Summary
Candida glabrata is an opportunistic fungal pathogen that ranks as the second most common cause of systemic candidiasis. Despite its genus name, this yeast is more closely related to the model yeast Saccharomyces cerevisiae than to other Candida pathogens, and hence its ability to infect humans is thought to have emerged independently. Moreover, C. glabrata has all the necessary genes to undergo a sexual cycle but is considered an asexual organism due to the lack of direct evidence of sexual reproduction. To reconstruct the recent evolution of this pathogen and find footprints of sexual reproduction, we assessed genomic and phenotypic variation across 33 globally distributed C. glabrata isolates. We cataloged extensive copy-number variation, which particularly affects genes encoding cell-wall-associated proteins, including adhesins. The observed level of genetic variation in C. glabrata is significantly higher than that found in Candida albicans. This variation is structured into seven deeply divergent clades, which show recent geographical dispersion and large within-clade genomic and phenotypic differences. We show compelling evidence of recent admixture between differentiated lineages and of purifying selection on mating genes, which provides the first evidence for the existence of an active sexual cycle in this yeast. Altogether, our data point to a recent global spread of previously genetically isolated populations and suggest that humans are only a secondary niche for this yeast.
Keywords: Candida glabrata, mating, evolution, population genomics, human fungal pathogens, adhesion
Highlights
-
•
Candida glabrata strains can be clustered into highly genetically divergent clades
-
•
Genetic structure suggests a recent global spread of previously isolated populations
-
•
The existence of sex in C. glabrata is supported by genomic footprints of selection
-
•
Mating-type switching occurs in C. glabrata natural populations but is error prone
Genome analyses of globally distributed isolates of the emerging fungal pathogen Candida point to a recent global spread of previously isolated populations, and suggest that humans are most likely a secondary niche for this yeast. Carreté et al. find evidence for the existence of recombination and mating in this purported “asexual” pathogen.
Introduction
The prevalence of infections by opportunistic pathogens (i.e., candidiasis) is increasing, partly owing to recent medical progress enabling the survival of susceptible individuals [1]. Main prevalent agents of candidiasis comprise three Candida species: Candida albicans, Candida glabrata, and Candida parapsilosis, generally in this order [2]. Phylogenetically, these species are only distantly related. C. glabrata belongs to the Nakaseomyces clade, a group that is more closely related to the baker’s yeast Saccharomyces cerevisiae than to C. albicans or C. parapsilosis [3]. Furthermore, both C. glabrata and C. albicans have closely related non-pathogenic relatives, and hence the ability to infect humans in these two lineages must have originated independently [4, 5]. Genome sequencing of non-pathogenic and mildly pathogenic relatives of C. glabrata has enabled tracing the genomic changes that correlate with the evolutionary emergence of pathogenesis in the Nakaseomyces group [3]. These analyses revealed that the ability to infect humans has most likely emerged at least twice independently in the Nakaseomyces, coinciding with parallel expansions of the encoded repertoire of cell-wall adhesins. Thus, increased—or more versatile—adherence may be implicated in the evolutionary emergence of virulence potential toward humans. In contrast, other virulence-related characteristics had a more ancient origin within the clade, and were also found in environmental relatives.
Our understanding of the evolution of C. glabrata at the species level is limited to analyses of natural variation in a few loci [6, 7, 8]. These studies have shown the existence of genetically distinct clades and generally suggested clonal, geographically structured populations. Geographically structured populations are also found in C. albicans, which is tightly associated with humans and which can undergo a parasexual cycle [9, 10, 11], and S. cerevisiae, for which some strains have been domesticated and which can undergo a full sexual cycle, usually involving self-mating [12, 13]. C. glabrata has been described as an asexual species despite the presence of homologs of S. cerevisiae genes involved in mating [14]. Here we undertook a genomics approach to shed light on several fundamental open questions on the recent evolution of this important opportunistic pathogen: namely (1) what is the genetic structure of the global C. glabrata population? (2) Does C. glabrata show patterns of co-evolution with human populations indicating an ancient association? (3) Is there evidence for active mating and mating-type switching systems in this species? (4) How dynamic is the C. glabrata genome and how does it underlie phenotypic diversity across strains? In order to answer these questions, we analyzed the genomes and phenotypes of 33 different clinical and colonizing C. glabrata isolates sampled from different human body sites and globally distributed locations, and chosen within genotyped collections in order to be representative of the previously explored population structure [8, 15] (Table 1). This sampling includes the extensively studied BG2 strain, as well as three pairs of strains, each isolated from a single patient.
Table 1.
Information About the 33 C. glabrata Isolates Analyzed in the Present Study, Including the Reference CBS138
| Sample ID | Synonymous ID | Mean Coverage | Site | Country | Mating Type | CC | RT | Source Data |
|---|---|---|---|---|---|---|---|---|
| BG2 | US01BG2Blo | 84.192 | blood | USA | a | 15 | 15 | [16] |
| CST34 | US000NY034 | 476.944 | blood | USA | alpha | 64 | 64 | [8] |
| CST35 | US003NY035 | 489.168 | blood | USA | alpha | 77 | 77 | [8] |
| E1114 | EB1114Mou | 120.349 | mouth | Belgium | a | 15 | 15 | [15] |
| EB0911Sto | − | 339.156 | stool | Belgium | alpha | 77 | 94 | [15] |
| EF0616Blo1 | − | 261.098 | blood | France | a | 52 | 52 | [8] |
| EF1237Blo1 | − | 301.835 | blood | France | a | 52 | 52 | [8] |
| EF1620Sto | − | 285.374 | stool | France | a | 52 | 98 | [15] |
| EI1815Blo1 | − | 301.624 | blood | Italy | alpha | 52 | 52 | [8] |
| EG01004Sto | − | 262.568 | stool | Germany | a | 15 | 17 | [15] |
| F03013 | EF0313Blo1 | 340.021 | blood | France | a | 15 | 13 | [8] |
| F11 | F11017, EF1117Blo1 | 69.306 | blood | France | a | NA | 88 | [17] |
| F15021 | EF1521Blo1 | 116.500 | blood | France | a | 15 | 15 | [8] |
| F15 | F15035, EF1535Blo1 | 80.737 | blood | France | a | 41 | 41 | [17] |
| M17 | US02Bal017 | 121.681 | blood | USA | a | 6 | 6 | [8] |
| P35_2 | P35-2 | 285.029 | mouth | Taiwan | a | NA | 106 | [18] |
| P35_3 | P35-3 | 245.637 | mouth | Taiwan | alpha | NA | 106 | [18] |
| B1012Ma | EB1012MouC | 302.046 | mouth | Belgium | alpha | NA | 103 | [15] |
| B1012Sa | EB1012StoC | 240.391 | stool | Belgium | alpha | 64 | 102 | [15] |
| BO101Sa | EB0101StoC | 312.022 | stool | Belgium | alpha | 64 | 104 | [15] |
| CST109 | US003NY109 | 294.590 | blood | USA | alpha | 64 | 66 | [8] |
| CST110 | US003NY110 | 235.403 | blood | USA | a | 15 | 15 | [8] |
| CST78 | US003NY078 | 266.426 | blood | USA | a | 6 | 8 | [8] |
| CST80 | US003NY080 | 241.972 | blood | USA | alpha | 64 | 64 | [8] |
| EB101Ma | EB0101MouC | 300.099 | mouth | Belgium | alpha | 64 | 104 | [15] |
| F1019 | EF1019Blo1 | 274.760 | blood | France | a | 6 | 6 | [8] |
| F1822 | EF1822Blo1 | 291.361 | blood | France | a | 6 | 10 | [8] |
| F2229 | EF2229Blo1 | 652.126 | blood | France | a | 6 | 7 | [8] |
| I1718 | EI1718Blo1 | 217.526 | blood | Italy | a | 6 | 5 | [8] |
| M12 | US02Bal012 | 268.030 | blood | USA | alpha | 6 | 11 | [8] |
| M6 | US02Bal006 | 247.431 | blood | USA | a | 15 | 15 | [8] |
| M7 | US02Bal007 | 314.610 | blood | USA | alpha | 64 | 65 | [8] |
| CBS138 | ATCC 2001 | NA | stool | Belgium | alpha | NA | 62 | [19] |
Columns indicate, in this order: strain name or ID; synonym (if any); mean sequencing coverage (if sequenced in this study); body site of isolation; country of isolation; mating type; CC (clonal complex); RT (repeat type); publication describing the source. NA, not assigned.
