Abstract
Despite the effectiveness of classic treatments and available diagnostic tools, cancer continues to be a leading world health problem, with devastating cancer-related death rates. Advances in oncolytic virotherapy have shown promise as potentially effective treatment options in the fight against cancer. The poxviruses have many features that make them an attractive platform for the development of oncolytic vectors, with some candidates currently in clinical trials. Here, we report the design and generation of a new oncolytic vector based on the vaccinia virus Western Reserve (WR) strain. We show that the WR-Δ4 virus, with the combined deletion of four specific viral genes that act on metabolic, proliferation, and signaling pathways (A48R, B18R, C11R, and J2R), has effective anti-tumor capabilities in vivo. In WR-Δ4-infected mice, we observed strong viral attenuation, reduced virus dissemination, and efficient tumor cell growth control in the B16F10 syngeneic melanoma model, with enhanced neutrophil migration and activation of tumor antigen-specific immune responses. This approach provides an alternative strategy toward ongoing efforts to develop an optimal oncolytic poxvirus vector.
Keywords: oncolytic vectors, vaccinia virus, virotherapy, immune responses
Introduction
Cancer is one of the main causes of mortality worldwide, with an estimated 8 million related deaths per year (based on data from the GLOBOCAN 2012 project). It thus continues to be necessary to develop more effective treatments to fight the group of diseases that cancer comprises. Oncolytic virotherapy is a promising experimental approach for cancer treatment; it has recently become a real option for oncologists following USA Food and Drug Administration (FDA) approval of IMLYGIC (Amgen), an oncolytic vector based on herpes simplex virus, for treatment of melanoma.1 Oncolytic virotherapy uses replicative viruses, naturally occurring or genetically modified, that selectively infect and lyse tumor cells while leaving healthy tissues unharmed.2 An important feature of these viral vectors is their potential for triggering an innate, followed by an adaptive, anti-tumor immune response.3, 4
Among the poxviruses, vaccinia virus (VACV) is one of the platforms boasting several features that meet requirements for oncolytic virotherapy: (1) it has a rapid replication cycle and lyses infected cells,5 (2) has broad cell tropism,6, 7 (3) has distinct types of viral particles used in different transmission pathways (mature virus [MV] and extracellular virus [EV]),8 (4) has the ability to stably incorporate large transgenes,9 (5) has a cytoplasmic viral cycle with no DNA integration,10 and (6) activates a strong CD8+ T cell response11 and induces the production of neutralizing antibodies.12 With the release of tumor antigens by cell lysis, these features generate protective antitumor immunity.13 A number of oncolytic poxviruses have been developed and are under study in preclinical and clinical trials;14 the most advanced candidates include JX-594 (Wyeth strain)4 and GL-ONC1 (Liverpool strain).15
Western Reserve (WR) is a replicative VACV strain with high lytic capacity and natural selectivity for tumor cells.16 WR can be modified to increase its safety in non-tumorigenic cells by deleting non-essential genes important for somatic infection. While on the one hand the effects derived from these gene deletions restrict mutant virus replication, on the other, the “hallmarks of cancer” within the tumor microenvironment (e.g., sustained proliferation, deregulated metabolism, and immune suppression)17 compensate for these effects, which leads to viral replication.
Here, we studied the effects of a combination of four gene deletions on safety, tumor growth control, and immune response activation. The genes selected were A48R, B18R, C11R, and J2R. A48R codes for thymidylate kinase, an enzyme that participates in nucleotide metabolism; its deletion attenuates WR virus.18 The B18R gene product is a soluble type I interferon (IFN) receptor, a glycoprotein that interferes with anti-viral responses; mutants that lack B18R show reduced virulence in murine model.19 The C11R gene product is known as vaccinia growth factor (VGF), a protein involved in cell proliferation that is homologous to cell epithelial growth factor (EGF).20 Finally, J2R codes for thymidine kinase, another enzyme for nucleotide metabolism, whose deletion attenuates virulence and induces selective in vivo replication in tumor cells.21, 22 The resulting mutant virus with four deletions, WR-Δ4, in which the luciferase gene replaced the J2R gene, had a more attenuated profile than the parental WR and the triple-deletion mutant (WR-Δ3). In the B16F10 syngeneic melanoma mouse model, the treatment with the WR-Δ4 virus led to a marked reduction in tumor growth and increased neutrophil infiltration. These results suggest that the viral vector WR-Δ4 is a potential candidate for tumor cell virotherapy and adds value to our understanding of the mechanisms of action of VACV oncolytic vectors.
Results
Generation of VACV Deletion Mutants WR-Δ3 and WR-Δ4
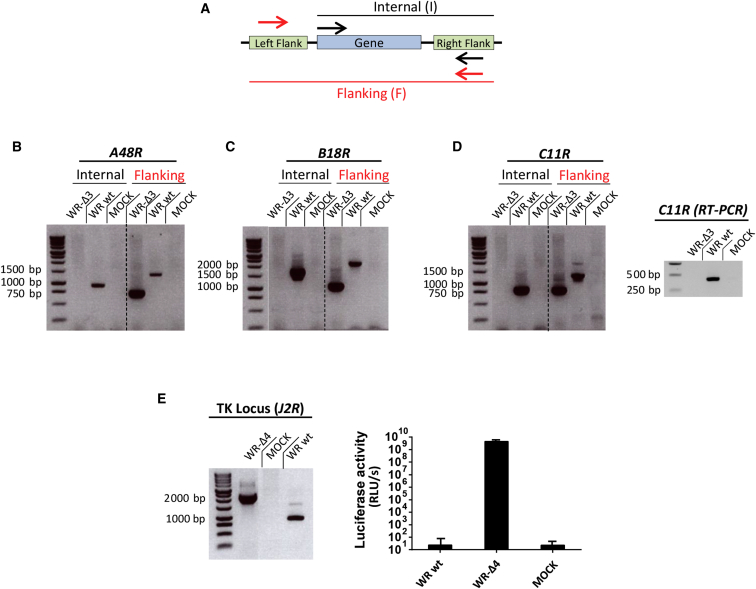
To generate the oncolytic VACV vectors used in this study, we first sequentially deleted selected viral genes. Correct deletion of the targeted VACV genes was confirmed by PCR using a double set of primers to cover both the internal and flanking regions (Figure 1A). Three genes, A48R, B19R, and C11R, were deleted from the WR genome, as indicated by absence of an amplification product for internal regions and by reduction in the PCR fragment obtained when using flanking primers in the mutant virus WR-ΔA48R-ΔB19R-ΔC11R (WR-Δ3) compared with the wild-type virus (Figures 1B–1D). Because the C11R gene is repeated at both ends of the VACV genome, we also analyzed WR-Δ3-infected cells by retro-transcriptase PCR (from total RNA samples), to ensure that both gene copies were completely deleted (Figure 1D, right) and lack of contamination by genomic DNA (data not shown). All deletion mutant viruses were tested by DNA sequencing to determine the correct deletions and absence of mutations in the flanking regions of the deleted genes.
Figure 1.
Generation of the WR-Δ3 and WR-Δ4 Viruses
Viral genes were deleted sequentially from the WR parental virus through homologous recombination using deletion plasmids. (A) PCR strategy was used to confirm the mutant viruses; arrows indicate the amplification regions termed “flanking” (red) and “internal” (black). (B–D) PCR analyses using internal and flanking primer sets were used to confirm deletion of (B) A48R (internal: WR WT, 1,031 bp; flanking: WR WT, 1,423 bp, WR-Δ3, 740 bp), (C) B18R (internal: WR WT, 1,469 bp; flanking: WR WT, 1,890 bp, WR-Δ3, 835 bp), and (D) C11R (internal: WR WT, 794 bp; flanking: WR WT, 1,172 bp, WR-Δ3, 750 bp; RT-PCR: WR WT, 422 bp). To delete the J2R gene, we used homologous recombination to replace the viral gene with the luciferase reporter gene. (E) Left: PCR with flanking primers was used to confirm J2R deletion/luciferase insertion (WR WT, 843 bp; WR-Δ4, 2,106 bp); right: luciferase activity was assayed using a luminometer. Data are shown as mean ± SD. Samples were obtained from infected BSC-40 cells (MOI 1 PFU/cell).
After generation of the WR-Δ3 virus, we eliminated the J2R (TK) gene and replaced it with the luciferase reporter gene. We evaluated correct J2R deletion and reporter gene insertion into the WR-Δ4 genome using PCR, which showed an increase in amplification product size compared with WR wild-type virus (WR WT) (Figure 1E, left). A luciferase assay confirmed correct luciferase expression in WR-Δ4-infected cells (Figure 1E, right). We thus generated recombinant WR-Δ3 virus with three specified gene deletions and WR-Δ4 virus with an additional deletion of the J2R gene that is replaced by fully active luciferase.
