Abstract
Glioma is the most common cancer in human brain system and seriously threatens human health. miRNA-320 has been demonstrated to be closely correlated with the development of glioma. However, its effect and molecular mechanism underlying radioresistance have not been fully elucidated in glioma. Here, RT-qPCR assay was used to assess the expressions of miR-320 and forkhead box protein M1 (FoxM1) mRNA in glioma tumor tissues and cells. The effects of miR-320, FoxM1 and sirtuin type 1 (Sirt1) on radiosensitivity in glioma cells were evaluated by clone formation assay, apoptosis assay, histone H2AX phosphorylation level (γH2AX) detection and caspase 3 activity analysis, respectively. The direct interaction between miR-320 and FoxM1 was detected by luciferase assay. The protein levels of FoxM1, Sirt1 and γH2AX were measured by western blot assay. We found that miR-320 expression was down-regulated and FoxM1 expression was up-regulated in radioresistant glioma tissues and IR-treated glioma cells. miR-320 overexpression dramatically enhanced radiosensitivity, promoted apoptosis, and improved γH2AX expression and caspase 3 activity in glioma cells. Luciferase reporter assay and western blot assay further validated that miR-320 suppressed FoxM1 expression by directly targeting 3’ UTR region of FoxM1. Moreover, miR-320 inhibited Sirt1 expression via targeting FoxM1 in glioma cells. Furthermore, overexpression of FoxM1 and Sirt1 strikingly attenuated miR-320-induced increase of radiosensitivity, apoptosis and γH2AX expression in glioma cells. In conclusion, miR-320 enhanced radiosensitivity of glioma cells through down-regulation of Sirt1 by directly targeting FoxM1.
Introduction
Glioma is the most prevalent malignant tumor in human central nervous systems (CNS) with the proportion of approximately 29% in all brain tumors [1]. Although much progress has been made in the diagnosis and therapy strategies including chemo/radiotherapy and surgery resection, the prognosis is still unsatisfactory, with an estimated 5-year survival rate of 15% in glioma patients [2], [3], [4]. Moreover, the existence and development of radiotherapy resistance markedly vitiates the therapy outcome [5]. Hence, a better understanding of the molecular mechanism underlying radioresistance in glioma cells is strikingly critical for the discovery of novel potential targets and more effective therapy strategies.
microRNAs (miRNAs), a class of small non-coding RNAs with the length of approximately 22 nucleotides, have been identified as critical mediators in various biological processes and carcinogenesis [6], [7]. Studies also manifested that miRNAs were closely associated with radioresistance of glioma cells [8]. miRNA-320, a member of miR-200 family, has been demonstrated to be down-regulated and function as a tumor suppressor in multiple cancers such as oral cancer [9] and gastric cancer [10]. Moreover, Wan et al. manifested that miR-320 enhanced radiosensitivity of colon cancer cells [11]. Yang et al. also demonstrated that deficiency of miR-320 decreased radiosensitivity of cervical cancer cells [12]. Also, recent studies indicated that dysregulated miR-320 was implicated in the development of glioma [13]. For instance, miRNA microarray results revealed that miR-320 expression level was significantly down-regulated in peripheral blood of glioma patients [14]. miR-320 overexpression suppressed glioma cell proliferation by targeting E2F Transcription Factor 1 (E2F1) [13]. However, the roles and molecular mechanisms of miR-320 involved in radiosensitivity in glioma have not been fully elucidated.
Forkhead box protein M1 (FoxM1), also named as HFH-11, MPP-2, WIN and Trident, is a transcription factor mapping to chromosome 12p13.3 [15], [16]. FoxM1 has been demonstrated to be associated with progression and treatment of cancers in a wide range of malignancies including breast cancer, liver cancer and nervous system cancers [17]. Increasing evidence suggested that FoxM1 might be a promising therapeutic target of cancers due to its significance in the development and therapy of cancers [18]. It is well documented that abnormal expression of FoxM1 was closely implicated in tumorigenicity and raidoresistance of glioma cells [19], [20]. Additionally, FoxM1 has been identified as an upstream regulator of sirtuin type 1 (Sirt1) in glioma cells [21]. More notably, FoxM1 was previously reported to be a target of miR-320 in colorectal cancer [11]. Consequently, we hypothesized whether miR-320 might affect radiosensitivity of glioma cells by regulating FoxM1 and Sirt1 expressions.
In the present study, we demonstrated that miR-320 expression was down-regulated while FoxM1 was up-regulated in radioresistant glioma tissues and IR-treated glioma cells. Moreover, miR-320 was inversely correlated with FoxM1 expression in glioma tissues. Functional analysis revealed that miR-320 enhanced radiosensitivity of glioma cells. Mechanistic studies revealed that miR-320 exerted its functions through down-regulation of Sirt1 by directly targeting FoxM1 in glioma cells.