Commensal strain.
By using genome-wide information on these 33 strains, we assessed the levels of genetic variation to infer the population structure of C. glabrata, its recent evolution, and its genomic plasticity. In addition, given the current consideration of C. glabrata as an asexual species despite the presence of the entire mating genetic toolkit, we used our dataset to search for evidence of mating at the genomic level. In order to do so, we looked for genomic footprints of recombination and mating-type switching, and we evaluated whether mating genes are under purifying or relaxed selection. Finally, we performed experiments to assess whether the observed genomic plasticity is reflected at the phenotypic level.
Results and Discussion
High Levels of Genetic Diversity between Clades and Lack of Strong Geographical Structure
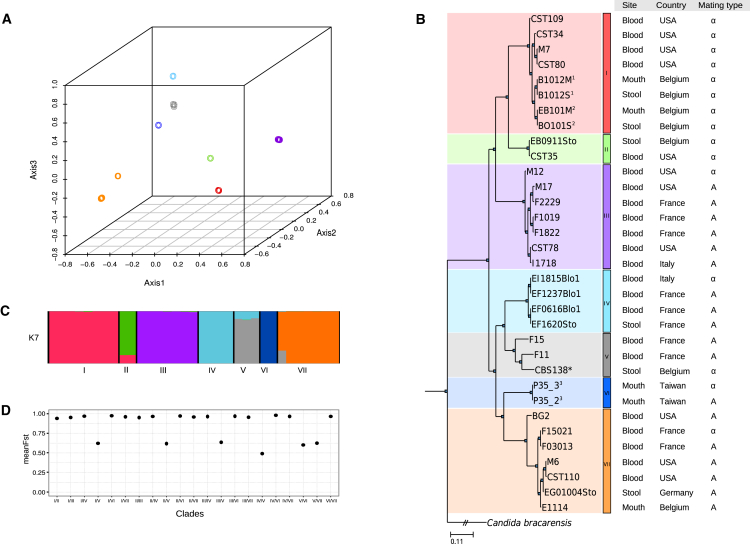
To characterize the genomic variability in the 33 studied strains of C. glabrata, we cataloged single-nucleotide polymorphisms (SNPs) and copy-number variations (CNVs) (STAR Methods) using a read-mapping strategy against the available reference genome [19]. Overall, we detected a range of 4.66–6.56 SNPs/kb per strain when compared to the reference, 0.04–7.23 SNPs/kb between pairs of strains from different patients, and 0.05–0.07 SNPs/kb between strains isolated from the same patient. The low variability between strains of the same individual is indicative that patients were colonized by a single strain that subsequently dispersed to different body sites. We used multiple correspondence analysis (MCA), maximum-likelihood (ML) phylogenetic reconstruction, and model-based clustering to establish the main relationships between all sequenced strains (Figure 1). Overall, these analyses support the existence of seven major clades, hereafter referred to as clade I through clade VII. Previous studies also classified different strains of C. glabrata in clades based on multilocus sequence typing (MLST) [6, 7]. We compared the topologies of strain phylogenies reconstructed from MLST or whole-genome data using the same set of strains. The two topologies overlap to a large extent but there are notable differences with respect to the relationships between clades (Figure S1). Notably, our model-based clustering of genetic variation suggested the existence of genetic admixture between different clades (particularly between clades I and II, IV and V, and V–VII). Phylogenetic reconstruction and fixation indices (FSTs) indicate that most clades diverged deeply within the C. glabrata lineage. Genetic distance between the two most distant clades (clade I and clade VII, 6.59–7.22 SNPs/kb) is only slightly higher than that between the most closely related ones (clade I, clade II, and clade III; 4.48–6 SNPs/kb), but up to two orders of magnitude larger than the amount of genetic divergence within clades (0.03–0.29 SNPs/kb for all clades except clade V, with 4.37–4.68 SNPs/kb). Comparatively, the level of variation between distant C. glabrata clades is higher than the amount of genetic variation among distant clades in human-associated C. albicans (average of 3.7 SNPs/kb) [10]. Most clades were present across distant locations and in different body sites, but they were generally enriched in one of the two mating types (Figure 1; Figure S2A).
Figure 1.
Population Structure of the 33 Strains of C. glabrata
Distribution and population structure of the 33 strains of C. glabrata based on SNP data analysis.
(A) 3D scatterplot of the multiple correspondence analysis (MCA), in which the different colors designate the seven clades detected.
(B) Phylogenetic tree computed using a ML approach. Super-indices indicate pairs of strains in which the two originate from the same patient (different body site or different isolation date; see Table 1 for more details). Clades from I to VII were designated using the same colors as in (A).
(C) Population admixture using STRUCTURE software with K = 7 using the same colors as in (A).
(D) Mean FST for all pairwise comparisons between the seven clades. Fisher test was used to analyze the association with geographical structure (p(country-clade) = 0.006), body site of isolation (p(site-clade) = 0.157), and mating type (p(mating-clade) = 6.064e-05).
See also Figures S1 and S2.
After these analyses were performed, several other studies reported the sequences of 20 additional strains [20, 21, 22]. To confirm the representativeness of our dataset, we repeated the population structure analyses on an expanded dataset of 53 strains, which include these 20 additional strains (Figure S2B). All the new strains clustered within our already delimited clades, indicating that they cover a large fraction of the diversity of C. glabrata clinical isolates. Of note, we observed that in the expanded dataset clade IV and clade V merged, justifying our previously discussed low FST values between these two clades. Importantly, the expanded dataset includes the pyruvate-producing strain C. glabrata CCTCC M202019 [20]. The parental of this strain has been isolated from fertile soil [23], and thus it represents the only available genome sequence from a strain not isolated from the human body. Our re-analysis of this strain (see STAR Methods) identifies relatively few SNPs when compared against the reference CBS138 (1,026 SNPs, 0.08 SNPs/kb), which situates this strain within clade V and as the most similar to the reference genome. Altogether, the current lack of a strong geographical structure of deeply divergent clades suggests recent global migration of human-associated C. glabrata strains. Finally, the appearance of a strain isolated from soil within a clade of human-associated strains suggests that strains with similar genetic backgrounds can colonize humans and the environment. Alternatively, this strain could actually represent a recent colonization of soil from a human source.