In Vitro Characteristics of the WR-Δ3 and WR-Δ4 Viruses
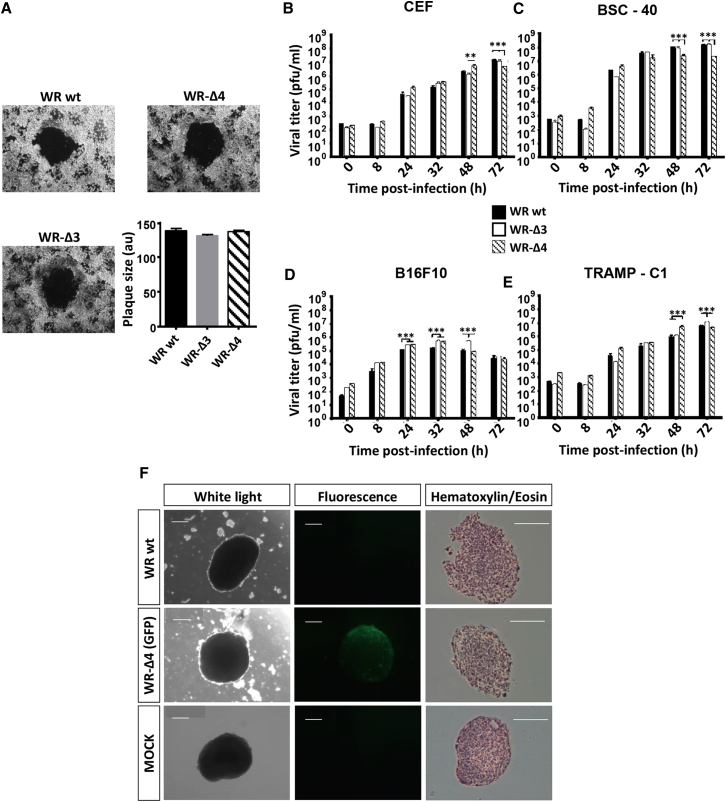
Because cell-to-cell viral spread and degree of infection are major factors in the development of oncolytic poxvirus vectors, we analyzed plaque size phenotype, virus production, and ability to infect spheroids to test whether the deleted genes affected viral growth. In infected African green monkey BSC-40 cells, plaque size and mean area were similar in WR wild-type (WT), WR-Δ3-, and WR-Δ4-infected monolayers, which implied that plaque formation was unaffected by the deletions (Figure 2A).
Figure 2.
Characterization of In Vitro Infection and Replication of the Mutant Deletion Viruses
(A) Plaque size was measured in a BSC-40 monolayer (solid agar medium) at 48 hpi (MOI 0.005 PFU/cell) and the area given in a.u. (B–E) Replication capacity was analyzed through viral growth curves by recovering infected monolayers (MOI 0.01 PFU/cell) at different time points (0, 8, 24, 32, 48, and 72 hpi) followed by titration; the cell lines used were (B) CEF, (C) BSC-40, (D) B16F10, and (E) TRAMP-C1. (F) Spheroids of TRAMP-C1 cells were infected (MOI 2 PFU/cell) and observed after 6 days. Left: white light; center: fluorescence filter for GFP; right: H&E-stained histological sections. Scale bars, 500 μm. Data are shown as mean ± SD. **p < 0.01; ***p < 0.005.
We evaluated the replicative capacity of the deletion mutant viruses by viral growth kinetics in several cell lines: primary (CEF; chick), immortalized (BSC-40), and murine tumor cells (B16F10 and TRAMP-C1). Cultured cells were infected and collected at various times post-infection (0, 8, 24, 32, 48, and 72 hr), viral plaques were stained with crystal violet, and virus yields were determined by titration in BSC-40 cells. Viral growth kinetics profiles for the WR-Δ3 and WR-Δ4 mutants and the parental WR WT were similar for the different cell types (Figures 2B–2E). B16F10 cells nonetheless showed increased lysis by all the WR viruses (Figure 2D), in contrast with the other cell lines. Infection of TRAMP-C1 cell spheroids with WR-Δ4/GFP (with the same deletions as in WR-Δ4 and expressing GFP) showed wide infection spread (as indicated by GFP expression) and a scattered phenotype in histological sections compared with mock-infected spheroids; these effects are attributed to cytolytic infection of the most exposed regions (Figure 2F). WR-Δ3 and WR-Δ4 virus replication capacities were unaffected in cell lines of various origins, including tumor cells and tumor-like spheroids.
Deletion Mutant Viruses WR-Δ3 and WR-Δ4 Show Decreased Virulence in Mice
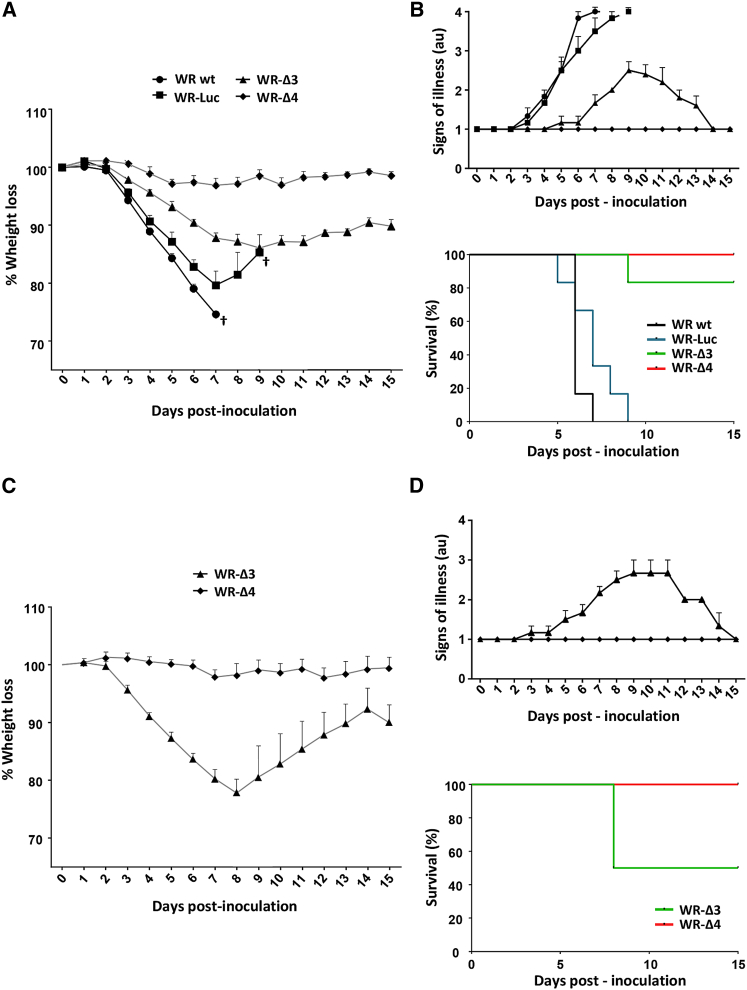
Vector safety is a crucial parameter for the therapeutic use of replication-competent viruses. To define the virulence of the oncolytic candidate vectors, we administered WR-Δ3 or WR-Δ4 intranasally (i.n.) to C56BL/6 mice (5 × 106 or 5 × 107 plaque-forming units [PFU]/mouse); control mice received 5 × 106 PFU/mouse of WR WT or WR-Luc (a single J2R deletion replaced with luciferase). Mice were monitored daily throughout the experiment for weight loss and signs of illness (loss of mobility, troubled breathing, hunched posture, absence of grooming, or inflammation of the eye membrane).
The 5 × 106 PFU/mouse virus dose was lethal for control groups infected with WR WT and WR-Luc, with severe loss of body weight within 7 days post-inoculation (Figure 3A), accompanied by associated signs of illness in infected animals (Figure 3B, top). Mice inoculated with WR-Δ3 showed a slight body weight reduction (up to 14%), with a peak at day 9 post-infection that coincided with the appearance of signs of illness (Figures 3A and 3B, top); thereafter, 80% of the mice recovered body weight and healed completely from the infection (given the absence of signs of illness at the end of the experiment). In contrast, all mice infected with WR-Δ4 showed stable weight and complete absence of symptoms attributable to the viral infection (Figures 3A and 3B, top). All mice inoculated with low WR WT and WR-Luc doses required sacrifice, whereas those that received WR-Δ3 and WR-Δ4 showed 80% and 100% survival, respectively (Figure 3B, bottom).
Figure 3.
Analysis of In Vivo Virulence of the Mutant Deletion Viruses
C57BL/6 mice were inoculated (i.n.) at various virus doses; after infection, weight and signs of illness were evaluated daily until day 15. (A and B) Mice infected with 5 × 106 PFU/mouse (WR WT, WR-Luc, WR-Δ3, and WR-Δ4) were evaluated for (A) percent weight loss, (B, top) signs of illness, and (B, bottom) percent survival. †Indicates that all mice in the group were sacrificed because of viral infection. (C and D) Mice infected with 5 × 107 PFU/mouse (WR-Δ3, WR-Δ4) were evaluated for (C) percent weight loss, (D, top), signs of illness, and (D, bottom) percent survival. Signs of illness were evaluated by assigning a score from 1 to 4 (1 = absence of illness and 4 = severe illness), considering loss of mobility, trouble breathing, hunched posture, absence of grooming, and inflammation of the eye membrane. Mice were sacrificed when they showed a body weight reduction >20% or had an illness score of 4. Data are shown as mean ± SD.
In mice infected with a high WR-Δ3 dose (5 × 107 PFU/mouse), we observed a decrease in body weight with time. At day 8, 50% of the mice had to be sacrificed, whereas the rest of the group recovered completely (Figures 3C and 3D, top). None of the mice infected with 5 × 107 PFU of WR-Δ4 showed weight loss or signs of disease related to the viral infection (Figures 3C and 3D, top). The lethal dose 50 (LD50) for WR-Δ3 was ∼5 × 107 PFU, whereas mice inoculated with the same dose of WR-Δ4 were unaffected (100% survival; Figure 3D, bottom). The four viral genes deleted in WR-Δ4 thus conferred 100% survival in infected mice in conditions in which all mice infected with 1 log lower dose of WR WT did not survive.