Materials and Methods
Tissues Samples
Tissues specimens were obtained from the glioma patients treated with radiotherapy for over 6 months after diagnosis at the First Affiliated Hospital of Zhengzhou University between 2014 and 2016. Our study received the approval of Ethics Committee of the First Affiliated Hospital of Zhengzhou University and obtained the informed consent from all patients. According to the radiosensitivity index model presented by previous studies [22], [23], glioma patients were dichotomized into radiosensitive and radioresistant groups (RSI index >0.5 means the patients were radioresistant). Briefly, RSI index was measured utilizing the probesets and rank-based linear regression algorithm in prior studies [22], [23].
Cell Culture
Human glioma U251 and U87 cells were obtained from American Tissue Culture Collection (ATCC, Manassas, VA, USA) and were maintained in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Invitrogen) and 1% penicillin/streptomycin (Invitrogen), at 37°C incubator containing 5% CO2.
Cell Transfection
miR-320 mimic and its scrambled control (miR-NC), miR-320 inhibitor and its control (inhibitor control) were synthesized by Genepharma Company (Shanghai, China). The full length sequences of FoxM1 or Sirt1 were amplified by PCR and subcloned into pcDNA3.1 vector (Invitrogen) to generate pcDNA-FoxM1 or pcDNA-Sirt1 plasmids. All plasmids or oligonucleotides were transfected into U251 and U87 cells using Lipofectamine 2000 (Invitrogen).
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
Total RNA was extracted from glioma tissues and cells using Trizol reagent (Invitrogen). For the detection of mRNA and miRNA expressions, 1 μg of total RNA was reversely transcribed into the first-strand cDNA by PrimeScript RT reagent kit (TaKaRa BIO, Shiga, Japan) or TaqMan MicroRNA Array kit (Applied Biosystems, Foster City, CA, USA). Then the expression patterns of FoxM1 were assessed using SYBR Premix Ex Taq™ II (TaKaRa BIO) reagent with GAPDH as the normalization. miR-320 relative expression level was determined by TaqMan Universal Master Mix II (Applied Biosystems) with U6 as the normalization. The primers were listed as follows: miR-320, 5’-ACA CTC CAG CTG GGA AAA GCT GGG TTG AGA-3′ (forward) and 5’-ACA CTC CAG CTG GGA AAA GCT GGG TTG AGA-3′ (reverse); U6, 5’-GCT TCG GCA GCA CAT ATA CTA AAA T-3′ (forward), 5’-CGC TTC ACG AAT TTG CGT GTC AT-3′ (reverse); FoxM1, 5’-TCT CAG CAC CAC TCC CTT G-3′ (forward) and 5′-GGA TCT TGC TGA GGC TGT C-3′ (reverse); GAPDH, 5′-TGG AAG GAC TCA TGA CCA CA-3′ (forward), 5’-TTC AGC TCA GGG ATG ACC TT-3′ (reverse).
Irradiation Exposure
Exponentially growing glioma cells were irradiated at the indicated dose using a 6 MV photon beam generated by an irradiation apparatus (2100C/D, VARIAN, Palo Alto, CA, USA) at a dose rate of 0.4 Gy/min at room temperature.
Colony Formation Assay
The survival fraction (SF) of U251 and U87 cells with different treatments was determined by clone formation assay. Briefly, cells were plated in 6-well plates and transfected with miR-NC, miR-320, or miR-320 + pcDNA-FoxM1. At 48 h post-transfection, transfected cells were transferred to 60 mm2 culture dish and then irradiated at 0, 2, 4, 6, 8 and 10 Gy using an irradiation apparatus (2100C/D, VARIAN). After irradiation, cells were cultured for another 9–14 days and then clones were fixed with 75% ethanol and stained with 0.5% (w/v) crystal violet solution (Sigma-Aldrich, St. Louis, MO, USA). Finally, colonies with more than 50 cells were counted by microscope (Leica Microsystems, Wetzlar, Germany). SF was calculated by the following formula: SF = formed colonies number / plated cells number × plating efficiency. Plating efficiency is the ratio of colonies number to plated cells number in the group without radiation treatment.
Cell Apoptosis Assay
Cell apoptosis rate was assessed by Annexin V-FITC/PI apoptosis detection kit (Abcam) referring the instructions of manufacturer. Briefly, 5 × 105 transfected cells with 2 Gy IR treatment were collected and re-suspended in 1 × Binding Buffer. Then cells were double-stained with Annexin V-FITC (5 μl) and PI (5 μl, 50 μg/ml) for 5 min at room temperature in the dark, followed by the quantification using the flow cytometry (BD Biosciences, San Jose, CA, USA). Caspase 3 activity was detected using Caspase 3 assay kit (Abcam, Cambridge, UK) according to the protocol of manufacturer and the optical density value was measured at the wavelength of 400 nm.