Genome Plasticity in C. glabrata: Extensive CNV and Presence of Aneuploidies and Re-arrangements
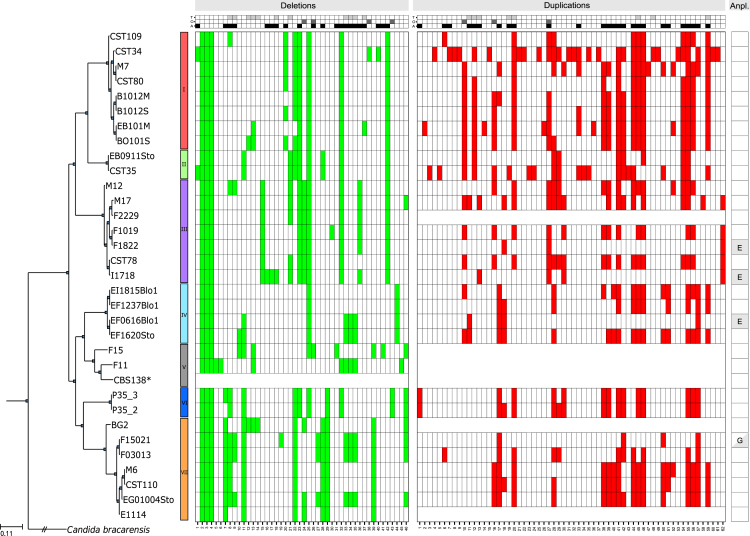
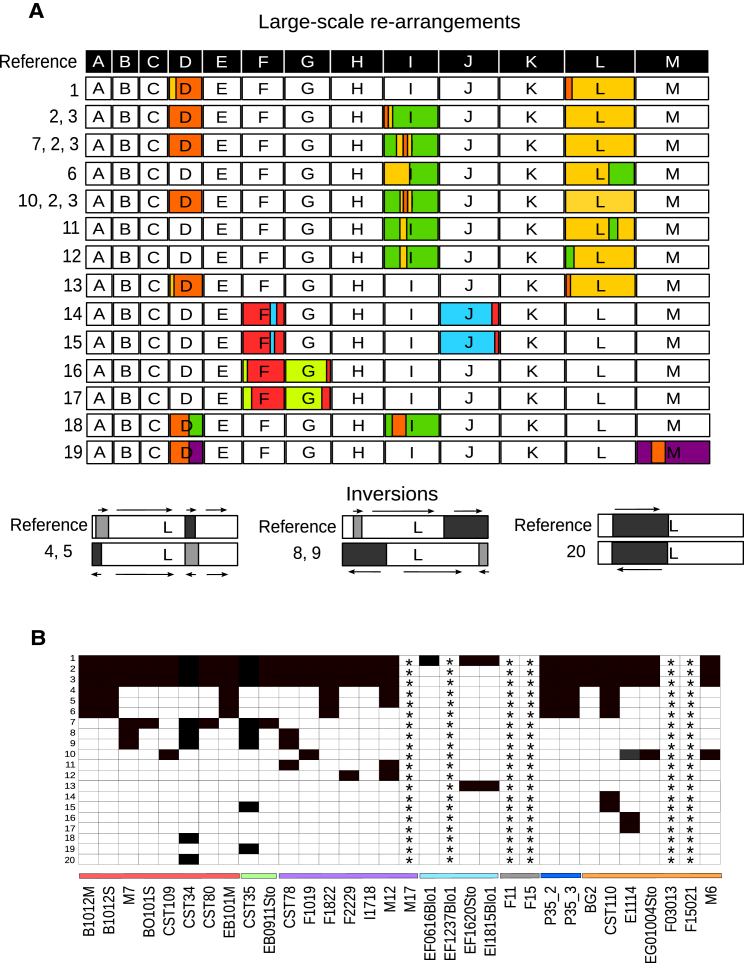
To evaluate the plasticity of the C. glabrata genome, we estimated the number of deletions and duplications in the 33 strains, using depth-of-coverage analyses (see STAR Methods). Overall, we detected a total of 46 deleted and 62 duplicated genes (Figure 2; Data S1). Of these, we experimentally confirmed a deletion covering three genes (see Figure S3). A significant fraction of the deleted (45.65%) or duplicated (41.94%) genes encoded glycosylphosphatidylinositol (GPI)-anchored adhesin-like proteins, as compared to the 1.3% that this functional category represents over the entire genome [24]. Taken together, analysis of biallelic SNPs, flow cytometry, and electrophoretic karyotyping indicate that all analyzed strains are haploid, albeit with variations in total DNA content and chromosome numbers and lengths (Figure S4). Depth-of-coverage analysis revealed aneuploidies involving a whole duplication of chromosome E, whose presence is interspersed in different clades, and one strain carrying a partial aneuploidy of chromosome G (Figure 2; Figure S5A). Aneuploid chromosomes had similar numbers of predicted heterozygous SNPs as other chromosomes when a diploid model was enforced in the SNP calling process (Figure S5B; see STAR Methods), suggesting the extra chromosomes diverged recently. Although all aneuploidies affect genes related to drug resistance, the aneuploid strains had normal sensitivity to tested antifungals (see below). Interestingly, our sequencing data indicated that a major duplication of chromosome J occurred spontaneously while growing one strain (F2229) in rich medium and in the absence of antifungals, as it was present only in about 50% of the cells at the time of sequencing (see Figure S5C). This underscores the plasticity of the C. glabrata genome even under laboratory conditions [25]. Karyotypes for each strain, assessed using pulsed-field gel electrophoresis (PFGE), revealed important variations in chromosome numbers and sizes (Figure S4B). We next assembled de novo and annotated the genomes for all the newly sequenced strains. Alignments of the newly sequenced strains with the reference revealed 20 different large re-arrangements grouped in 17 conformations and affecting 26 different strains, including 14 translocations and 3 inversions (Figure 3), some of which confirmed previous reports based on electrophoretic karyotyping and comparative genome hybridization [17].
Figure 2.
Structural Variations in the Analyzed Strains of C. glabrata
Heatmap showing the deletions, duplications, and aneuploidies (Anpl.) detected in the analyzed strains of C. glabrata sorted by clade. Reference (CBS138) and chromosomes with aneuploidies in affected strains (see below) or genomes with unstable coverage are not shown. The heatmap at the top of the figure designates gene information: light gray, gray, and black represent genes in a tandem duplication (T), orphan genes (O), and genes encoding GPI-anchored adhesin-like proteins (A), respectively. The heatmap colored in green designates the 46 genes affected by deletions, and the heatmap colored in red designates the 62 genes affected by duplications. Aneuploidies are indicated with a light gray background with the letter of the chromosome affected. Fisher test was used to test the significant enrichment in genes encoding GPI-anchored adhesin-like proteins (p < 1.4e-26 and 5.2e-31 in deletions and duplications, respectively), orphan proteins (p < 0.051 and 0.436 in deletions and duplications, respectively), and genes in a tandem duplication (p < 1.5e-09 and 3.0e-05 in deletions and duplications, respectively). See Data S1 for the complete list of genes affected.
See also Figures S3–S5.
Figure 3.
Chromosomal Re-arrangements
(A) Diagram showing the 20 different large re-arrangements found in C. glabrata. Re-arrangements are grouped in 17 different conformations, including 14 translocations and 3 inversions. Chromosomes are indicated in letters from A to M and each one in a different color. Re-arrangement in a chromosome is indicated with the colors of the chromosomes affected. Arrows near the inversion plots indicate the relative orientation of the indicated fragments.
(B) Heatmap showing the distribution of annotated re-arrangements (1–20) and the 26 affected strains. Asterisks indicate strains not included in the analysis due to high fragmentation of the assembly.
See also Figure S4.
We next compared all genomes in terms of their protein-coding content, using a gene similarity-based clustering approach, excluding four strains whose assemblies were deemed of low quality (see STAR Methods). We found that 580 protein-coding genes from the reference strain (CBS138) are unique to this genome. Similarly, a range of 302–580 predicted genes (average 342) are unique for each strain, totaling 9,915 strain-specific genes among the 29 strains considered. Although most of these genes are most likely the result of spurious automated annotations or artifacts from the clustering approach, some may constitute recently emerged genes. 3,603 protein-coding gene clusters are present in all analyzed strains. Given the likely incompleteness of the de novo assemblies, we consider this a lower bound to the C. glabrata core genome. Widespread genes, present in at least 20 of the 29 considered genomes, comprised 4,726 gene clusters. The remaining 252 gene clusters, which comprise 32 genes in CBS138, had more restricted distributions, being present in 2–19 C. glabrata strains, and often being clade specific.
Evidence for Genomic Recombination between Distinct Clades
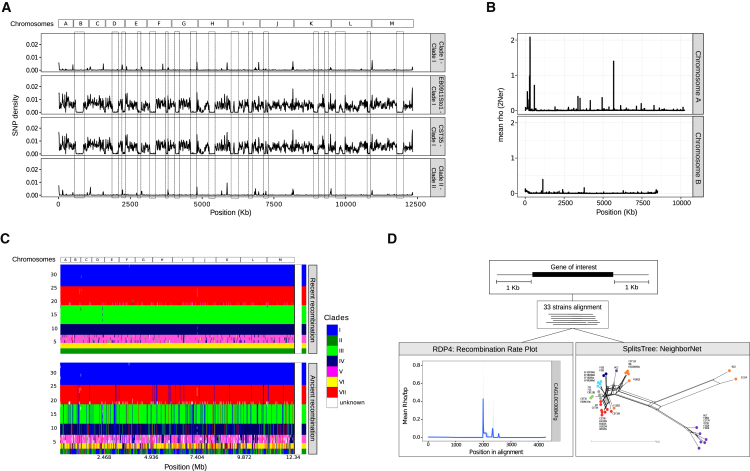
As mentioned above, model-based clustering of genetic variation provided indication of genetic admixture between different clades. In particular, individuals of clade II may have undergone extensive recombination with clade I, as indicated by the presence of large interspersed regions without SNPs when comparing pairwise differences in SNP density (Figure 4A). The presence of these regions is indicative of recombination, because under a scenario of shared ancestral variation we would expect to find this shared variation dispersed across the genome and not organized in large blocks, as is the case. We estimated recombination rates (rho = 2Ner for haploid species) between pairs of SNPs in each chromosome (Figure 4B; Figure S6). Despite overall low mean values (ranging from 0.008 to 0.003), we found evidence of recombination in all chromosomes, and a quite heterogeneous distribution both between and within chromosomes. We also used fastGEAR software to elucidate whether recombination predates the diversification of strains within a clade (i.e., ancestral) or is subsequent to it (i.e., recent) (Figure 4C). This software first classifies the strains in lineages and subsequently calculates the number of ancestral and recent recombination events and tests for their significance. Of note, when using all concatenated chromosomes, the software classified the strains in the same way as STRUCTURE (see above). Importantly, we consistently found that the levels of ancestral recombination are much higher than the recent, and the test of significance suggests that some degree of recent recombination is still occurring in all chromosomes. We also found several striking cases of deletions, and large re-arrangements that were shared between distantly related isolates (Figures 2 and 3), despite the fact that, overall, the distribution of most CNVs and large-scale re-arrangements described above agreed with the defined clades. In a few cases, specific CNVs were found across individuals and populations and may be explained by shared ancestral variation. However, given the close correspondence of the predicted boundaries of some re-arrangements, we considered it unlikely that most of these patterns emerged independently, and suspected the existence of recombination events between distinct lineages. To further confirm the presence of recombination around CNVs shared between distant strains, we performed a detailed analysis of 19 such cases, estimating recombination rates and reconstructing phylogenetic networks (see STAR Methods). Seventeen out of 19 deletions showed a recombination rate higher than 0.05 (which is indicative of a recombination hotspot) or strains from different clades clustered together in the phylogenetic network, suggesting that those deletions are most likely the result of genetic exchange mediated through genomic recombination (Figure 4D). Overall, our results indicate that C. glabrata is still currently able to recombine, and that recombination impacts the genetic variation across chromosomes but also its structure. Finally, the finding of recombination between different strains necessarily implies the existence of some type of mating in C. glabrata.