Deletions in WR-Δ4 Restrict Viral Replication in Mice
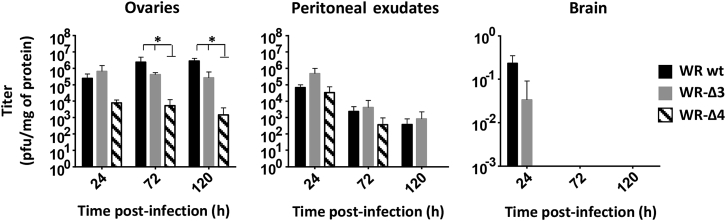
To study the tissue distribution of the deletion mutant viruses after systemic delivery, we inoculated C57BL/6 mice intraperitoneally (i.p.) with 2 × 107 PFU/mouse of WR WT, WR-Δ3, or WR-Δ4 and analyzed viral load at 24, 72, and 120 hr post-infection (hpi) in mouse ovary, peritoneal exudate cells (PECs), and brain tissue. In ovaries, virus titers were similar in WR WT- and WR-Δ3-infected mice, whereas ovaries from WR-Δ4-infected mice showed a significantly lower (p < 0.05) viral titer than that in the other two groups: WR WT and WR-Δ3 (Figure 4). For PEC titers, all three infected mouse groups had comparable virus levels at 24 and 72 hpi. At 120 hpi, we were unable to detect virus in PEC of WR-Δ4-infected mice (Figure 4). In brain, we observed very low WR WT titers, and there was no detectable virus in the WR-Δ3 or WR-Δ4 groups after 24 hr of infection (Figure 4). The combined deletions in WR-Δ4 thus led to a virus whose tissue distribution profile was similar to that of WR WT, whereas WR-Δ4 titers were lower in ovaries and PECs.
Figure 4.
Characterization of Biodistribution of WR-Δ3 and WR-Δ4 Viruses
For the biodistribution assay, C57BL/6 mice received a dose of 2 × 107 PFU/mouse (i.p.); were sacrificed at 24, 72, and 120 hpi; and ovaries, peritoneal exudates, and brain were processed and titrated. Data are shown as mean ± SD. *p < 0.05.
WR-Δ3 and WR-Δ4 Elicit Cell Migration in Mice
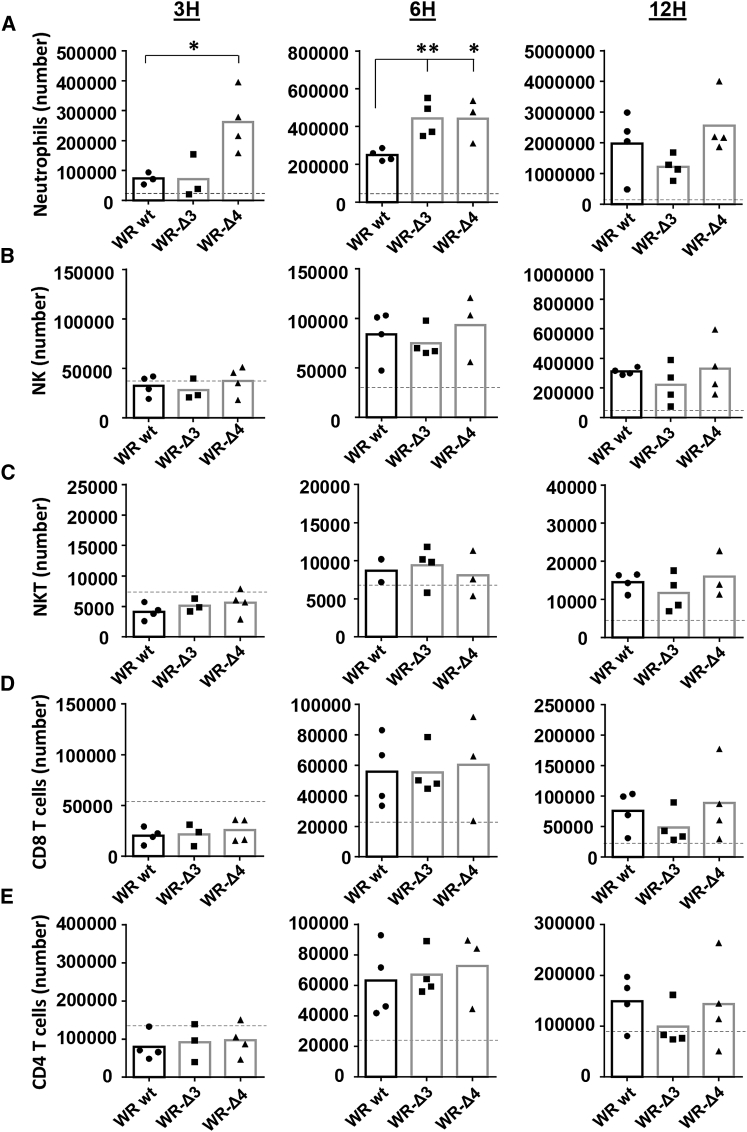
Because in the context of a poxvirus infection migration of neutrophils to the site of virus infection can affect CD8+ T cell activation,23 we next characterized the migration profile of murine innate immune cells elicited by WR-Δ3 and WR-Δ4 viruses; we injected WR WT, WR-Δ3, WR-Δ4 (1 × 107 PFU/mouse, i.p.), or PBS, and evaluated in the peritoneal cavity absolute numbers of cell populations 3, 6, and 12 hpi. This virus dose was lower than for intratumoral to limit the cytopathic effects of the virus in cells of the peritoneal cavity and maintain their integrity. Of the populations analyzed, we found a significant early (3 hpi) increase in the number of neutrophils recruited in WR-Δ4-infected mice, whereas levels were similar in WR WT and WR-Δ3 groups (Figure 5A). This enhanced neutrophil number was maintained at 6 hpi, when WR-Δ3 and WR-Δ4 reached similar levels, higher than those in WR WT (Figure 5A). At 12 hpi, the overall number of neutrophils recruited to the infection site increased ∼10-fold compared with the numbers at 3 hpi, with no significant differences between all virus groups (Figure 5A).
Figure 5.
Characterization of Innate Immune Cell Population Recruitment by WR-Δ3 and WR-Δ4 Viruses In Vivo
For analysis of innate immune cell responses, mice were inoculated with 1 × 107 PFU/mouse (i.p.), and peritoneal cavity cell populations were analyzed at 3, 6, and 12 hpi. (A–E) Absolute numbers are shown for (A) neutrophils, (B) NK cells, (C) NKT cells, (D) CD8 T lymphocytes, and (E) CD4 T lymphocytes. Dashed lines indicate mean values for PBS-inoculated mice in each case. *p < 0.05; **p < 0.01.
Of other innate immune cells analyzed (NK, NKT, CD8, and CD4 T cells), all groups showed a comparable profile at all three time points, with recruitment levels of these cells resembling that of PBS-control mice at 3 hpi, with higher levels at 6 and 12 hpi (Figures 5B–5E). These data indicate that neutrophil recruitment to the infection site was enhanced early in infection by WR-Δ4, an effect not observed for any virus group for other immune system cells (natural killer [NK], NK T [NKT], CD8, and CD4 T cells). Immune system cells such as NKs or CD8 and CD4 T lymphocytes, which have anti-tumor-associated roles, thus remained unaffected.24, 25
WR-Δ4 Shows Potent Oncolytic Activity against Syngeneic B16F10 Tumors
To evaluate the anti-tumor effectiveness of the mutant viruses, we injected melanoma B16F10 cells intradermally (i.d.) into C57BL/6 mice, followed 7 days later by intratumor (i.t.) inoculation of 1 × 108 PFU/mouse of WR WT, WR-Δ3, WR-Δ4, or PBS. A single virus dose was used to define a priming effect on tumor growth; the high virus dose was to provide a sufficient amount of infective particles to ensure infection of the tumor with about 50-mm3 volume. General mouse well-being, weight, and tumor volume were followed up daily.
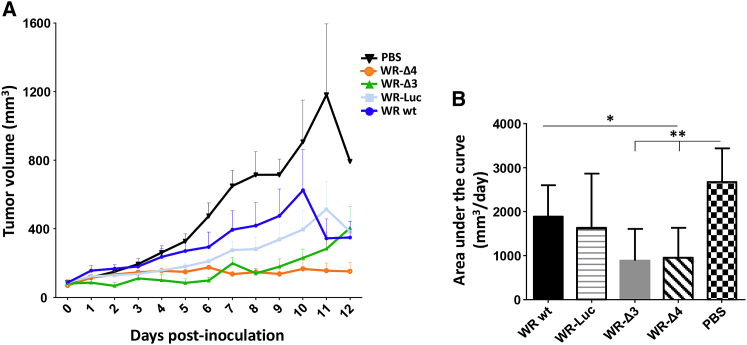
Infection of tumors with WR WT or WR-Luc led to a slight reduction in tumor growth compared with the PBS-treated group (Figure 6A), which can be attributed to intrinsic VACV oncolytic capacity. When mice were infected with WR-Δ3 or WR-Δ4 viruses, a strong reduction in tumor proliferation was sustained throughout the experiment in the case of WR-Δ4 and until day 8 post-treatment in the WR-Δ3 group (Figure 6A). None of the mice showed negative effects on state of health as a result of the tumor or the virus treatment (data not shown).