Western Blot Assay
The total protein was extracted using RIPA buffer (Beyotime, Shanghai, China) containing cocktail (Roche Diagnostics, Basel, Switzerland) and quantified using a BCA protein assay kit (Thermo Scientific, Rockford, IL, USA). Then equal amount of protein (50 μg) was separated by 10% SDS-PAGE gel and transferred to polyvinylidene fluoride membrane (PVDF; Millipore, Billerica, MA, USA). Next, the membrane was blocked in 5% non-fat milk for 1 h and incubated with primary antibodies against FoxM1 (1:2000), γH2AX (1:2000), Sirt1 (1:800) and β-actin (1:5000) (Cell Signaling Technology, Inc., Beverly, MA, USA) overnight at 4°C. Subsequently, the membrane was probed with horseradish peroxidase (HRP)-conjugated second antibody (1:1000; Santa Cruz Biotech, Santa Cruz, CA, USA) for 1 h at room temperature. Finally, the protein signal was determined by ECL reagent (Millipore, Billerica, MA, USA).
Luciferase Reporter Assay
The 3’UTR sequences of FoxM1 containing wide-type or mutant-type miR-320 binding sites were amplified by PCR and subcloned into pGL3-control vectors (Promega, Madison, WI, USA) to generate FoxM1-wt or FoxM1-mut luciferase reporter vectors. Then the constructed reporter vector was respectively cotransfected with pRL-TK vectors (Promega) and miRNA (miR-NC or miR-320) into U251 and U87 cells. After transfection 48 h, luciferase activity was detected using dual-luciferase reporter assay system (Promega). Luciferase ratio = firefly luciferase activity/Renilla luciferase activity.
Statistical Analysis
All data were displayed as mean ± standard deviation (mean ± SD) from three independent experiments. The comparison of data in different groups was performed by Student's t-test or one-way ANOVA. Differences were considered as statistically significant when P < .05.
Results
miR-320 was Down-Regulated and FoxM1 was Up-Regulated in Radioresistant Glioma Tissues
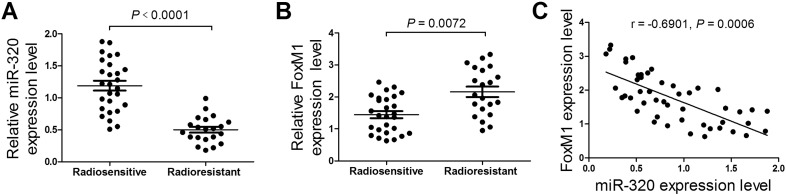
We firstly investigated the expression patterns and correlation between miR-320 and FoxM1 in radiosensitive (n = 27) and radioresistant glioma patient tumors (n = 21). The results showed that miR-320 expression level was strikingly decreased (Figure 1A) and FoxM1 was significantly increased (Figure 1B) in radioresistant glioma tumors when compared with that in radiosensitive tumors. Notably, miR-320 was inversely correlated with FoxM1 expression in glioma patient tumors (Figure 1C). These data indicated miR-320 and FoxM1 might be associated with radioresistance in glioma.
Figure 1.
miR-320 was down-regulated and FoxM1 was up-regulated in radioresistant glioma tissues. The expression profiles of miR-320 (A) and FoxM1 (B) in radiosensitive (n = 27) and radioresistant (n = 21) glioma patient tumors. (C) The correlation analysis between miR-320 and FoxM1 in glioma patients tumors tissues (n = 48). *P < .05.
miR-320 Expression was Gradually Down-Regulated and FoxM1 was Progressively Up-Regulated in Glioma Cells Following Exposure to IR
Subsequently, the effect of irradiation on the expressions of miR-320 and FoxM1 was also detected every 2 h for 24 h after 2 Gy irradiation in glioma cells. As shown in Figure 2, A and B, compared with cells without radiation treatment, miR-320 expression level was markedly reduced after irradiation treatment in U251 and U87 glioma cells. On the contrary, FoxM1 expression at mRNA (Figure 2, C and D) and protein (Figure 2, E and F) levels were both strikingly up-regulated after irradiation treatment in U251 and U87 glioma cells. These results suggested miR-320 and FoxM1 were reversely expressed in glioma cells exposing to IR.
Figure 2.
miR-320 expression level was gradually down-regulated and FoxM1 level was progressively up-regulated in glioma cells following exposure to IR. U251 and U87 cells were treated with 2 Gy IR for different time, followed by qRT-PCR detection of the expression patterns of miR-320 (A and B) and FoxM1 mRNA (C and D) every 2 h for 24 h. (E and F) FoxM1 protein level every 4 h for 24 h was measured by western blot after exposure to 2 Gy in U251 and U87 cells. *P < .05.