Figure 4.
Recombination Analyses
(A) Profile of SNP densities obtained when comparing the genomes of strains in clade I and clade II using non-overlapping 10-kb windows along the entire genome. The bar at the top indicates the order and relative length of C. glabrata chromosomes in CBS138. The first profile indicates SNP density between the two strains from clade I (M7 versus B1012M). Second and third profiles indicate SNP density between EB0911Sto and CST35 from clade II versus clade I (using B1012M as a reference for this clade). The fourth profile indicates SNP density between the two strains of clade II. Boxes indicate regions without SNPs, which is indicative of recombination.
(B) Distribution of the recombination rate (rho) across chromosomes, estimated from SNP data using the interval program implemented in the LDhat v2.2 package. For this figure, we selected chromosome A and chromosome B as an example (recombination rate 0.008 and 0.004, respectively). A complete illustration including all chromosomes is found in Figure S6.
(C) Visual representation of the population genetic structure and recombination events inferred by fastGEAR. The bar at the top indicates the order and relative length of C. glabrata chromosomes. We provide two panels corresponding to ancestral recombinations (occurred before the most recent common ancestor of both clusters) and recent recombinations (occurred after the diversification of the clusters). In each panel, the different colors designate each lineage; rows correspond to sequences and columns to positions.
(D) Analysis of the region surrounding the deletion in CAGL0C00847 g gene is shown as an example. First, we selected regions containing the gene of interest and 1-kb flanking regions in the 33 strains. Second, we estimated recombination rates (rho/bp) in the selected regions and computed phylogenetic networks (see STAR Methods). Bottom left: plot showing recombination rates along the genomic region. Values higher than 0.05 indicate a recombination hotspot. Bottom right: NeighborNet splits network showing gene flow between strains and the phylogenetic signal in the region. Clades are indicated as dots of different colors, and the length of the edges is proportional to the weight of the associated split. Strains of different clades cluster together, suggesting a much closer genetic relationship than expected from the genome-wide analysis, which is indicative of recombination between different clades.
Genes Involved in Mating Are Evolutionarily Constrained at the Species Level
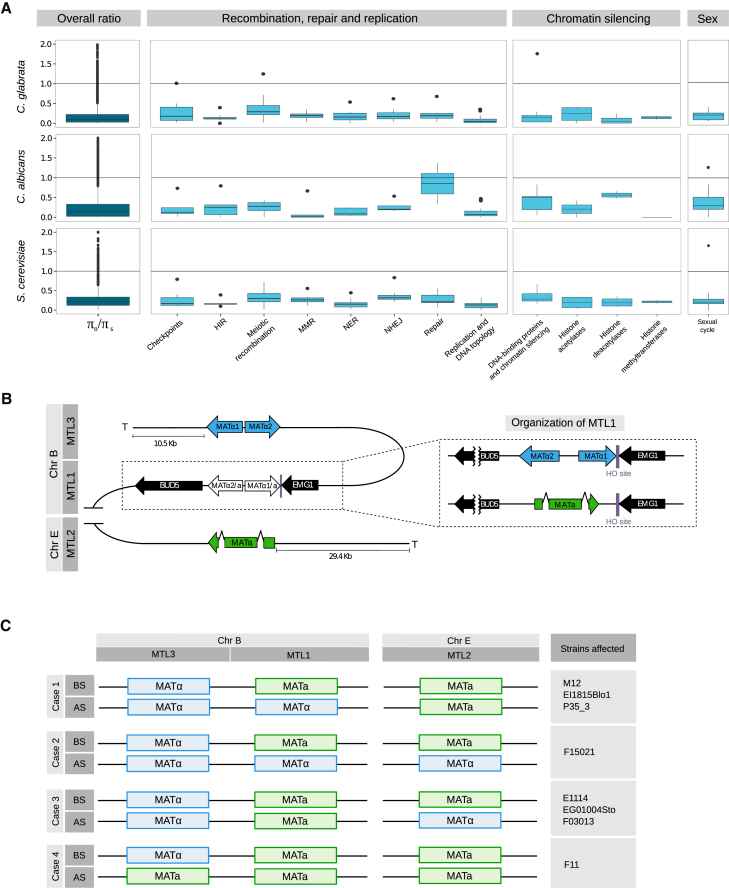
The above results suggest that mating does occur in C. glabrata. If mating has played a role in C. glabrata adaptation, we expect genes involved in mating to show hallmarks of selective constraints at the species level. We assessed levels of genetic variation using nucleotide diversity in C. glabrata genes, and compared these with those obtained from re-analyzing published data in C. albicans and S. cerevisiae, which show parasexual and sexual cycles, respectively (see STAR Methods; Table S1). At the genome-wide level, the three species show overall similar levels of constraints (Figure 5A). We next focused on three different classes of genes involved in mating and recombination: (1) genes involved in the first steps of the sexual cycle [14]; (2) genes involved in chromatin silencing of sexual genes and regulation of mating-type cassettes [14]; and (3) genes involved in replication, repair, and recombination [26] (Figure 5A). All three classes showed signatures of constraints in the three species. Of note, although some classes are more constrained than others, similar patterns are observed in all species. For instance, genes involved in meiotic recombination and repair had signs of relaxation of selection as compared to genes involved in other cellular processes (p = 2.7e−05), whereas genes involved in the sexual cycle are more constrained compared to S. cerevisiae and C. albicans (p = 0.009 and p = 4.3e−05, respectively). We searched for genes with nucleotide diversity higher than the one in C. glabrata as compared to S. cerevisiae and C. albicans, indicating an excess of non-synonymous variations (Table S2). This uncovered the orthologs of S. cerevisiae genes ESC1, MEI4, REC114, and RAD9, which are involved in silencing, meiotic double-strand break formation, meiotic recombination, and DNA damage repair, respectively. Importantly, Esc1p interacts with Sir4p, which is involved in telomere silencing of the HML and HMR cassettes in S. cerevisiae [27]. Altogether, our results show that C. glabrata genes involved in mating and meiosis have comparable levels of selective constraints as those found in C. albicans and S. cerevisiae, providing support for the existence of a sexual or parasexual cycle in C. glabrata. Of note, the anomalous excess of non-synonymous mutation in the C. glabrata ortholog ESC1, suggestive of a recent functional shift, may provide a clue for the observed differences in silencing of mating loci between S. cerevisiae and C. glabrata [28].
Figure 5.
Ratio of Non-synonymous and Synonymous Nucleotide Diversity and Mating-type Switching in C. glabrata
(A) Ratio of non-synonymous and synonymous nucleotide diversity (πN/πS) in genes involved in mating and recombination in C. glabrata, and in their one-to-one orthologs in C. albicans and S. cerevisiae. Dark blue plots show overall πN/πS values in each category, and light blue plots show specific groups of genes included in each category. The most distant outliers are not shown, as the length of the y axis was limited to 2. NHEJ, non-homologous end joining.