Figure 6.
Anti-tumor Efficacy Analysis Using the Syngeneic B16F10 Transplant Model
To evaluate viral effectiveness against tumors, C57BL/6 mice received 5 × 105 B16F10 melanoma cells/mouse (i.d.). When the tumor reached a volume of 50 mm3, mice were treated (i.t.) with PBS or 1 × 108 PFU/mouse WR WT, WR-Luc, WR-Δ3, or WR-Δ4. (A) Tumor volume was measured daily, and mice were sacrificed when the tumor reached 1,500 mm3. Data are shown as mean ± SEM. (B) Area under the curve was calculated considering the tumor volume of each mouse during the experiment. Data are shown as mean ± SD. *p < 0.05; **p < 0.01.
To quantify the results, we calculated and compared the area under the curve for tumor volume for each treated mouse. WR-Δ3- and WR-Δ4-treated groups both showed a significant reduction in cubic millimeters (mm3) × day values compared with the PBS-treated group; the reduction for the WR-Δ4 group was also significant relative to that of the WR WT group (Figure 6B). It was therefore clear that the combination of gene deletions in the oncolytic vectors WR-Δ3 and WR-Δ4 translated to increased anti-tumor effectiveness in the B16F10 syngeneic model.
Analysis of the Anti-tumor Response in WR-Δ4-Treated Mice
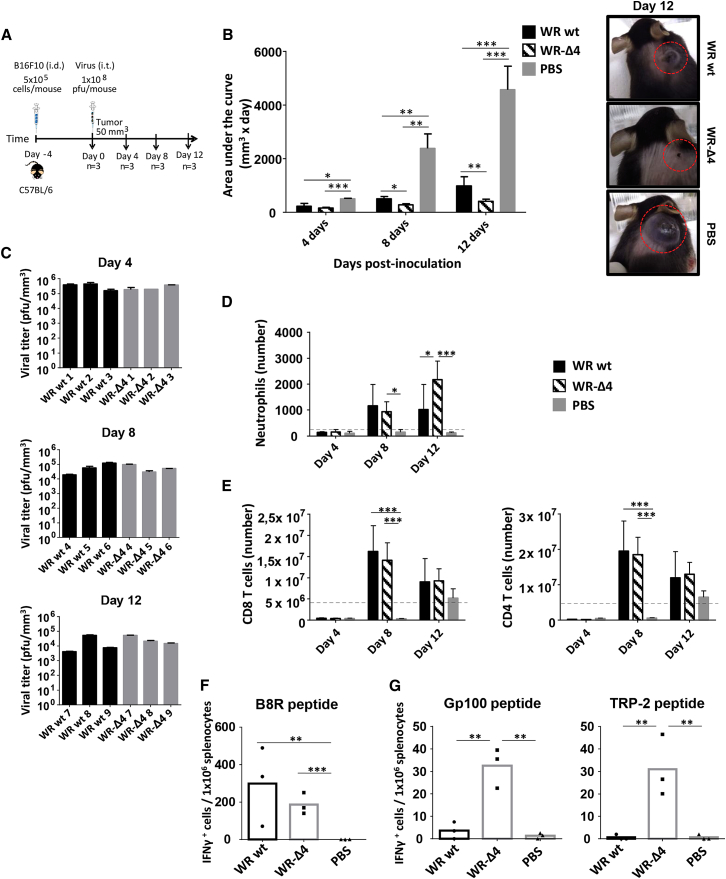
Because WR-Δ4 showed the most potent oncolytic activity and a greater reduction in virulence, we further characterized this virus for its oncolytic potential and possible correlates of protection. We injected B16F10 cells (i.d.) into C57BL/6 mice and infected the tumor (i.t.) 4 days later with WR-Δ4 (1 × 108 PFU). Other mouse groups with tumors received inoculations of WR WT or with PBS (control). At various times post-infection (days 4, 8, and 12), we determined the extent of tumor growth, presence of infectious virus in tumors, and innate and tumor-specific adaptive immune responses (Figure 7A). An examination of tumor proliferation showed significant differences in tumor size between virus- and PBS-treated mice at various time points (Figure 7B). Comparison of WR WT and WR-Δ4 showed a significantly greater reduction in tumor growth in the deletion mutant (Figure 7B). The extent of tumor reduction is observed in a representative photograph (Figure 7B) on day 12, with potent inhibition of tumor size in the WR-Δ4-treated mice versus WR WT and PBS groups. The presence of infectious virus in the treated tumors was characterized by virus plaque titration; excised tumors were treated with collagenase, processed, and titrated (titers were adjusted to tumor volume in each case). The virus-treated groups showed similar yields of WR WT and WR-Δ4 at days 4, 8, and 12 (Figure 7C), maintaining high levels of infectious virus until day 12.
Figure 7.
Characterization of WR-Δ4 Virus Anti-tumor Features
(A) Scheme of the immunization protocol. C57BL/6 mice received 5 × 105 melanoma B16F10 cells/mouse (i.d.); when tumor volume reached 50 mm3, mice were treated (i.t.) with PBS or 1 × 108 PFU/mouse WR WT or WR-Δ4. Mice were sacrificed 4, 8, and 12 days after for characterization of viral replication, and innate and adaptive immune responses elicited. Tumor volume was measured daily. (B) Area under the curve was calculated considering the tumor volume of each mouse during the experiment. (C) Viral titer of homogenized tumors relative to tumor volume for each mouse. (D and E) The innate immune response is given as absolute numbers (relative to tumor volume for each mouse) of (D) neutrophils in tumor and (E) CD8 and CD4 T lymphocytes in spleen at 4, 8, and 12 days after treatment. Dashed lines indicate mean values for pretreated mice (day 0) in tumors or in spleen. (F and G) Adaptive immune response evaluated using ELISpot on splenocytes from mice at 12 days after stimulation with (F) B8R peptide and (G, left) gp100 and (G, right) TRP-2 peptides. Data are shown as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.005.
To help explain the oncolytic potential of the WR WT and WR-Δ4 vectors, we used flow cytometry to analyze immune cell populations in the tumors and spleens of treated mice. The WR-Δ4-infected tumor showed an increase in infiltrated neutrophil numbers throughout the infection period; neutrophil recruitment to the tumor was significantly higher at day 12 and surpassed the values for the WR WT group (p < 0.05) (Figure 7D), whereas infiltrated neutrophil numbers in PBS-treated mice remained low. For the remaining immune cells analyzed (NK, NKT, CD8 and CD4 T cells, macrophages, monocytes, B cells, and dendritic cells), all mice bearing virus-treated tumors had variable levels of immune cells, lower than values for PBS-treated mice (data not shown). In the case of immune cell populations in the spleen, a central organ in adaptive immune response development, we observed a significant increase in the number of CD8 and CD4 T cells that migrated to the spleen in WR-Δ4-treated mice at day 8 compared with PBS-treated mice (Figure 7E). At day 12, values decreased to levels slightly higher than those in the PBS group, but still higher than the initial numbers of CD8 and CD4 T cells in spleen (Figure 7E). The remaining spleen immune cells analyzed showed no differences between the assayed groups at various time points (data not shown).
To determine whether WR WT-, WR-Δ4-, or PBS-treated mice with tumors develop specific immune responses to tumor antigens and to the virus, we performed an IFNγ ELISpot assay using splenocytes at day 12 post-treatment. We used B8R as a VACV-specific peptide stimulus, and glycoprotein (gp)100 and TRP-2 peptides as B16F10 CD8-specific tumor antigens. Splenocytes from both virus-treated groups showed a specific IFNγ response to the VACV peptide, which was not seen in the PBS group (Figure 7F). Only WR-Δ4-treated mice had an IFNγ response to tumor-specific gp100 and TRP-2 peptides, which is significant relative to the WR WT and PBS groups (Figure 7G). The tumor-specific immune response in splenocytes from the WR WT group was negligible, similar to that of the PBS group (Figure 7G).
Discussion
The basis for development of an optimal oncolytic viral vector includes requirements for safety, ability to infect and destroy tumor and stromal cells, and capacity to trigger tumor-specific immune responses.4, 26 To develop replication-competent oncolytic VACV vectors, we defined and characterized a novel vector, WR-Δ4. This vector is based on the VACV WR strain and has a unique set of four deletions of the viral genes A48R, B19R, C11R, and J2R, some of which are reported to be effective in oncolytic vectors.27, 28, 29 The deletions in WR-Δ4 did not affect its infection and replication capacity in cultured cells of various origins, and its plaque phenotype is comparable with that of the WR WT. Despite the deletions, WR-Δ4 maintains infection capacity, replication, and spread characteristics in cell monolayers, all critical factors for an effective anti-tumor response.13 Spheroids are a valuable tool with which to infer viral infection and spread in the context of a solid tumor mass in vitro and provide reliable information before in vivo analyses.5, 30 In our study, WR-Δ4 showed a high capacity to infect these 3D tumors, with broad homogeneous infection and lysis of exposed regions of murine prostate TRAMP-C1 cell spheroids, as also observed for the WR WT virus.