miR-320 Overexpression Enhanced Radiosensitivity of Glioma Cells
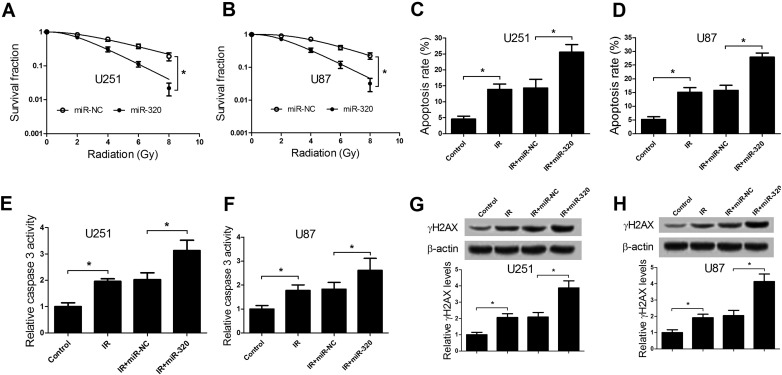
As is known to all, IR treatment can hinder cell proliferation and trigger cell apoptosis along with DNA break. Therefore, we further assessed the effects of miR-320 overexpression on proliferation, apoptosis and γH2AX expression in glioma cells under IR treatment by transfecting with miR-320 or miR-NC. As presented in Figure 3, A and B, colony formation assay manifested that miR-320 overexpression resulted in a marked decrease of survival fraction in U251 (Figure 3A) and U87 cells (Figure 3B), suggesting that ectopic expression of miR-320 enhanced radiosensitivity of glioma cells. Moreover, we further demonstrated that cells apoptotic rate was significantly increased after 2 Gy treatment and miR-320 overexpression strikingly enhanced IR-induced apoptosis in U251 cells (Figure 3C) and U87 cells (Figure 3D), as illustrated by flow cytometry analysis. To further verify this conclusion, caspase 3 activity was also evaluated. As expected, 2 Gy treatment remarkably increased caspase 3 activity, which was dramatically reinforced by forced expression of miR-320 in U251 (Figure 3E) and U87 (Figure 3F) cells. It is generally accepted that DNA repair enhanced resistance of glioma to radiotherapy [24]. Hence, DNA double-stranded break (DSB) induced by IR were detected by measuring histone H2AX phosphorylation (γH2AX) level in U251 and U87 cells. As shown in Figure 3, G and H, IR triggered an obvious increase in γH2AX expression, which was conspicuously augmented by miR-320 restoration. Together, these data revealed that ectopic expression of miR-320 could enhance radiosensitivity of glioma cells.
Figure 3.
miR-320 enhanced radiosensitivity of glioma cells. U251 and U87 cells were transfected with miR-NC or miR-320 for 48 h. (A and B) The transfected U251 and U87 cells were exposed to different doses of IR (0, 2, 4, 6, 8 Gy), survival fraction was detected using colony formation assay. (C and D) The transfected U251 and U87 cells were treated with 2 Gy, followed by the measurement of cells apoptosis rate by flow cytometry analysis. (E and F) Caspase 3 activity was measured in transfected U251 and U87 cells with 2 Gy treatment. (G and H) Western blot analysis of γH2AX protein levels in transfected U251 and U87 cells exposing to 2 Gy. *P < .05.
miR-320 Inhibited Sirt1 Expression by Targeting FoxM1
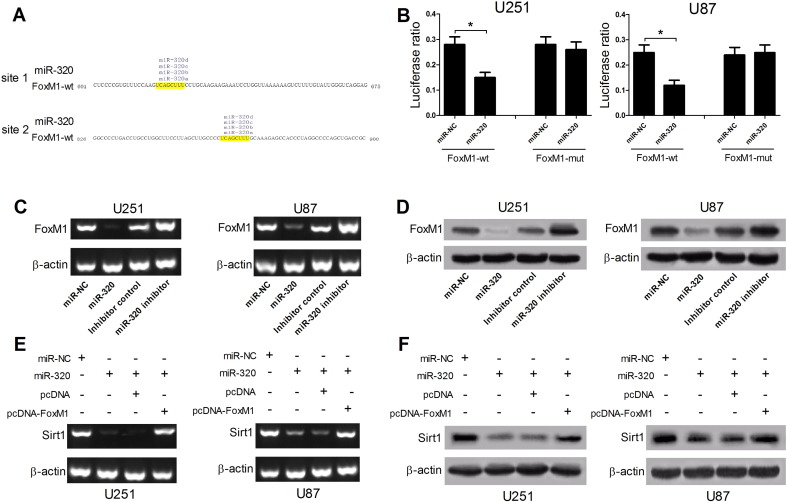
Increasing studies demonstrated that miRNAs exerted function by regulating its target mRNAs expression [25]. Hence, TargetScan, microRNA.org, PicTar or miRbase softwares were employed to search for the potential targets of miR-320. The prediction results showed that there existed two complementary binding regions between FoxM1 and miR-320, indicating FoxM1 was a potential target of miR-320, as shown in Figure 4A. To further confirm this prediction, 3’UTR sequences of FoxM1 containing two miR-320 binding sites were cloned into luciferase reporter vectors to generate FoxM1-wt reporter. Moreover, FoxM1-mut reporter with the corresponding mutant miR-320 binding sites was also constructed. The following luciferase assay revealed that relative luciferase activity of FoxM1-wt was significantly decreased by miR-320 up-regulation in U251 and U87 cells but had no obvious inhibitory effect on luciferase activity of FoxM1-mut (Figure 4B), indicating that FoxM1 interacted with miR-320 by the putative binding sites. Moreover, we further demonstrated that ectopic expression of miR-320 inhibited both FoxM1 mRNA and protein expression, while miR-320 inhibitor facilitated FoxM1 expression in U251 and U87 cells (Figure 4, C and D). Additionally, FoxM1 was reported to be an upstream regulator of Sirt1 and could facilitate Sirt1 expression in glioma cells through binding with Sirt1 promoter region [21]. Thereby, we explored whether miR-320 could affect Sirt1 expression by regulating FoxM1 expression in glioma cells. As expected, miR-320 overexpression strikingly suppressed both Sirt1 mRNA and protein levels in U251 and U87 cells, while ectopic expression of FoxM1 markedly abrogated miR-320-mediated inhibition on Sirt1 expression (Figure 4, E and F). Therefore, these results indicated that miR-320 hindered Sirt1 expression by targeting FoxM1.