(B) Organization of mating-type loci in C. glabrata: MTL1 in white, MTL2 in green, and MTL3 in blue. MTL1 is shown enlarged on the right, encoding either a- or alpha-type genes.
(C) Diagram of the four cases of mating-type switching events likely to have occurred in sequenced strains. BS, before switching; AS, after switching; MMR, mismatch repair; NER, nucleotide excision repair.
Illegitimate Mating-type Switching
Both the evidence of recombination and constraints detected in genes involved in mating support the existence of mating-type switching, albeit very limited, in C. glabrata populations, consistent with earlier observations [8]. Similar to S. cerevisiae, the C. glabrata genome encodes the two mating types (a and alpha) in three different loci called MTL1 (MAT), MTL2 (HMR), and MTL3 (HML), and the HO gene, which encodes the endonuclease responsible for gene conversion-based mating-type switching. MTL2 and MTL3 encode a and alpha information, respectively, and they are close to telomeres. The MTL1 locus encodes either a or alpha, and this information determines the mating-type identity of the cell (Figure 5B). To unveil whether mating-type switching occurred in the studied strains, we analyzed in detail the mating-type loci. Our analysis revealed eight strains that present gene conversion events of four different types (Figure 5C; Figure S7). In three cases, a normal conversion event at MTL1 switched the mating type from a to alpha. The five remaining cases represent cases of aberrant conversions. In one case, a-to-alpha switching at MTL1 is accompanied by illegitimate conversion at MTL2, resulting in a triple-alpha strain. In three cases, illegitimate MTL2 conversion occurred in the apparent absence of MTL1 switching. A final case represents illegitimate conversion of MTL3 in the apparent absence of MTL1 switching, leading to a triple-a strain. In all aberrant cases, the correspondence of the conversion track with an HO cutting site strongly suggests that this switching is mediated by illegitimate cuts in MTL2 or MTL3. These results show that aberrant conversions, which had so far only been observed when induced experimentally [28, 29], can occur in natural populations.
Genomic Plasticity Enables Large Phenotypic Differences between and within Clades
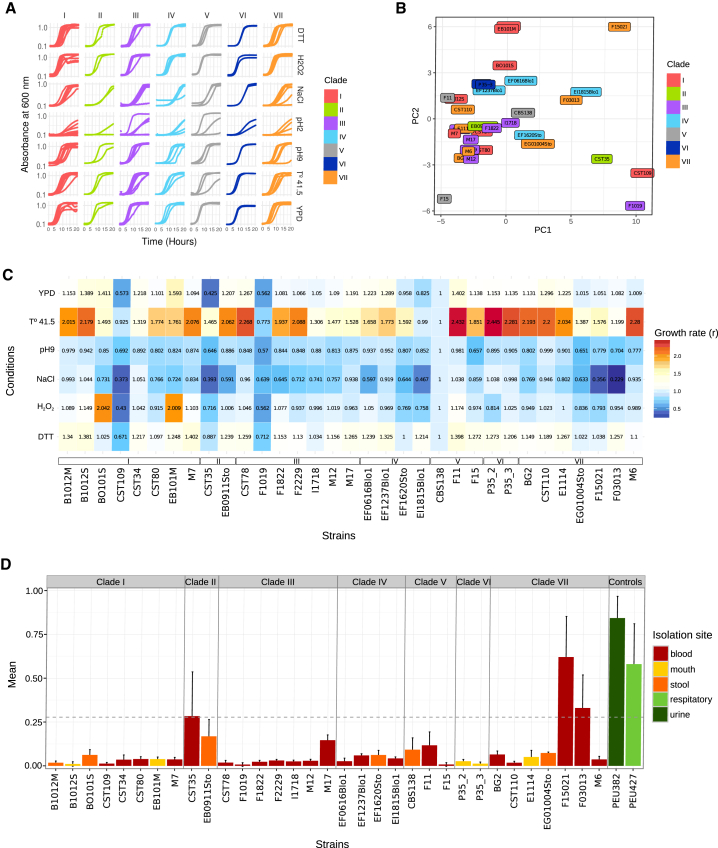
To assess whether the observed genomic plasticity was reflected at the phenotypic level, we measured several relevant phenotypes of the sequenced strains. Specifically, we tested biofilm formation properties and antifungal drug susceptibility, and measured growth under stress conditions such as high and low pH, high temperature, and presence of DTT, sodium chloride, or hydrogen peroxide (see STAR Methods, Figure 6, and Table S3). Most conditions showed important differences between some strains of the same clade (Figure 6). In fact, for most conditions and most clades, intra-clade variation was of a similar range as inter-clade variation (Figures 6A and 6C). Using the R package Growthcurver, we obtained a table with the main growth curve parameters (Table S3). A principal component analysis using those values showed that clades do not cluster by phenotype, underscoring the high phenotypic plasticity even within a similar genetic background (Figure 6B). We next surveyed private mutations, that is, SNPs and CNVs present in a single strain of a clade (Data S2). This resource may help to identify the genetic bases of phenotypic differences in strains behaving drastically different from their close relatives, as well as identifying common mutations in distant strains that show similar behaviors in a given condition. Three strains (M6, M7, and M17) showed reduced sensitivity to one or more antifungal drugs, among eight tested (Table S4). Each carried a unique, private mutation in PDR1 (leading to amino acid exchanges I390K in M6, I378T in M7, and N306S in M17), a known regulator of pleiotropic drug response [31]. Recently, it has been claimed that prevalent mutations in the mismatch repair gene MSH2 found in clinical isolates promote drug resistance through a mutator phenotype [32]. Fifteen (45%) of the 33 analyzed strains carry non-synonymous SNPs in that gene, which include only one (M17) of the three strains with reduced antifungal susceptibility. These 15 strains carried four different MSH2 variants, of which two correspond to variants previously proposed to be loss-of-function mutator genotypes (V239L/A942T and V239L). However, these SNPs were shared by all strains in the same clade and correspond to fixed mutations in other yeast species, and strains with these genotypes did not show unusual patterns of non-synonymous or synonymous variations (see Table S5). Altogether, our results suggest that these mutations represent natural genetic variation and are most likely not related to a mutator phenotype. We next focused on differences in adherence properties, a virulent trait that may vary depending on the repertoire of proteins attached to the cell wall. We noted that three strains showed high (F03013) or moderately high (CST35 and F15021) ability to form biofilm on polystyrol (Figure 6D). These three biofilm-forming strains shared independent duplications of PWP4 and deletions of AWP13, two GPI-anchored adhesins [24]. Although these and other genotype-phenotype relationships enabled by the current dataset can provide useful hints, further experiments are needed to assess what genomic alterations underlie a given phenotypic variation.
Figure 6.
Phenotype Analysis Testing the Growth Rate Using Seven Different Conditions
(A) Growth curves for the 7 different conditions. The first condition was YPD as a normal medium for growth. Following conditions were H2O2, NaCl, DTT, high temperature (41.5 T°), basic pH (pH 9), and acid (pH 2). Unless indicated otherwise, all growth curves were carried out at 37°C. The y axis shows the optical density (OD) for each clade, and the x axis shows time (in hours).
(B) Principal component analysis (PCA) showing the relationship between the statistics values for the growth curves and the distribution of the strains.
(C) Heatmap with growth rate value (r) for all strains and normalized values with the growth rate from the reference strain.
(D) Results represent the averages of three independent replicas of four technical repeats each. Positive controls are well-characterized clinical isolates from urine and respiratory material, respectively, with known high-adherence phenotypes [30]. Error bars indicate SD from mean values.
Conclusions
Our results show that human-associated C. glabrata isolates belong to (at least) seven genetically distinct clades, some of which present levels of genetic diversity comparable to that found in the global C. albicans population. In addition, the absence of a strong geographical structure and the deep genetic divergence between the clades suggest a model of ancient geographical differentiation with recent global dispersion, most likely mediated by humans. This recent dispersion has most likely put in contact C. glabrata clades that had been separated over a long period of time. Importantly, our results show that this admixture has resulted in genetic exchange between distinct clades. The existence of some form of sexual cycle is also strongly supported by similar patterns of evolutionary constraints in reproduction-related genes in C. glabrata, C. albicans, and S. cerevisiae. Our results are consistent with previous reports of successful mating-type switching in MTL1 from a to alpha, but also reveal frequent illegitimate recombination at the other MTL loci. Importantly, the illegitimate recombinations most likely result from cutting of the HO endonuclease sites present in MTL2 and MTL3, which are generally not targeted in S. cerevisiae. This difference may relate to the conformational or epigenetic status of these genomic regions. Our finding of an excess of non-synonymous variation of the C. glabrata ortholog of ESC1, encoding a protein that participates in telomere silencing, may provide the first clue to this fundamental difference, as a functional shift in this protein may have directly impacted the structural or epigenetic organization of C. glabrata telomeres and subtelomeric regions.