Attenuation is a determining factor when a replication-competent virus is used in therapy for patients, most of whom are immunocompromised;13, 31 we therefore studied viral pathogenesis in mice infected with the mutant viruses. A major advantage of attenuation is to limit the spread of the virus to normal tissues other than to the tumor, thus avoiding complications of a systemic infection and reducing immune responses to the vector while enhancing those to the tumor antigens. WR-Δ4 shows increased virus attenuation, attributable to the specific combination of deletions, some of which are associated with reduced virulence (ΔA48R, ΔJ2R18, 21, 32). Compared with the parental virus, WR-Δ4 shows reduced infection of ovaries (a main VACV target tissue33, 34, 35) with lower titers in peritoneal exudates and is totally absent in the brain, thus avoiding the neurotropism of the WR strain.36, 37 Because the innate immune response constitutes one of the first defenses in the response to infection and to tumors, and the type of response elicited depends on the infectious agent involved,38 we determined the effect of WR-Δ4 on immune system cells. Analysis of innate immune cell migration showed that the four WR-Δ4 deletions not only resulted in a significant increase in neutrophils that migrated to the infection site, but also maintained the number of cells with roles in activating immune responses to infectious agents and to tumors (NK, NKT, CD8, and CD4 T cells).
In the B16F10 syngeneic mouse model, we observed that WR-Δ3 and WR-Δ4 viruses caused comparable reductions in in vivo tumor growth, which surpassed the limited reductions by the parental WR WT or WR-Luc. These results also confirm the absence of a contribution to tumor growth reduction by the luciferase reporter gene or the single J2R deletion, as reported previously.39 The reason why the WR-Δ3 virus did not gave the same potency as the WR-Δ4 virus has to do with the additional deletion of the TK gene and the combination of the four deleted virus genes, A48R, B19R, C11R, and J2R. Elsewhere we will show that modulation of immune responses of a poxvirus vector depends on the combination of virus gene deletions (C.E. Gomez, B. Perdiguero, C.O.S. Sorzano, M.E., personal communication).
To explain the anti-tumor phenotype of the WR-Δ4 virus, we considered several factors that might correlate with the reduced tumor proliferation. WR-Δ4 infects tumors efficiently and replicates at a rate similar to that of WR WT, which allows the virus to take over the tumor with time, shown by the sustained presence of infectious virus at day 12 post-treatment. Recruitment of immune system cells during virus infection of the tumor contributes to an effective anti-tumor response.4 In our study, WR-Δ4-treated B16F10 tumor-bearing mice showed improved ability to recruit tumor-infiltrating neutrophils (as also observed in the peritoneal cavity). This observation coincides with the importance of neutrophils as one of the first immune cells to arrive and combat infection.40 These cells have a critical role during VACV infection,41 because they act as antigen-presenting cells and improve the specific immune response;23 in addition, they are linked to oncolytic efficiency.42, 43 The anti-tumorigenic influence of virus-stimulated neutrophils is also reported in the B16F10 syngeneic tumor model.44
Another factor is WR-Δ4 induction of tumor-specific immune responses, as seen in splenocytes from tumor-bearing mice. Tumor-associated antigens (TAAs) are overexpressed proteins related to tumor cell specifically. As self-proteins, they are tolerated by the immune system, and their specific interactions with immune cells are restricted to low-avidity interactions. Generation of an adaptive immune response specific for TAAs is thus one of the most desirable factors for any anti-tumor therapy.25, 45, 46 We found stimulation of a specific IFNγ+ immune response to the B16F10 TAA gp100 and TRP-2 peptides exclusively in splenocytes from the WR-Δ4-treated group. Because the tumor cells were the only source of these TAAs for all groups, the WR-Δ4 virus appears able to break immune tolerance to tumor antigens and to elicit a specific adaptive immune response. This might assist and reinforce the anti-tumor effects associated to WR-Δ4 treatment, following administration of a single dose of virus.
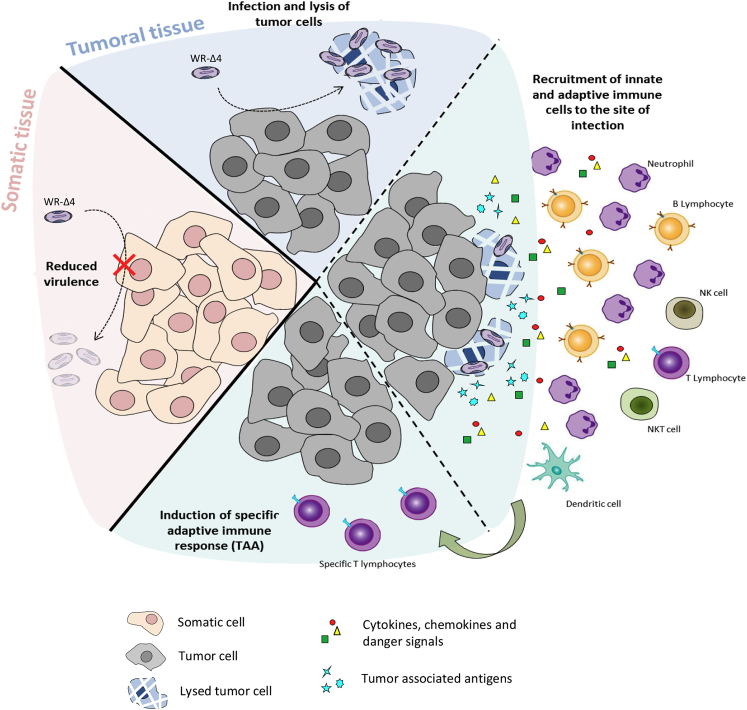
In clinical trials, oncolytic poxviral vectors have shown anti-tumor activity through various strategies that include combination of deletions, such as JX-594/Pexa-Vec (ΔJ2R + GM-CSF), GL-ONC1 (ΔJ2R/ΔA56R/ΔF14.5L), and WR-vvDD (ΔJ2R/ΔC11R).14 Based on studies of the immunoregulatory role of specific gene deletions in the poxvirus vectors MVA and NYVAC, we established that deletions could augment the induction of specific immune responses to foreign antigens.23, 47, 48 The nature and combination of these deletions is an important criterion in the design of optimal VACV vectors for vaccination. We used various viral deletions that contribute to the oncolytic phenotype in several viral vectors (B18R, C11R, and J2R) and combined them with ablation of the A48R function (previously unreported for oncolytic use) in the single WR-Δ4 vector. A48R, also implicated in nucleotide metabolism, was deleted in combination with the other viral genes to take advantage of tumor microenvironmental properties, given the abundance of nucleotides within the tumor that promote mutant virus growth. We found that these deletions did not affect viral infection or replication in vitro and increased safety, given the reduction in in vivo virulence and pathogenicity. This deletion mutant proved to have marked characteristics as an oncolytic vector, as it shows anti-tumor efficacy, sustained replication in the tumor, ability to induce neutrophil recruitment to the tumor, and capacity to break immune tolerance by activating tumor-specific immune responses. These properties are illustrated in a scheme that includes the “safety/oncolytic” equilibrium of the specific combination of the four deleted viral genes (Figure 8). Whether our vectors had oncolytic advantages over those in clinical development (like JX594 or GL-ONC1) cannot be determined unless head-to-head comparisons are performed.
Figure 8.
Model of Action and Biological Features Triggered by the Oncolytic Vector WR-Δ4
The VACV-based oncolytic vector WR-Δ4 combines four deletions (A48R, B18R, C11R, and J2R) that result in reduced virulence in healthy tissues (increased safety) and an anti-tumor response through infection and replication in tumor cells (with associated release of cytokines, chemokines, danger signals, and tumor-associated antigens), and stimulate innate and adaptive (TAA-specific) immune responses.
These results, using the WR-Δ4 virus deletion mutant in a single-dose immunization protocol to treat the melanoma tumor, highlight the potential of this vector for tumor therapy. Although improvements remain, such as transgene insertion to boost the anti-tumor response (TAAs, cytokines, suicide genes, stimulatory molecules4, 31), or the combination with anti-PD1/CTLA-4 antibodies used in immunotherapy and/or with co-stimulatory molecules.49, 50 Our study contributes to the growing field of oncolysis by laying the foundations of a new vector for anti-cancer treatment.
Materials and Methods
Cells
Cercopithecus aethiops kidney cells (BSC-40; American Type Culture Collection [ATCC]), B16F10 melanoma cells (ATCC, Barcelona, Spain), and chicken embryo fibroblast (CEF) primary cells (Intervet, Salamanca, Spain) were cultured in DMEM supplemented with 10% (v/v) newborn calf serum (NCS), or with fetal calf serum (FCS) for CEF cells. Murine TRAMP-C1 prostate cancer cells (ATCC, Barcelona, Spain) were cultured as described previously.51 Mouse splenocytes were cultured in RPMI 1640 medium (Sigma-Aldrich Chemie, Steinheim, Germany) supplemented with 10% FCS (v/v) and 50 μM β-mercaptoethanol. Cells were cultured at 37°C at 80% humidity and 5% CO2.
Plasmids
For deletion of the A48R, B18R, and C11R viral genes, pGEM-RG-ΔA48R, pGEM-RG-ΔB18R, and pGEM-RG-ΔC11R were generated by cloning the flanking region of each gene into the pGEM-RG plasmid, to delete the genes by homologous recombination. The plasmids bore the GFP gene between two left flanking regions, and this reporter gene was eliminated through consecutive passages.