Figure 4.
miR-320 inhibited Sirt1 expression by targeting FoxM1. (A) The putative binding sites of miR-320 in 3’UTR region of FoxM1. (B) Luciferase reporter assay was performed to detect luciferase activity in U251 and U87 cells cotransfected with miR-320 or miR-NC and FoxM1-wt or FoxM1-mut. (C and D) The FoxM1 mRNA and protein levels in U251 and U87 cells transfected miR-320, miR-320 inhibitor, or matched controls. (E and F) Sirt1 mRNA and protein levels were detected in U251 and U87 cells transfected with miR-NC, miR-320, miR-320 + pcDNA3.1, or miR-320 + pcDNA-FoxM1. *P < .05.
miR-320 Enhanced Radiosensitivity of Glioma Cells Through Down-Regulation of Sirt1 by Directly Targeting FoxM1
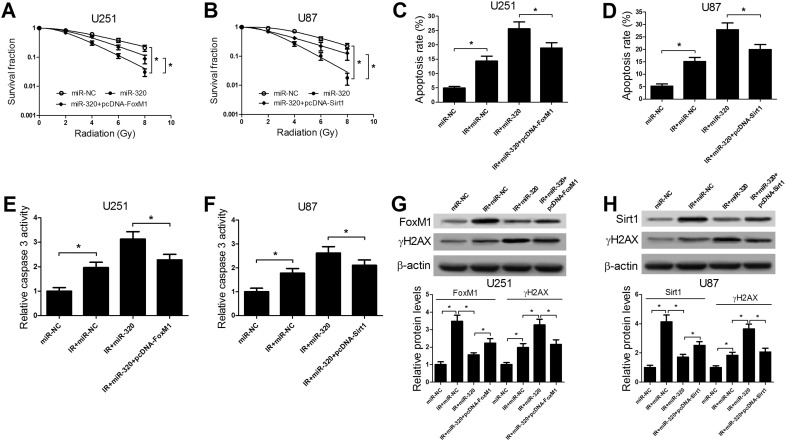
Subsequently, rescue experiments were performed to investigate the effect of FoxM1 and Sirt1 on miR-320-mediated radiosensitivity in glioma cells. Colony formation assay demonstrated that the reintroduction of FoxM1 (Figure 5A) or Sirt1 (Figure 5B) markedly weakened the inhibitory effect of miR-320 on the survival fraction of U251 or U87 cells, indicating that overexpression of FoxM1 or Sirt1 abated miR-320-mediated enhancement effect on radiosensitivity in glioma cells. Meanwhile, we further demonstrated that the exogenous expressions of FoxM1 (Figure 5, C and E) or Sirt1 (Figure 5, D and F) strikingly attenuated the promotive effects on apoptosis and caspase 3 activity conferred by miR-320 overexpression in U251 and U87 cells under IR treatment, as illustrated by flow cytometry analysis and caspase 3 activity assay. Furthermore, miR-320 overexpression-induced increase of the expression of DSB indicator γH2AX was also notably suppressed by forced expression of FoxM1 or Sirt1 in U251 (Figure 5G) and U87 (Figure 5H) cells after IR treatment. Taken together, we further validated that miR-320 enhanced radiosensitivity of glioma cells by modulating FoxM1 and Sirt1 expression.
Figure 5.
miR-320 enhanced radiosensitivity of glioma cells through down-regulation of Sirt1 by directly targeting FoxM1. U251 cells were transfected with miR-NC, miR-320, miR-320 + pcDNA-FoxM1 and U87 cells were transfected with miR-NC, miR-320, miR-320 + pcDNA-Sirt1. (A and B) Colony formation assay was performed to detect survival fraction of transfected U251 and U87 cells, followed by exposure to various doses of IR (0, 2, 4, 6, 8 Gy). (C and D) Cells apoptosis was detected by flow cytometry analysis in transfected U251 and U87 cells treated with 2 Gy. (E and F) Caspase 3 activity was measured by caspase 3 activity assay in transfected U251 and U87 cells treated with 2 Gy. (G and H) FoxM1 or Sirt1 and γH2AX protein levels were detected by western blot analysis in transfected U251 and U87 cells treated with 2 Gy. *P < .05.