We report extensive phenotypic and genetic variation, even between closely related strains, indicating fast evolutionary dynamics and a potential for fast adaptation in C. glabrata. Genetic variation particularly affects cell-wall proteins involved in adhesion. A species-specific increase in adhesins has been purported as a key step in the emergence of the ability to infect humans in the C. glabrata lineage [3]. Our finding of a highly dynamic genetic repertoire of adhesins and large differences in adhesion capabilities suggests that there is a large degree of standing variation of this trait, which may be the subject of ongoing directional selection. Most of these genes are encoded in subtelomeric regions. The above-mentioned difference in ESC1 could also be related to such dynamism. Indeed, null mutants of S. cerevisiae ESC1 show higher chromosome instability and increased transposition of transposable elements. Thus, it is tempting to speculate that a functional shift in ESC1 may have impacted both mating-type switching and the dynamics of subtelomeric genes, and could also be related to the plasticity of chromosomal structure in C. glabrata.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Penicillin / Streptomycin solution | THERMO FISHER SCIENTIFIC, S.L.U. | 15070063 |
| Chloroform-isoamylalcohol (24:1) | Sigma-Aldrich | C0549-1PT |
| Isopropanol | Merck | LOT0476145 |
| Ethanol | SIGMA-ALDRICH QUIMICA S.L | 51976-500ML-F |
| Ribonuclease A from bovine pancreas | SIGMA-ALDRICH QUIMICA S.L | R6513-10MG |
| T4 DNA polymerase | New England Biolabs | M0201L |
| dATP | New England Biolabs | N0440S |
| 3′-5′-exo- Klenow fragment | New England Biolabs | M0212L |
| T4 DNA ligase | New England Biolabs | M0202L |
| Phusion DNA polymerase | Finnzymes | F530S |
| dNTPs | This study | R0181 |
| Hydrogen Peroxide | SIGMA-ALDRICH QUIMICA S.L. | 16911-250ML-F |
| Dithiothreitol | LIFE TECHNOLOGIES S.A. | R0861 |
| 0.1% (w/v) crystal violet | Fisher Scientific | 2479-4 |
| Sodium dodecyl sulfate 10% SDS, 100 mL | SIGMA-ALDRICH QUIMICA S.L. | 71736-100ML |
| Sabouraud agar plates | Oxoid | PO0410 |
| Fluconazole | SIGMA-ALDRICH QUIMICA S.L. | F8929-100MG |
| Isavuconazole | CLINISCIENCES SL | A15783-2 |
| Posaconazole | SIGMA-ALDRICH QUIMICA S.L. | 32103-25MG |
| Voriconazole | SIGMA-ALDRICH QUIMICA S.L. | PZ0005-5MG |
| Micafungin | MOLPORT | MolPort-035-789-689 |
| Caspofungin | SIGMA-ALDRICH QUIMICA S.L. | SML0425-5MG |
| 5-Fluorcytosine | SIGMA-ALDRICH QUIMICA S.L. | F7129-1G |
| Amphotericin B | SIGMA-ALDRICH QUIMICA S.L | A4888-100MG |
| Methanol (Reag. Ph. Eur.) for analysis, ACS, ISO | PANREAC QUIMICA SLU | 1310911211 |
| D-(+)-Glucose anhydrous, free-flowing, Redi-Dri, ≥ 99.5% | SIGMA-ALDRICH QUIMICA S.L. | RDD016-1KG |
| MOPS ≥ 99.5% (titration), 250 g | SIGMA-ALDRICH QUIMICA S.L. | M1254-250G |
| Antibiotic Broth for microbiology (AM 3) | SIGMA-ALDRICH QUIMICA S.L. | 70184-500G |
| RPMI-1640 Medium With L-glutamine, without sodium bicarbonate, powder, suitable for cell culture | SIGMA-ALDRICH QUIMICA S.L. | R6504-10L |
| Agarose | Cultek, SL | H350000 |
| Ethidium Bromide | ThermoFisher Scientific | 15585011 |
| Glass beads | SIGMA-ALDRICH QUIMICA S.L. | G8772-100G |
| Critical Commercial Assays | ||
| QIAquick PCR purification kit | QIAGEN | 50928106 |
| MinElute spin columns | QIAGEN | 28004 |
| MasterPure Yeast DNA Purification Kit | EPICENTRE | MPY80200 |
| Pfu DNA polymerase | PROMEGA | M7745 |
| Deposited Data | ||
| Sequence data | This study | PRJNA361477 |
| Sequence data | [20] | PRJNA222546 |
| Sequence data | [21] | PRJNA310957 |
| Sequence data | [22] | PRJNA297263 |
| Experimental Models: Organisms/Strains | ||
| Candida glabrata BG2 | This study | BG2 |
| Candida glabrata CST34 | This study | CST34 |
| Candida glabrata CST35 | This study | CST35 |
| Candida glabrata E1114 | This study | E1114 |
| Candida glabrata EB0911Sto | This study | EB0911Sto |
| Candida glabrata EF0616Blo1 | This study | EF0616Blo1 |
| Candida glabrata EF1237Blo1 | This study | EF1237Blo1 |
| Candida glabrata EF1620Sto | This study | EF1620Sto |
| Candida glabrata EI1815Blo1 | This study | EI1815Blo1 |
| Candida glabrata EG01004Sto | This study | EG01004Sto |
| Candida glabrata F03013 | This study | F03013 |
| Candida glabrata F11 | This study | F11 |
| Candida glabrata F15021 | This study | F15021 |
| Candida glabrata F15 | This study | F15 |
| Candida glabrata M17 | This study | M17 |
| Candida glabrata P35_2 | This study | P35_2 |
| Candida glabrata P35_3 | This study | P35_3 |
| Candida glabrata B1012M | This study | B1012M |
| Candida glabrata B1012S | This study | B1012S |
| Candida glabrata BO101S | This study | BO101S |
| Candida glabrata CST109 | This study | CST109 |
| Candida glabrata CST110 | This study | CST110 |
| Candida glabrata CST78 | This study | CST78 |
| Candida glabrata CST80 | This study | CST80 |
| Candida glabrata EB101M | This study | EB101M |
| Candida glabrata F1019 | This study | F1019 |
| Candida glabrata F1822 | This study | F1822 |
| Candida glabrata F2229 | This study | F2229 |
| Candida glabrata I1718 | This study | I1718 |
| Candida glabrata M12 | This study | M12 |
| Candida glabrata M6 | This study | M6 |
| Candida glabrata M7 | This study | M7 |
| Candida glabrata reference genome CBS138 | [19] | CBS138 |
| Oligonucleotides | ||
| FWD1: TTGGTCTGTTCCTGAGCCGG | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| FWD2: ACGAACTGGATAGCACCTCC | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| FWD3: ATACTGTGACCTTCCCTGTT | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| REV1: CTCAGCATTGGCAGTAGTGG | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| REV2: CTTCGCTCCGTGGGTAAACA | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| REV3: CTTCAGATTGGCAGTGTCGG | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| MTL1_Forward: CGGTCTGATGGTGCAATTGT | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| MTL1_Reverse: TTGAGTCAAGTGTCGAGGCT | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| MTL2_Forward: GCTCTTCACTCAACGTACTCC | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| MTL2_Reverse: TTTACAAACCCACACCGAGG | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| MTL3_Forward: GTGAGCACTTTGGACCTTCA | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| MTL3_reverse: ACCATAGTCAGACCACCGAC | SIGMA-ALDRICH QUIMICA S.L. | N/A |
| Software and Algorithms | ||
| Trimmomatic v0.36 | [34] | http://www.usadellab.org/cms/?page=trimmomatic |
| SOAPdenovo2 r240 | [35] | https://github.com/aquaskyline/SOAPdenovo2 |
| SPAdes v3.1.1 | [36] | http://bioinf.spbau.ru/spades |
| AUGUSTUS v3.2.3 | [37] | http://bioinf.uni-greifswald.de/augustus/ |
| OrthoMCL v2.0.9 | [38] | http://orthomcl.org |
| BWA 0.7.12 | [39] | http://bio-bwa.sourceforge.net/ |
| Wgsim, v0.3.1 | N/A | https://github.com/lh3/wgsim |
| GATK v3.3 | [40, 41, 42] | https://software.broadinstitute.org/gatk/ |
| SAMtools | [43, 44] | http://samtools.sourceforge.net/ |
| LDhat v2.2 | [45] | http://ldhat.sourceforge.net/ |
| fastGEAR | [46] | https://mostowylab.com/2017/02/26/fastgear/ |
| RDP4 v4.15 | [47, 48] | http://web.cbio.uct.ac.za/∼darren/rdp.html |
| SplitTree v4 | [49] | http://www.splitstree.org/ |
| Mauve v2.4.0 | [50] | http://darlinglab.org/mauve/mauve.html |
| BlastN | [51] | https://blast.ncbi.nlm.nih.gov/ |
| Mugsy v1.2.3 | [52] | http://mugsy.sourceforge.net/ |
| TrimAl v.1.4 | [53] | https://github.com/scapella/trimal |
| RAxML v7.3.5 | [54] | https://sco.h-its.org/exelixis/web/software/raxml/ |
| STRUCTURE v2.3.4 | [55] | https://web.stanford.edu/group/pritchardlab/structure.html |
| PopGenome, R package | [56] | https://CRAN.R-project.org/package=PopGenome |
| Growthcurver v0.2.1, R package | [57] | https://CRAN.R-project.org/package=growthcurver |
| ggplot2, R package | [58] | https://CRAN.R-project.org/package=ggplot2 |
| ade4, R package | [59] | https://CRAN.R-project.org/package=ade4 |
| Other | ||
| dbSNP | NCBI | https://www.ncbi.nlm.nih.gov/SNP/ |
| MICROPLATE, 96 WELL, PS, F-BOTTOM, CLEAR, STERILE, 2 PCS./BAG | Greiner Bio-One North America | 655161 |
| LID, PS, HIGH PROFILE (9 MM), CLEAR, STERILE | Greiner Bio-One North America | 656161 |
Contact for Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Toni Gabaldón (toni.gabaldon@crg.eu).