The pLZAW1-Luc plasmid (generated in our laboratory by Dr. Mauro Di Pilato) was used to insert the luciferase gene into the thymidine kinase (TK) locus (J2R).
Viruses and Virus Generation
All viruses used in this study are based on the WR strain. The WR wild-type (WT) virus was kindly provided by Dr. Rostom Bablanian (State University of New York, Brooklyn, NY, USA). The WR-Luc virus bearing the luciferase reporter gene (Photinus pyralis) under the control of the early/late p7.5 promoter replacing the viral J2R gene was generated in our laboratory.32
For the WR-Δ3 deletion mutant virus (with deletions in A48R, B18R, and C11R), monkey BSC-40 monolayers were infected with the parental virus (MOI 0.01 PFU/cell); at 1 hpi (hours post-infection), cells were transfected with 8 μg of the deletion plasmid (one for each gene targeted) for 5 hr. At 48 hr after the infection/transfection (or when a patent cytopathic effect was observed), monolayers were collected in 1 mL of fresh serum-free DMEM, subjected to three freeze/thaw cycles, and serially diluted for infection of new BSC-40 monolayers. After several passages of plaque purification in cell culture, the GFP and DS-Red fluorescence reporter genes were used to select and isolate the deletion mutant virus, as reported previously.52 Finally, a deletion mutant virus without the target viral gene or any reporter genes was obtained. Each gene was deleted sequentially.
A similar protocol was followed to generate the WR-Δ4 virus, except that the luciferase reporter gene (under the control of the late/early optimized LEO160 [pLEO160] promoter53) was inserted to replace the J2R gene. In this case, the insertion plasmid pLZAW1-Luc was used instead of a deletion plasmid, and the mutant virus was selected and isolated with the reporter gene LacZ (β-galactosidase), as reported previously.54 Luciferase activity was measured in lysed BSC-40-infected cells using the Luciferase Assay System (Promega, Madison, WI, USA) and a Lumat LB 9501 luminometer (Berthold Technologies). Virus stocks for in vivo use were grown, purified from infected BSC-40 cells through two sucrose cushions (36% w/v), and titrated in BSC-40 cells as described previously.55
PCR and RT-PCR
PCR analysis of genomic viral DNA extracted by the SDS/proteinase K/phenol method from infected BSC-40 cells was used to confirm correct deletion of the target genes A48R, B18R, and C11R. Primers used were: A48R left flank: 5′-GTCATTGGGCCCTCGATCATGAATGG-3′, A48R right flank: 5′-CTATACAGGATCCAATTTCATTGTCG-3′, A48R internal-R: 5′-CCAGTAACCGTG TGTATAGC-3′, B18R left flank: 5′-CGGGCCCATTTTCAAAATACATGTCG-3′, B18R right flank: 5′-CGGGATCCTACTAGTTGTGTACTTTGATC-3′, B18R internal-R: 5′-CCACGACATTTATATGT ATTACC-3′, C11R left flank: 5′-CGGGCCCTCGTTTATTCAGATCGCAGTG-3′, C11R right flank: 5′-CGGGATCCAACAGGAATATAGCATGGGAC-3′, C11R internal-R: 5′-GCACAACCATATCTT GTATAGG-3′. TK-L and TK-R primers were used as described previously54 to confirm elimination of the J2R gene and its replacement with the luciferase gene.
For retrotranscription analyses, total RNA was extracted from infected cells using the RNeasy Mini kit (QIAGEN, Germantown, MD, USA) and treated with DNase I (RNase free; Roche Diagnostics, Mannheim, Germany). cDNA was obtained using SuperScript III reverse transcriptase (Invitrogen, Life Technologies Europe, Bleiswijk, the Netherlands) with Oligo dT12–18 (Invitrogen,) following manufacturer’s recommendations. The cDNA obtained was used in PCR to test for C11R expression; primers used were C11R internal-F: 5′-GATGTTGTTGTTCGCTGCTATG-3′ and C11R internal-R: 5′-GCACAACCATATCTTGTATAGG-3′.
Evaluation of the Size of Virus Plaques in Infected Cells
In order to determine the plaque size from the various viruses assayed, we infected BSC-40 monolayers at an MOI of 0.005 PFU/cell in solid agar medium (DMEM 2× and 1.9% agar in PBS, 1:1 v/v ratio). After 48 hr of infection, the solid agar medium was removed and the visualized virus plaques were photographed with a ZEISS AXIO Vert.A1 microscope using the technology “Nikon Digital Sight DS-2Mv.” The size of the plaques was measured using a scale with a.u., and the area for comparison was calculated using the formula: Area = (π) × (radius of the minor axis) × (radius of the largest axis).
Spheroid Generation from TRAMP-C1 Prostate Cancer Cells
To M12 multiwell plates, 1 mL 0.95% (w/v) agarose in PBS was added and allowed to solidify, followed by 1 mL TRAMP-C1 cell medium/well and incubation (2 hr, 37°C). Medium was removed and 2.5 × 105 TRAMP-C1 cells were added per well and mixed at low speed on an orbital shaker (37°C incubator, 80% humidity, 5% CO2) following an adapted protocol.56, 57 The spheroids were infected when they reached a volume of 0.5–1 mm3, and size was monitored daily under a microscope.
Histochemistry
Infected spheroids were fixed in 4% (v/v) paraformaldehyde in PBS (12 hr, 4°C), washed three times with PBS, and stored in 70% ethanol (v/v) at 4°C. Spheroids were dehydrated, embedded in paraffin blocks, sectioned (5 μm), and H&E stained (Histology Service, Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas [CNB-CSIC]). Images of sections and spheroids were acquired on a Zeiss Axio Vert.A1 microscope using Nikon Digital Sight DS-2Mv technology.
Mice and Immunization Protocols
C57BL/6 female mice (5–7 weeks old) were purchased from Harlan (Envigo, Barcelona, Spain). For the survival assay, mice received WR WT, WR-Luc, WR-Δ3, or WR-Δ4 virus (5 × 106 or 5 × 107 PFU/mouse i.n.). Mice were inspected daily for weight loss and signs of illness (using a scale from 1 to 4; 1 = absence of illness and 4 = severe illness including loss of mobility, trouble breathing, hunched posture, absence of grooming, and inflammation of the eye membrane), and were sacrificed when body weight loss was greater than 20% or when they showed stage 4 signs of illness. For the biodistribution experiment, mice were inoculated (i.p.) with 2 × 107 PFU/mouse of each virus and sacrificed 24, 72, and 120 hpi; ovaries, peritoneal exudate, and brain were collected, each processed with a homogenizer (VDI 12 homogenizer with a S12N-5S dispersion element; VWR) and titrated in BSC-40 cells. For analysis of innate immune cell populations, mice were inoculated (i.p.) with 1 × 107 PFU/mouse of each virus and sacrificed at 3, 6, and 12 hpi; peritoneal exudate cells were obtained by peritoneal washing and stained for cytometry. For anti-tumor effectiveness studies, tumor-bearing mice were inoculated (i.t.) with 1 × 108 PFU/mouse of each virus. All mouse experiments were approved by the Ethical Committee of Animal Experimentation of the National Center for Biotechnology (CNB-CSIC) in conformity with national and international guidelines for animal experimentation and with Royal Decrees RD1201/2005 and RD53/2013 (CEEA-CNB permit numbers 12074 and 011/15).
Immunization, Processing, and Analysis in the Syngeneic B16F10 Melanoma Model
To analyze anti-tumor activity, we inoculated (i.d.) mice with 5 × 105 B16F10 cells/mouse in the upper dorsal flank; once tumors reached a volume of 40–50 mm3, mice were treated (i.t.) with PBS or 1 × 108 PFU/mouse of virus. Daily follow-up included calculating tumor volume (volume = length × [width]2) and observation of general well-being. Mice were sacrificed when tumor volume reached 1,500 mm3.
For processing, tumors were excised and epithelial tissue removed and minced (1-mm2 pieces) in a plate with PBS. The fragmented tumors were incubated in 2 mL of each dissociation buffer (DMEM with 5% [v/v] FCS, 200 U/mL collagenase VIII, 100 μg/mL DNase I) for 30 min in a tube rotator at 37°C with intermittent vortex every 10 min. After dissociation, tumors were centrifuged (1,500 rpm, 5 min) and washed with PBS with 2% (v/v) FCS. The final suspension was passed through a cell strainer (40 μm) before use. One part of each sample was used for staining and flow cytometry and the other for VACV titration.
ELISpot
The ELISpot assay was used to determine IFNγ-specific T lymphocytes against a stimulus.58 Spleens were collected from tumor-bearing and treated mice, processed in a cell strainer (40 μm), treated with NH4Cl (0.1 M, 5 min) to deplete erythrocytes, and washed three times with RPMI with 10% (v/v) FCS. Splenocytes (1 × 106 splenocytes/condition) were incubated in nitrocellulose-bottom 96-multiwell plates (Millipore, Merck Chemicals & Life Science, Madrid, Spain) coated with anti-mouse IFNγ monoclonal antibody (R4-6A2; Pharmingen, San Diego, CA, USA) in RPMI with B8R20–27 (H-2Kb, TSYKFESV; 10 μg/mL), gp10025–33 (H-2Db, KVPRNQDWL; 1 μg/mL), or TRP-2180–188 peptides (H-2Db, SVYDFFVWL; 1 μg/mL) for 48 hr. The plate was then washed three times with PBS (0.05% [v/v] Tween 20), incubated with 2 μg/mL biotinylated rat anti-mouse IFNγ antibody (XMG1.2, Pharmingen), followed by avidin-peroxidase (1:800; Sigma-Aldrich, St. Louis, MO, USA), and developed with staining solution (1 μg/mL 3,3′-diaminobenzidine (Sigma) in 50 mM Tris-HCl [pH 7.5], with 0.015% [v/v] H2O2). IFNγ+ spots were quantified and analyzed on an ELR02 ELISpot Reader (AID/Autoimmun Diagnostika) using AID ELISpot software (Vitro).