Discussion
Emerging evidence has indicated that miRNA dysregulation was closely associated with the development, progression and chemo/radioresistance of cancers [26]. Furthermore, accumulating evidence showed that miRNAs were involved in the regulation of radioresistance of glioma [8]. For instance, miR-181a enhanced U87MG glioma cell radiosensitivity by targeting Bcl-2 [27]. On the contrary, miR-21 knockdown conferred glioma cell radiosensitivity by inhibiting phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) pathway and promoting autophagy [28]. Moreover, overexpression of miR-17-5p improved the xenograft tumor sensitivity to irradiation in glioma [29]. In summary, previous studies manifested that miRNAs exerted diverse functions in regulating radioresistance of glioma.
miR-320 has been identified as a tumor suppressor in several tumors [30]. For example, miR-320 suppressed ovarian cancer oncogenicity by targeting Twist homolog 1 (TWIST1) [31]. miR-320 was down-regulated in cervical cancer tissues and inhibited cervical cancer progression by down-regulation of Mcl-1 [30]. miR-320 was down-regulated in Osteosarcoma (OsC) and heterogeneous expression of miR-320 inhibited cell proliferation and induced cell cycle arrest by targeting E2F transcription factor 1 (E2F1) [32]. FoxM1 has demonstrated to be a vital mediator in cancer initiation, progression and drug resistance [17]. Moreover, miR-320 was demonstrated to enhance radiosensitivity of colon cancer and cervical cancer cells [11], [12]. FoxM1 enhanced glioma radioresistance by modulating SRY-box 2 (Sox2) and signal activator and transducer or transcription (STAT3) [20], [33]. All these studies showed that miR-320 and FoxM1 were closely related with radioresistance of cancer.
In the present study, we demonstrated that miR-320 was down-regulated and FoxM1 was up-regulated in radioresistant glioma tissues and glioma cells exposed to IR. Moreover, miR-320 was inversely correlated with FoxM1 expression in glioma tissues, indicating that miR-320 and FoxM1 might be associated with radioresistance of glioma cells. Moreover, miR-320 overexpression strikingly enhanced radiosensitivity of glioma cells by promoting IR-induced apoptosis and enhancing γH2AX expression. Previous study also demonstrated that DNA repair enhanced resistance of glioma to chemotherapeutics and radiotherapy [24]. Hence, γH2AX as a indicator of DNA DSB was employed to assess DNA repair [34].
Accumulating evidence shows miRNAs exert function by regulating target mRNAs expression [35]. Accordingly, bioinformatics analysis was performed to discover the potential targets of miR-320. The prediction results indicated that FoxM1 was a potential target of miR-320, which was further validated by the following luciferase assay and western blot assay. FoxM1 as a target of miR-320, also has been validated in colorectal cancer [11]. Chetty et al. verified that IR treatment induced FoxM1 expression and FoxM1 knockdown suppressed DNA repair and facilitated IR-induced DNA breaks in lung cancer cells [36], indicating the close association between FoxM1 and radioresistance. Moreover, FoxM1 has been identified as a mediator of radioresistance in glioma [20]. Sirt1, a NAD+-dependent histone deacetylase, plays a critical role in various biological processes [37]. Previous studies manifested that Sirt1 functioned as an oncogene, accompanied by the high-level expression in several tumors such as breast cancer [38], and colon cancer [39]. It was previously suggested that FoxM1 facilitated Sirt1 expression in glioma cells through binding with Sirt1 promoter region [21]. Therefore, we further explored the effect of miR-320 on Sirt1 expression. As expected, miR-320 repressed Sirt1 expression in glioma cells by targeting FoxM1. Mechanistic studies further revealed that miR-320 enhanced radiosensitivity of glioma cells through down-regulation of Sirt1 by targeting FoxM1, which was in accordance with a previous report that miR-320 improved radiosensitivity of human colon cancer cells by targeting FoxM1 [11]. Our study demonstrated that miR-320 could affect Sirt1 expression by miR-320/FoxM1/Sirt1 regulation pathway, while Sirt1 has been demonstrated to be direct target of miR-320 in lung cancer [40]. That is to say, miR-320 may regulate Sirt1 expression by direct or indirect interaction. Moreover, FoxM1 and Sirt1 were linked with p53 signaling [41], [42], indicating that FoxM1 and Sirt1 may participate in development of various cancers due to the vital roles of p53 in multiple biological processes.
In conclusion, our study firstly demonstrated that miR-320 facilitated radiosensitivity of glioma cells through down-regulation of Sirt1 by directly targeting FoxM1. The discovery of regulating mechanism contributed to developing more effective targets and therapy strategies to reverse radioresistance of glioma cells.
Conflicts of Interest
None.