Experimental Model and Subject Details
Strains
The collection of 33 C. glabrata strains used for the analyses in this study are listed in Table 1. Additional 20 C. glabrata strains were obtained at short read archive under accession PRJNA222546, PRJNA310957 and PRJNA297263 [20, 21, 22].
DNA extraction
C. glabrata cultures were grown overnight in an orbital shaker (200 rpm, 30°C) in 2 mL YPD (Yeast Peptone Dextrose) medium (0.5% yeast extract, 1% peptone, 1% glucose) supplemented with 1% penicillin- streptomycin solution (Sigma). Subsequently, cells were centrifuged (3000 rpm, 5 min) and washed twice with 1x sterile PBS. The pellet was resuspended in 500 μL lysis buffer (1 w/V% SDS, 50mM EDTA, 100 mM TRIS pH = 8), afterward 500 μL of glass beads were added to the cells which then were disrupted by using a vortex for 3 min. 275 μL 7M ammonium-acetate were added (65°C, 5 min) and the samples were cooled on ice for 5 min. Then 500 μL of chloroform-isoamylalcohol (24:1) were added to the mixture, which was then centrifuged for 10 min at 13000 rpm. The upper phase of the solution was transferred to a new microcentrifuge tube, and the previous step was repeated. 500 μL isopropanol was mixed with the upper phase of the solution in a new microcentrifuge tube, and the mixture was held in a refrigerator at −20°C for 5 min. The solution was centrifuged at 13000 rpm for 10 min. The supernatant was discarded, and the pellet was washed twice with 500 μL 70% ethanol. After the second washing step the pellet was dried and resuspended in 100 μL bi-distilled water containing RNase (Sigma).
Method Details
Sequencing
The genome sequences for all the strains were obtained at the Ultra-sequencing core facility of the CRG, using Illumina HiSeq2000 sequencing machines. Paired-end libraries were prepared. For this, DNA was fragmented by nebulization or in Covaris to a size of ∼600 bp. After shearing, the ends of the DNA fragments were blunted with T4 DNA polymerase and Klenow fragment (New England Biolabs). DNA was purified with a QIAquick PCR purification kit (QIAGEN). 3′-adenylation was performed by incubation with dATP and 3′-5′-exo- Klenow fragment (New England Biolabs). DNA was purified using MinElute spin columns (QIAGEN) and double-stranded Illumina paired-end adapters were ligated to the DNA using rapid T4 DNA ligase (New England Biolabs). After another purification step, adaptor-ligated fragments were enriched, and adapters were extended by selective amplification in an 18-cycle PCR reaction using Phusion DNA polymerase (Finnzymes). Libraries were quantified and loaded into Illumina flow-cells at concentrations of 7–20 pM. Cluster generation was performed in an Illumina cluster station. Sequence runs of 2x100 cycles were performed on the sequencing instrument. Base calling was performed using Illumina pipeline software. In multiplexed libraries, we used 4 bp internal indexes (5′ indexed sequences). De-convolution was performed using the CASAVA software (Illumina). All sequence data has been deposited in SRA and will be available upon publication.
Genome assembly
Reads were pre-processed previous to assembly to trim at the first undetermined base or at the first base having PHRED quality below 10 using Trimmomatic v0.36 [32]. The pairs with reads shorter than 31 bases after trimming were not included from the assembly process. SOAPdenovo2 [35] and SPAdes v3.1.1 [36] with default parameters was used to assemble paired-ends reads into chromosomes. AUGUSTUS software [37] was used to predict genes and after that, were clustered based on gene similarity using orthoMCL software [38] in order to obtain core genome analysis. Strains CST34, CST35, M17 and F2229 were removed from the analysis because of the low quality of the raw reads.
SNP calling
Reads were aligned onto the reference assembly of the CBS138 strain [19]þusing BWA, with the BWA-MEM algorithm with 16 as number of threads [39]. As no raw reads are publicly available for the recently-sequenced pyruvate-producing strain C. glabrata CCTCC M202019 [20], the Wgsim software v0.3.1-r13 (https://github.com/lh3/wgsim) was used to simulate reads from the assembled genome sequence using defaults parameters, except for the rate of mutations and fraction of indels that was set to 0 and number of read pairs that was set to 100000000.
We identified SNPs using GATK v3.3 [40, 41, 42]þwith an haploid model, filtering out clusters of 5 variants within 20 bases and low quality variants, and using thresholds for mapping quality and read depth (> 40 and > 30 respectively). To confirm ploidy levels and assess heterozygosity in duplicated chromosomes we repeated the SNP calling analysis enforcing a diploid model. Thereafter, variants were divided into homozygous and heterozygous categories.
Structural variants
To detect structural variants we used deviations from the expected depth of coverage. Calling deletions or duplications at genomic regions with variable coverage is a widely accepted methodology [60]. For every C. glabrata strain we calculated the number of genes deleted and duplicated using depth of coverage analysis from Samtools [43, 44]. After mapping the reads of each strain to the reference genome a gene was considered missing in the strain if less than 90% of the length of a given gene was covered by reads. For duplications and large scale structural variants, we normalized the number of reads per gene and a duplication was called if the median coverage of that gene was 1.8 times or higher than the median coverage of the chromosome. All these structural variants were manually curated and one deletion comprising three genes was validated experimentally (see below).
