi A, La Yun B, Moon WK. Correlation of perfusion parameters on dynamic contrast-enhanced MRI with prognostic factors and subtypes of breast cancers. J Magn Reson Imaging. 2012;36(1):145–151. doi: 10.1002/jmri.23635. [DOI] [PubMed] [Google Scholar]
- 33.Partridge SC, Vanantwerp RK, Doot RK, Chai X, Kurland BF, Eby PR, Specht JM, Lisa K, Schubert EK, Lehman CD. Association between serial dynamic contrast-enhanced MRI and dynamic 18F-FDG PET measures in patients undergoing neoadjuvant chemotherapy for locally advanced breast cancer. J Magn Reson Imaging. 2010;32(5):1124–1131. doi: 10.1002/jmri.22362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee M-C, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35(2):228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Padhani AR, Liu G, Mu-Koh D, Chenevert TL, Thoeny HC, Takahara T, Dzik-Jurasz A, Ross BD, Van Cauteren M, Collins D. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia. 2009;11(2):102–125. doi: 10.1593/neo.81328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eriksson D, Stigbrand T. Radiation-induced cell death mechanisms. Tumour Biol. 2010;31(4):363–372. doi: 10.1007/s13277-010-0042-8. [DOI] [PubMed] [Google Scholar]
- 37.Hambrock T, Somford DM, Huisman HJ, van Oort IM, Witjes JA, Hulsbergen-van de Kaa CA, Scheenen T, Barentsz JO. Relationship between apparent diffusion coefficients at 3.0-T MR imaging and Gleason grade in peripheral zone prostate cancer. Radiology. 2011;259(2):453–461. doi: 10.1148/radiol.11091409. [DOI] [PubMed] [Google Scholar]
- 38.Rosenkrantz AB, Niver BE, Fitzgerald EF, Babb JS, Chandarana H, Melamed J. Utility of the apparent diffusion coefficient for distinguishing clear cell renal cell carcinoma of low and high nuclear grade. Am J Roentgenol. 2010;195(5):W344–W351. doi: 10.2214/AJR.10.4688. [DOI] [PubMed] [Google Scholar]
- 39.Bozgeyik Z, Onur MR, Poyraz AK. The role of diffusion weighted magnetic resonance imaging in oncologic settings. Quant Imaging Med Surg. 2013;3(5):269–278. doi: 10.3978/j.issn.2223-4292.2013.10.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore WA, Khatri G, Madhuranthakam AJ, Sims RD, Pedrosa I. Added value of diffusion-weighted acquisitions in MRI of the abdomen and pelvis. Am J Roentgenol. 2014;202(5):995–1006. doi: 10.2214/AJR.12.9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill DK, Kim E, Teruel JR, Jamin Y, Widerøe M, Søgaard CD, Størkersen Ø, Rodrigues DN, Heindl A, Yuan Y. Diffusion-weighted MRI for early detection and characterization of prostate cancer in the transgenic adenocarcinoma of the mouse prostate model. J Magn Reson Imaging. 2016;43(5):1207–1217. doi: 10.1002/jmri.25087. [DOI] [PubMed] [Google Scholar]
- 42.Gibbs P, Liney GP, Pickles MD, Zelhof B, Rodrigues G, Turnbull LW. Correlation of ADC and T2 measurements with cell density in prostate cancer at 3.0 Tesla. Invest Radiol. 2009;44(9):572–576. doi: 10.1097/RLI.0b013e3181b4c10e. [DOI] [PubMed] [Google Scholar]
- 43.Van Rijswijk CSP, Kunz P, Hogendoorn PCW, Taminiau AHM, Doornbos J, Bloem JL. Diffusion-weighted MRI in the characterization of soft-tissue tumors. J Magn Reson Imaging. 2002;15(3):302–307. doi: 10.1002/jmri.10061. [DOI] [PubMed] [Google Scholar]
- 44.Biaglow JE, Mitchell JB, Held K. The Seventh International Conference on Chemical Modifiers of Cancer Treatment Part 2: the importance of peroxide and superoxide in the X-ray response. Int J Radiat Oncol Biol Phys. 1992;22(4):665–669. doi: 10.1016/0360-3016(92)90499-8. [DOI] [PubMed] [Google Scholar]
- 45.Xiao X, Luo H, Vanek KN, LaRue AC, Schulte BA, Wang GY. Catalase inhibits ionizing radiation-induced apoptosis in hematopoietic stem and progenitor cells. Stem Cells Dev. 2015;24(11):1342–1351. doi: 10.1089/scd.2014.0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yi L, Li L, Yin J, Hu N, Li G, Ding D. Proteomics analysis of liver tissues from C57BL/6J mice receiving low-dose 137Cs radiation. Environ Sci Pollut Res. 2016;23(3):2549–2556. doi: 10.1007/s11356-015-5494-3. [DOI] [PubMed] [Google Scholar]
- 47.Guppy M. The hypoxic core: a possible answer to the cancer paradox. Biochem Biophys Res Commun. 2002;299(4):676–680. doi: 10.1016/s0006-291x(02)02710-9. [DOI] [PubMed] [Google Scholar]
- 48.Oddstig J, Bernhardt P, Lizana H, Nilsson O, Ahlman H, Kölby L, Forssell-Aronsson E. Inhomogeneous activity distribution of 177Lu-DOTA0-Tyr3-octreotate and effects on somatostatin receptor expression in human carcinoid GOT1 tumors in nude mice. Tumour Biol. 2012;33(1):229–239. doi: 10.1007/s13277-011-0268-0. [DOI] [PubMed] [Google Scholar]
- 49.Foryś U, Mokwa-Borkowska A. Solid tumour growth analysis of necrotic core formation. Math Comput Model. 2005;42(5):593–600. [Google Scholar]
- 50.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 51.Haq NF, Kozlowski P, Jones EC, Chang SD, Goldenberg SL, Moradi M. A data-driven approach to prostate cancer detection from dynamic contrast enhanced MRI. Comput Med Imaging Graph. 2015;41:37–45. doi: 10.1016/j.compmedimag.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gustafsson O, Montelius M, Starck G, Ljungberg M. Impact of prior distributions and central tendency measures on Bayesian intravoxel incoherent motion model fitting. Magn Reson Med. 2017 doi: 10.1002/mrm.26783. [DOI] [PubMed] [Google Scholar]