References
- 1.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro-Oncology. 2015;17(Suppl. 4):iv1. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen PY, Reardon DA. Neuro-oncology in 2015: Progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016;12(2):69–70. doi: 10.1038/nrneurol.2015.242. [DOI] [PubMed] [Google Scholar]
- 3.Lou X, Chen T, Huang X, Zheng J, Zheng X, Zhang H, Wu H, Guo J. Radiotherapy plus chemotherapy in the treatment of malignant glioma: a systematic review and meta-analysis. Int J Clin Exp Med. 2016;9(11):20519–20530. [Google Scholar]
- 4.Ho VK, Reijneveld JC, Enting RH, Bienfait HP, Robe P, Baumert BG, Visser O. Changing incidence and improved survival of gliomas. Eur J Cancer. 2014;50(13):2309–2318. doi: 10.1016/j.ejca.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 5.Kim JJ, Tannock IF. Repopulation of cancer cells during therapy: an important cause of treatment failure. Nat Rev Cancer. 2005;5(7):516–525. doi: 10.1038/nrc1650. [DOI] [PubMed] [Google Scholar]
- 6.Osada H, Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis. 2007;28(1):2–12. doi: 10.1093/carcin/bgl185. [DOI] [PubMed] [Google Scholar]
- 7.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer--a brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Besse A, Sana J, Fadrus P, Slaby O. MicroRNAs involved in chemo- and radioresistance of high-grade gliomas. Tumor Biol. 2013;34(4):1969–1978. doi: 10.1007/s13277-013-0772-5. [DOI] [PubMed] [Google Scholar]
- 9.Ishan H. MicroRNA-320 regulates beta-catenin expression and functions in oral cancer progression. Cancer Res. 2008;68:5035. [Google Scholar]
- 10.Yue Z, Dong Q, Wang E. MicroRNA-320 inhibits invasion and induces apoptosis by targeting CRKL and inhibiting ERK and AKT signaling in gastric cancer cells. OncoTargets Ther. 2017;10:1049–1058. doi: 10.2147/OTT.S123324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan LY, Deng J, Xiang XJ, Zhang L, Yu F, Chen J, Sun Z, Feng M, Xiong JP. miR-320 enhances the sensitivity of human colon cancer cells to chemoradiotherapy in vitro by targeting FOXM1. Biochem Biophys Res Commun. 2015;457(2):125–132. doi: 10.1016/j.bbrc.2014.11.039. [DOI] [PubMed] [Google Scholar]
- 12.Yang C, Zhang S, Li J, Yang B, Ouyang W, Mei Z, Chen J, Dai J, Ke S, Zhou F. MicroRNA-320 regulates the radiosensitivity of cervical cancer cells C33AR by targeting β-catenin. Oncol Lett. 2016;12(6):4983–4990. doi: 10.3892/ol.2016.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun JY, Xiao WZ, Wang F, Zhu YH, Wu YF, Miao ZL, Lin YC. MicroRNA-320 inhibits cell proliferation in glioma by targeting E2F1. Mol Med Rep. 2015;12(2):2355–2359. doi: 10.3892/mmr.2015.3657. [DOI] [PubMed] [Google Scholar]
- 14.Dong L, Li Y, Han C, Wang X, She L, Zhang H. miRNA microarray reveals specific expression in the peripheral blood of glioblastoma patients. Int J Oncol. 2014;45(2):746–756. doi: 10.3892/ijo.2014.2459. [DOI] [PubMed] [Google Scholar]
- 15.Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, Major ML, Gusarova GA, Yoder HM, Costa RH. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66(4):2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 16.Juan WU, Zhao J, Yang LN, Yu LI, Yang H. FoxM1 Associated with Cancer Mechanism of Splicing. Prog Mod Biomed. 2015;15(01):171–173. [Google Scholar]
- 17.Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta. 2012;1819(1):28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu TP, Yin L, Zhang EB, De W, Shu YQ. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J Hematol Oncol. 2015;8:50–57. doi: 10.1186/s13045-015-0146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, Huang H, Xue J, Liu M, Wang Y. FoxM1 Promotes β-Catenin Nuclear Localization and Controls Wnt Target-Gene Expression and Glioma Tumorigenesis. Cancer Cell. 2011;20(4):427–442. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y, Kang HK, Dong GK, Cho HJ, Kim Y, Rheey J, Shin K, Yun JS, Choi YS, Lee JI. FoxM1 Promotes Stemness and Radio-Resistance of Glioblastoma by Regulating the Master Stem Cell Regulator Sox2. PLoS One. 2015;10(10):e0137703. doi: 10.1371/journal.pone.0137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu GY, Shi BZ, Li Y. FoxM1 regulates Sirt1 expression in glioma cells. Eur Rev Med Pharmacol Sci. 2014;18(2):205–211. [PubMed] [Google Scholar]
- 22.Ahmed KA, Prakash C, Fulp WJ, Steven E, Torres-Roca JF, Caudell, JJ. The radiosensitivity index predicts for overall survival in glioblastoma. Oncotarget. 2015;6(33):34414–34422. doi: 10.18632/oncotarget.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eschrich SA, Pramana J, Zhang H, Zhao H, Boulware D, Lee JH, Bloom G, Rochalima C, Kelley S, Calvin DP. A gene expression model of intrinsic tumor radiosensitivity: Prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys. 2009;75(2):489–496. doi: 10.1016/j.ijrobp.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frosina G. DNA repair and resistance of gliomas to chemotherapy and radiotherapy. Mol Cancer Res. 2009;7(7):989–999. doi: 10.1158/1541-7786.MCR-09-0030. [DOI] [PubMed] [Google Scholar]