Flow Cytometry
Peritoneal exudates and tumors were processed and the cells pretreated with Fc Block (mouse anti-CD16/CD32; 2.4G2; BD Pharmingen) and stained for surface markers using anti-CD11c (HL3), -Ly6G (1A8), -CD19 (1D3), and -I-A/I-E (2G9) (BD Pharmingen); anti-CD11b (M1/70), -CD45 (30-F11), -CD8 (53-6.7), and -CD335 (29A1.4) (BioLegend, San Diego, CA, USA); anti-F4/80 (BM8) and -CD3 (17A2) (eBioscience, Bleiswijk, the Netherlands); and anti-CD4 (GK1.5) (Beckman Coulter, Brea, CA, USA). Samples were acquired on a GALLIOS flow cytometer (Beckman Coulter), and data were analyzed using FlowJo software (v.8.5.3; Tree Star, Ashland, OR, USA).
For analysis of immune cell populations in peritoneal exudates, spleen, and tumors, CD45+ cells were selected and different cell types gated as neutrophils (CD11b+/Ly6G+), monocytes (F4/80med/CD11bmed), macrophages (F4/80high/CD11bhigh), dendritic cells (CD11c+/I-A/I-E+), NK cells (CD335+/CD3−), NKT cells (CD335+/CD3+), CD4 T lymphocytes (CD4+/CD3+), CD8 T lymphocytes (CD8+/CD3+), and B lymphocytes (CD11b+/CD19+).
Statistical Analysis
Statistical analyses were carried out using GraphPad Prism (v. 6.01; GraphPad, La Jolla, CA, USA). Significance for comparison between groups was determined using a one-way ANOVA test with Bonferroni correction for multiple comparisons. Differences were considered statistically significant when *p < 0.05; **p < 0.01; ***p < 0.005.
Author Contributions
E.M.-P. and M.E. conceived and designed the project. E.M.-P. and L.C.-F. developed the methodology. E.M.-P. and L.C.-F. acquired and analyzed the data. E.M.-P., L.C.-F., and M.E. interpreted and discussed the data, and wrote and reviewed the manuscript.
Acknowledgments
We thank V. Jiménez for tissue culture and virus growth, as well as the CNB Cytometry and Histology Services, and I. Jareño from the CNB Animal Facility for their help and assistance, and C. Mark for excellent editorial assistance. E.M.-P. was supported by an FPU fellowship from the Spanish Ministry of Health and Education and L.C.-F. by a postdoctoral fellowship from the Consejo Nacional de Ciencia y Tecnología (CONACYT), Mexico. This work was supported by a grant from the MINECO/FEDER Spain (SAF2013-45232R).
References
- 1.Greig S.L. Talimogene laherparepvec: first global approval. Drugs. 2016;76:147–154. doi: 10.1007/s40265-015-0522-7. [DOI] [PubMed] [Google Scholar]
- 2.Thorne S.H., Hermiston T., Kirn D. Oncolytic virotherapy: approaches to tumor targeting and enhancing antitumor effects. Semin. Oncol. 2005;32:537–548. doi: 10.1053/j.seminoncol.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman H.L., Kohlhapp F.J., Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat. Rev. Drug Discov. 2015;14:642–662. doi: 10.1038/nrd4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keller B.A., Bell J.C. Oncolytic viruses—immunotherapeutics on the rise. J. Mol. Med. (Berl.) 2016;94:979–991. doi: 10.1007/s00109-016-1453-9. [DOI] [PubMed] [Google Scholar]
- 5.Wein L.M., Wu J.T., Kirn D.H. Validation and analysis of a mathematical model of a replication-competent oncolytic virus for cancer treatment: implications for virus design and delivery. Cancer Res. 2003;63:1317–1324. [PubMed] [Google Scholar]
- 6.Dimitrov D.S. Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McFadden G. Poxvirus tropism. Nat. Rev. Microbiol. 2005;3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirn D.H., Wang Y., Liang W., Contag C.H., Thorne S.H. Enhancing poxvirus oncolytic effects through increased spread and immune evasion. Cancer Res. 2008;68:2071–2075. doi: 10.1158/0008-5472.CAN-07-6515. [DOI] [PubMed] [Google Scholar]
- 9.Smith G.L., Moss B. Infectious poxvirus vectors have capacity for at least 25 000 base pairs of foreign DNA. Gene. 1983;25:21–28. doi: 10.1016/0378-1119(83)90163-4. [DOI] [PubMed] [Google Scholar]
- 10.Moss B. Lippincott-Raven; 2007. Poxviridae: The Viruses and Their Replication; pp. 2637–2671. [Google Scholar]
- 11.Miller J.D., van der Most R.G., Akondy R.S., Glidewell J.T., Albott S., Masopust D., Murali-Krishna K., Mahar P.L., Edupuganti S., Lalor S. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Pütz M.M., Midgley C.M., Law M., Smith G.L. Quantification of antibody responses against multiple antigens of the two infectious forms of Vaccinia virus provides a benchmark for smallpox vaccination. Nat. Med. 2006;12:1310–1315. doi: 10.1038/nm1457. [DOI] [PubMed] [Google Scholar]
- 13.Kirn D.H., Thorne S.H. Targeted and armed oncolytic poxviruses: a novel multi-mechanistic therapeutic class for cancer. Nat. Rev. Cancer. 2009;9:64–71. doi: 10.1038/nrc2545. [DOI] [PubMed] [Google Scholar]
- 14.Chan W.M., McFadden G. Oncolytic poxviruses. Annu. Rev. Virol. 2014;1:119–141. doi: 10.1146/annurev-virology-031413-085442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mell L.K., Brumund K.T., Daniels G.A., Advani S.J., Zakeri K., Wright M.E., Onyeama S.J., Weisman R.A., Sanghvi P.R., Martin P.J., Szalay A.A. Phase I trial of intravenous oncolytic Vaccinia virus (GL-ONC1) with cisplatin and radiotherapy in patients with locoregionally advanced head and neck carcinoma. Clin. Cancer Res. 2017;23:5696–5702. doi: 10.1158/1078-0432.CCR-16-3232. [DOI] [PubMed] [Google Scholar]
- 16.Thorne S.H., Hwang T.H., O’Gorman W.E., Bartlett D.L., Sei S., Kanji F., Brown C., Werier J., Cho J.H., Lee D.E. Rational strain selection and engineering creates a broad-spectrum, systemically effective oncolytic poxvirus, JX-963. J. Clin. Invest. 2007;117:3350–3358. doi: 10.1172/JCI32727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Hughes S.J., Johnston L.H., de Carlos A., Smith G.L. Vaccinia virus encodes an active thymidylate kinase that complements a cdc8 mutant of Saccharomyces cerevisiae. J. Biol. Chem. 1991;266:20103–20109. [PubMed] [Google Scholar]
- 19.Symons J.A., Alcamí A., Smith G.L. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 20.de Magalhães J.C., Andrade A.A., Silva P.N., Sousa L.P., Ropert C., Ferreira P.C., Kroon E.G., Gazzinelli R.T., Bonjardim C.A. A mitogenic signal triggered at an early stage of vaccinia virus infection: implication of MEK/ERK and protein kinase A in virus multiplication. J. Biol. Chem. 2001;276:38353–38360. doi: 10.1074/jbc.M100183200. [DOI] [PubMed] [Google Scholar]
- 21.Buller R.M., Smith G.L., Cremer K., Notkins A.L., Moss B. Decreased virulence of recombinant vaccinia virus expression vectors is associated with a thymidine kinase-negative phenotype. Nature. 1985;317:813–815. doi: 10.1038/317813a0. [DOI] [PubMed] [Google Scholar]
- 22.Puhlmann M., Brown C.K., Gnant M., Huang J., Libutti S.K., Alexander H.R., Bartlett D.L. Vaccinia as a vector for tumor-directed gene therapy: biodistribution of a thymidine kinase-deleted mutant. Cancer Gene Ther. 2000;7:66–73. doi: 10.1038/sj.cgt.7700075. [DOI] [PubMed] [Google Scholar]
- 23.Di Pilato M., Mejías-Pérez E., Zonca M., Perdiguero B., Gómez C.E., Trakala M., Nieto J., Nájera J.L., Sorzano C.O., Combadière C. NFκB activation by modified vaccinia virus as a novel strategy to enhance neutrophil migration and HIV-specific T-cell responses. Proc. Natl. Acad. Sci. USA. 2015;112:E1333–E1342. doi: 10.1073/pnas.1424341112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vesely M.D., Kershaw M.H., Schreiber R.D., Smyth M.J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011;29:235–271. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 25.Gajewski T.F., Schreiber H., Fu Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breitbach C.J., Paterson J.M., Lemay C.G., Falls T.J., McGuire A., Parato K.A., Stojdl D.F., Daneshmand M., Speth K., Kirn D. Targeted inflammation during oncolytic virus therapy severely compromises tumor blood flow. Mol. Ther. 2007;15:1686–1693. doi: 10.1038/sj.mt.6300215. [DOI] [PubMed] [Google Scholar]
- 27.McCart J.A., Ward J.M., Lee J., Hu Y., Alexander H.R., Libutti S.K., Moss B., Bartlett D.L. Systemic cancer therapy with a tumor-selective vaccinia virus mutant lacking thymidine kinase and vaccinia growth factor genes. Cancer Res. 2001;61:8751–8757. [PubMed] [Google Scholar]