- 25.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Quintana V, Palma-Berre L, Campos-Parra AD, Lopez-Urrutia E, Peralta-Zaragoza O, Vazquez-Romo R, Perez-Plasencia C. MicroRNAs are involved in cervical cancer development, progression, clinical outcome and improvement treatment response. Oncol Rep. 2007;35(1):3–12. doi: 10.3892/or.2015.4369. [DOI] [PubMed] [Google Scholar]
- 27.Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu, P, Hu W. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep. 2010;23(4):997–1003. doi: 10.3892/or_00000725. [DOI] [PubMed] [Google Scholar]
- 28.Gwak HS, Kim TH, Jo GH, Kim YJ, Kwak HJ, kim JH, Yin J, Yoo H, Lee SH, Park JB. Silencing of MicroRNA-21 Confers Radio-Sensitivity through Inhibition of the PI3K/AKT Pathway and Enhancing Autophagy in Malignant Glioma Cell Lines. PLos One. 2012;7(10):e47449. doi: 10.1371/journal.pone.0047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou W, Song L, Zhao Y, Liu Q, Zhang S. Inhibition of Beclin-1-Mediated Autophagy by MicroRNA-17-5p Enhanced the Radiosensitivity of Glioma Cells. Oncol Res. 2017:43–53. doi: 10.3727/096504016X14719078133285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang T, Zou P, Wang T, Xiang J, Cheng J, Chen D, Zhou J. Down-regulation of miR-320 associated with cancer progression and cell apoptosis via targeting Mcl-1 in cervical cancer. Tumour Biol. 2016;37(7):8931–8940. doi: 10.1007/s13277-015-4771-6. [DOI] [PubMed] [Google Scholar]
- 31.Li C, Duan P, Wang J, Lu X, Cheng J. miR-320 inhibited ovarian cancer oncogenicity via targeting TWIST1 expression. Am J Transl Res. 2017;9(8):3705–3713. [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, Li W, Zhang M, Zhu S, Zhang D, Wang X. Inhibitory roles of miR-320 in osteosarcoma via regulating E2F1. J Cancer Res Ther. 2016;12(Supplement):68–71. doi: 10.4103/0973-1482.191635. [DOI] [PubMed] [Google Scholar]
- 33.Maachani UB, Shankavaram U, Kramp T, Tofilon, PJ, Camphausen K, Tandle AT. FOXM1 and STAT3 interaction confers radioresistance in glioblastoma cells. Oncotarget. 2016;7(47):77365–77377. doi: 10.18632/oncotarget.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerić M, Gajski G, Garaj-Vrhovac V. γ-H2AX as a biomarker for DNA double-strand breaks in ecotoxicology. Ecotoxicol Environ Saf. 2014;105(1):13–21. doi: 10.1016/j.ecoenv.2014.03.035. [DOI] [PubMed] [Google Scholar]
- 35.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 36.Chetty C, Bhoopathi P, Rao JS, Lakka SS. Inhibition of matrix metalloproteinase-2 enhances radiosensitivity by abrogating radiation-induced FoxM1-mediated G2/M arrest in A549 lung cancer cells. Int J Cancer. 2009;124(10):2468–2477. doi: 10.1002/ijc.24209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim CS. Human SIRT1: A potential biomarker for tumorigenesis? Cell Biol Int. 2007;31(6):636–637. doi: 10.1016/j.cellbi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Elangovan S, Ramachandran S, Venkatesan N, Ananth S, Gnanaprakasam JP, Martin PM, Browning DD, Schoenlein PV, Prasad PD, Ganapathy V. SIRT1 is essential for oncogenic signaling by estrogen/estrogen receptor α in breast cancer. Cancer Res. 2011;71(21):6654–6664. doi: 10.1158/0008-5472.CAN-11-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabra N, Li Z, Chen L, Li B, Zhang X, Wang C, Yeatman T, Coppola D, Chen J. SirT1 is an inhibitor of proliferation and tumor formation in colon cancer. J Biol Chem. 2009;284(27):18210–18217. doi: 10.1074/jbc.M109.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang K, Gao B, Wei W, Li Z, Pan L, Zhang J, Zhao Q, Chen W, Xu Z. Changed profile of microRNAs in acute lung injury induced by cardio-pulmonary bypass and its mechanism involved with SIRT1. Int J Clin Exp Pathol. 2015;8(2):1104–1115. [PMC free article] [PubMed] [Google Scholar]
- 41.Millour J, De Olano N, Horimoto Y, Monteiro LJ, Langer JK, Aligue R, Hajji N, Lam EW. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol Cancer Ther. 2011;10(6):1046–1058. doi: 10.1158/1535-7163.MCT-11-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JS, Park JR, Kwon OS, Lee TH, Nakano I, Miyoshi H, Chun KH, Park MJ, Hong JL, Kim SU. SIRT1 is required for oncogenic transformation of neural stem cells and for the survival of “cancer cells with neural stemness” in a p53-dependent manner. Neuro Oncol. 2015;17(1):95–106. doi: 10.1093/neuonc/nou145. [DOI] [PMC free article] [PubMed] [Google Scholar]