- 28.Kim J.H., Oh J.Y., Park B.H., Lee D.E., Kim J.S., Park H.E., Roh M.S., Je J.E., Yoon J.H., Thorne S.H. Systemic armed oncolytic and immunologic therapy for cancer with JX-594, a targeted poxvirus expressing GM-CSF. Mol. Ther. 2006;14:361–370. doi: 10.1016/j.ymthe.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Kirn D.H., Wang Y., Le Boeuf F., Bell J., Thorne S.H. Targeting of interferon-beta to produce a specific, multi-mechanistic oncolytic vaccinia virus. PLoS Med. 2007;4:e353. doi: 10.1371/journal.pmed.0040353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam J.T., Hemminki A., Kanerva A., Lee K.B., Blackwell J.L., Desmond R., Siegal G.P., Curiel D.T. A three-dimensional assay for measurement of viral-induced oncolysis. Cancer Gene Ther. 2007;14:421–430. doi: 10.1038/sj.cgt.7701028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lichty B.D., Breitbach C.J., Stojdl D.F., Bell J.C. Going viral with cancer immunotherapy. Nat. Rev. Cancer. 2014;14:559–567. doi: 10.1038/nrc3770. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez D., Rodriguez J.R., Rodriguez J.F., Trauber D., Esteban M. Highly attenuated vaccinia virus mutants for the generation of safe recombinant viruses. Proc. Natl. Acad. Sci. USA. 1989;86:1287–1291. doi: 10.1073/pnas.86.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y., Adams Y.F., Croft M. Preferential replication of vaccinia virus in the ovaries is independent of immune regulation through IL-10 and TGF-β. Viral Immunol. 2011;24:387–396. doi: 10.1089/vim.2011.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benning N., Hassett D.E. Vaccinia virus infection during murine pregnancy: a new pathogenesis model for vaccinia fetalis. J. Virol. 2004;78:3133–3139. doi: 10.1128/JVI.78.6.3133-3139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karupiah G., Coupar B., Ramshaw I., Boyle D., Blanden R., Andrew M. Vaccinia virus-mediated damage of murine ovaries and protection by virus-expressed interleukin-2. Immunol. Cell Biol. 1990;68:325–333. doi: 10.1038/icb.1990.44. [DOI] [PubMed] [Google Scholar]
- 36.Tschannen R., Steck A.J., Schäfer R. Mechanisms in the pathogenesis of post-infectious vaccinia virus encephalomyelitis in the mouse. Neurosci. Lett. 1979;15:295–300. doi: 10.1016/0304-3940(79)96129-9. [DOI] [PubMed] [Google Scholar]
- 37.Tschannen R., Schafer R. Strain and host-cell dependence of vaccinia virus proteins. Eur. J. Biochem. 1980;111:145–150. doi: 10.1111/j.1432-1033.1980.tb06086.x. [DOI] [PubMed] [Google Scholar]
- 38.Thaiss C.A., Zmora N., Levy M., Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 39.Tiffen J.C., Bailey C.G., Ng C., Rasko J.E., Holst J. Luciferase expression and bioluminescence does not affect tumor cell growth in vitro or in vivo. Mol. Cancer. 2010;9:299. doi: 10.1186/1476-4598-9-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Duffy D., Perrin H., Abadie V., Benhabiles N., Boissonnas A., Liard C., Descours B., Reboulleau D., Bonduelle O., Verrier B. Neutrophils transport antigen from the dermis to the bone marrow, initiating a source of memory CD8+ T cells. Immunity. 2012;37:917–929. doi: 10.1016/j.immuni.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 42.Gentschev I., Adelfinger M., Josupeit R., Rudolph S., Ehrig K., Donat U., Weibel S., Chen N.G., Yu Y.A., Zhang Q. Preclinical evaluation of oncolytic vaccinia virus for therapy of canine soft tissue sarcoma. PLoS ONE. 2012;7:e37239. doi: 10.1371/journal.pone.0037239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parviainen S., Ahonen M., Diaconu I., Kipar A., Siurala M., Vähä-Koskela M., Kanerva A., Cerullo V., Hemminki A. GMCSF-armed vaccinia virus induces an antitumor immune response. Int. J. Cancer. 2015;136:1065–1072. doi: 10.1002/ijc.29068. [DOI] [PubMed] [Google Scholar]
- 44.Chang C.Y., Tai J.A., Li S., Nishikawa T., Kaneda Y. Virus-stimulated neutrophils in the tumor microenvironment enhance T cell-mediated anti-tumor immunity. Oncotarget. 2016;7:42195–42207. doi: 10.18632/oncotarget.9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bos R., Marquardt K.L., Cheung J., Sherman L.A. Functional differences between low- and high-affinity CD8(+) T cells in the tumor environment. OncoImmunology. 2012;1:1239–1247. doi: 10.4161/onci.21285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makkouk A., Weiner G.J. Cancer immunotherapy and breaking immune tolerance: new approaches to an old challenge. Cancer Res. 2015;75:5–10. doi: 10.1158/0008-5472.CAN-14-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.García-Arriaza J., Esteban M. Enhancing poxvirus vectors vaccine immunogenicity. Hum. Vaccin. Immunother. 2014;10:2235–2244. doi: 10.4161/hv.28974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Pilato M., Mejías-Pérez E., Sorzano C.O.S., Esteban M. Distinct roles of vaccinia virus NF-κB inhibitor proteins A52, B15, and K7 in the immune response. J. Virol. 2017;91 doi: 10.1128/JVI.00575-17. e00575-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swaika A., Hammond W.A., Joseph R.W. Current state of anti-PD-L1 and anti-PD-1 agents in cancer therapy. Mol. Immunol. 2015;67(2 Pt A):4–17. doi: 10.1016/j.molimm.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 50.Funt S.A., Page D.B., Wolchok J.D., Postow M.A. CTLA-4 antibodies: new directions, new combinations. Oncology (Williston Park) 2014;28(Suppl 3):6–14. [PubMed] [Google Scholar]
- 51.Yang D., Holt G.E., Velders M.P., Kwon E.D., Kast W.M. Murine six-transmembrane epithelial antigen of the prostate, prostate stem cell antigen, and prostate-specific membrane antigen: prostate-specific cell-surface antigens highly expressed in prostate cancer of transgenic adenocarcinoma mouse prostate mice. Cancer Res. 2001;61:5857–5860. [PubMed] [Google Scholar]
- 52.Perdiguero B., Gómez C.E., Di Pilato M., Sorzano C.O., Delaloye J., Roger T., Calandra T., Pantaleo G., Esteban M. Deletion of the vaccinia virus gene A46R, encoding for an inhibitor of TLR signalling, is an effective approach to enhance the immunogenicity in mice of the HIV/AIDS vaccine candidate NYVAC-C. PLoS ONE. 2013;8:e74831. doi: 10.1371/journal.pone.0074831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Pilato M., Sánchez-Sampedro L., Mejías-Pérez E., Sorzano C.O., Esteban M. Modification of promoter spacer length in vaccinia virus as a strategy to control the antigen expression. J. Gen. Virol. 2015;96:2360–2371. doi: 10.1099/vir.0.000183. [DOI] [PubMed] [Google Scholar]
- 54.Gómez C.E., Nájera J.L., Jiménez V., Bieler K., Wild J., Kostic L., Heidari S., Chen M., Frachette M.J., Pantaleo G. Generation and immunogenicity of novel HIV/AIDS vaccine candidates targeting HIV-1 Env/Gag-Pol-Nef antigens of clade C. Vaccine. 2007;25:1969–1992. doi: 10.1016/j.vaccine.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 55.Ramírez J.C., Gherardi M.M., Esteban M. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J. Virol. 2000;74:923–933. doi: 10.1128/jvi.74.2.923-933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodríguez-Enríquez S., Gallardo-Pérez J.C., Avilés-Salas A., Marín-Hernández A., Carreño-Fuentes L., Maldonado-Lagunas V., Moreno-Sánchez R. Energy metabolism transition in multi-cellular human tumor spheroids. J. Cell. Physiol. 2008;216:189–197. doi: 10.1002/jcp.21392. [DOI] [PubMed] [Google Scholar]
- 57.Hirschhaeuser F., Menne H., Dittfeld C., West J., Mueller-Klieser W., Kunz-Schughart L.A. Multicellular tumor spheroids: an underestimated tool is catching up again. J. Biotechnol. 2010;148:3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 58.Miyahira Y., Murata K., Rodriguez D., Rodriguez J.R., Esteban M., Rodrigues M.M., Zavala F. Quantification of antigen specific CD8+ T cells using an ELISPOT assay. J. Immunol. Methods. 1995;181:45–54. doi: 10.1016/0022-1759(94)00327-s. [DOI] [PubMed] [Google Scholar]