Experimental validation of CNVs
The deletion of a region comprising three genes (corresponding to deletion numbers 33, 34 and 35 (Data S1) was experimentally confirmed by means of PCR and Sanger sequencing in the following strains: EF1620Sto, F11, F15, EG01004Sto, F03013, F15021 and CBS138. Because the investigated fragment was 8,134 base pairs long, four different sets of primers were designed to be able to capture the whole fragment (Figure S4A). First PCRs were performed with CBS138 (control strain) and one of the investigated strains (EF1620Sto) and primers FWD1:REV1, FWD2:REV2, FWD3:REV3 to validate the absence of the deletion in the control strain and the feasibility of the primers and PCR reactions (Figure S4B). In this case amplicons from the three primer pairs are expected only in the control strain. Then the absence of the fragment was tested with the results of the PCR with FWD1:REV3 (Figure S4C). With this primer, the absence of a band in the CBS138 control strain indicates that the deletion is not present, and the amplification of a fragment of approximately 2 kbp long in the other strains confirms the existence of the deletion. DNA extraction was performed with the MasterPure Yeast DNA Purification Kit from EPICENTRE according to the manufacturer’s protocol. PCRs were carried out by using Pfu DNA polymerase from PROMEGA. The reaction mixture included primer concentration of 0.4 μM, 5 μL of Pfu polymerase 10X buffer with MgSO4, 200 μM of dNTPs each, 1.2 U of Pfu DNA polymerase, 100 ng of DNA and water up to a final volume of 50 μL. Standard PCR protocol was used for primers: FWD1:REV1, FWD3:REV3 and FWD1:REV3. Here, initial denaturation was performed at 95°C for 2min, followed by 30 cycles of 30 s at 92°C, 30 s at either 60.3°C (FWD1:REV1), 59.3°C (FWD3:REV3) or 60.3°C (FWD1:REV3); 190 s (FWD1:REV1) or 120 s (FWD3:REV3) or 220 s (FWD1:REV3) at 72°C. It was finished with final extension for 5 min at 72°C and cooled to 4°C. The touchdown PCR was performed for FWD2:REV2. Cycling condition began with 2 min at 95°C, followed by 15 cycles of 30 s at 95°C, 15 s at the annealing temperature of 61.3°C (decreasing 0.5°C each cycle) and 4 min 20 s at 72°C. Then, other 20 cycles of 30 s at 95°C, 15 s at the annealing temperature of 54.3°C and 4 min 20 s at 72°C were set up, with a final extension step at 72°C for 5 min. All PCR products were visualized by 1% agarose gel electrophoresis (Figures S3B and S3C) and were then purified using the QIAquick PCR Purification Kit (QIAGEN) for subsequent Sanger sequencing. Sequences of the primers used (5′- > 3′) were: FWD1: TTGGTCTGTTCCTGAGCCGG; FWD2: ACGAACTGGATAGCACCTCC, FWD3: ATACTGTGACCTTCCCTGTT; REV1: CTCAGCATTGGCAGTAGTGG; REV2: CTTCGCTCCGTGGGTAAACA and REV3: CTTCAGATTGGCAGTGTCGG.
PCR amplification of mating-type regions and sequencing
In order to validate mating-type switching in C. glabrata, we performed Sanger sequencing of the three different loci MTL1, MTL2 and MTL3 encoding for the two a and alpha mating types in 14 different strains: E1114, M6, CST110, EG01004Sto, F15021, F03013, BG2, P35_2, P35_3, M12, EI1815Blo, F11, EF1237Blo1 and the reference CBS138. DNA extraction and PCRs were performed as indicated in the previous section. Primers used (5′- > 3′) and expected amplicon sizes (bp) are as follows: MTL1_Forward: CGGTCTGATGGTGCAATTGT, MTL1_Reverse: TTGAGTCAAGTGTCGAGGCT (1760 bp); MTL2_Forward: GCTCTTCACTCAACGTACTCC, MTL2_Reverse: TTTACAAACCCACACCGAGG (1305 bp); MTL3_Forward: GTGAGCACTTTGGACCTTCA, MTL3_reverse: ACCATAGTCAGACCACCGAC (1908 bp). Briefly, each reaction included primer concentration of 0.4 μM, 5 μL of Pfu polymerase 10X buffer with MgSO4, 200 μM of dNTPs each, 1.2 U of Pfu DNA polymerase, 100 ng of DNA and water up to a final volume of 50 μL. Cycling condition began with a warm-up step of 2 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at the corresponding annealing temperature (55.5°C, 58.4°C and 58.3°C for MTL1, MTL2 and MTL3, respectively) and an elongation step at 72°C for 3 min 50 s, 2 min 20 s, and 3 min 20 s for MTL1, MTL2 and MTL3, respectively, with a final elongation step of 72°C for 5 min. PCR products were confirmed by 1.5% agarose gel electrophoresis, were then purified using QIAquick PCR purification kit according to manufacturer’s instructions (QIAGEN) and finally sequenced with Sanger using the same set of primers.
Recombination estimates
We used the interval program implemented in the LDhat v2.2 package [45] to estimate population-scale recombination rates for each chromosome separately. The program was executed for 5 million iterations with sampling every 2500 iterations as recommended in the user manual. The output of interval was summarized using stats software from LDhat v2.2 as indicated by the authors. The amount of ancestral and recent recombination for all chromosomes was estimated using fastGEAR software [46] using default parameters. To specifically estimate recombination in the deleted regions and nearby, we used a two-steps pipeline. First, we select deletions that appeared in more than one clade. Second, for each selected deletion we extracted genomic regions located 1Kb up- and downstream of the affected gene. Then, we used RDP4 v4.15 [47, 48] to identify footprints of homologous recombination, and to produce recombination rate plots [61]. Finally, we used SplitTree v4 [49] to reconstruct phylogenetic networks and to detect gene flow.
Chromosomal rearrangements
We identified the presence of chromosomal arrangements using two steps. First, we obtained de novo assemblies of all genome from the 32 C. glabrata strains. Second, we reordered them using CBS138 as a reference applying Mauve Contig Mover from Mauve [50]. Final contigs with rearrangements were confirmed using BlastN [51]
Phylogenetic analysis
We reconstructed a species tree including the 32 sequenced Candida glabrata strains, the reference strain CBS138 and Candida bracarensis (CBS10154) [3] as the outgroup. By using the previously annotated SNPs for the 32 strains, we reconstructed the sequence of each strain by replacing the reference nucleotide for a given SNP. Then, these 34 genomes were aligned using Mugsy v1.2.3 [52]. The resulting alignment was trimmed using TrimAl v.1.4 [53]þto delete positions with more than 50% gaps. Finally, a phylogenetic tree was reconstructed from the trimmed alignment using RAxML v7.3.5, model Protgammalg [54].
For comparison with the phylogenetic tree based in MLST, we used the MLST sequences from the 32 sequenced strains and the reference CBS138. Following the same steps, we reconstructed the sequences of each strain by replacing the reference nucleotide for a given SNP, then we replaced nucleotides with a coverage lower than 30 with gaps. Finally, the final sequences were aligned and trimmed using Mugsy v1.2.3 and TrimAl v.1.4 respectively. The final phylogenetic tree was reconstructed using RAxML v7.3.5 as used during the whole genome tree.
Population Genomics
We used the software STRUCTURE v2.3.4 to study the genetic structure of the population [55]. In addition, we used popGenome to estimate FST between different clades [56]. We recorded the number of SNPs in C. glabrata population using the 33 strains. We obtained the number of SNP also using 32 different strains from C. albicans, as indicated above for C. glabrata, and we obtained SNP data from Saccharomyces cerevisiae using dbSNP [62]þas of October 2015. We calculated the ratio of non-synonymous and synonymous nucleotide diversity (πN/πS) assuming that ¾ of all sites are non-synonymous, and ¼ synonymous.
Phenotypic analyses: Growth curves
Each strain was recovered from our glycerol stock collection and grown for 2 days at 37°C on a YPD agar plate. First, single colonies were cultivated in 15 mL YPD medium in an orbital shaker (37°C, 200 rpm, overnight). Second, each sample was diluted to an optical density (OD) at 600 nm of 0.2 in 3 mL of YPD medium and grown for 3 h more in the same conditions (37°C, 200 rpm). Then, dilutions were made again to have an OD at 600 nm of 0.5 in 1 mL of YPD medium in order to start all the experiments with approximately the same amount of cells. The samples were centrifuged for 2 min at 3000 g, washed with 1 mL of sterile water, and centrifuged again for 2 min at 3000 g for a final resuspension of the pellet in 1 mL of sterile water. Finally, 5 μL of each sample was inoculated in 95 μL of the corresponding medium in a 96-well plate. All experiments were run in triplicate.
A total of six different growth conditions were tested: the oxidative stress was assessed by the growth of the cultures on YPD medium supplemented with 10 mM H2O2, reductive stress with 2.5 mM DTT and osmotic stress with 1 M NaCl. We also measured the impact of elevated temperature (41.5°C), pH = 2 and pH = 9 along with the control growth on YPD itself. Cultures were grown in 96-well plates at 37°C or 41.5°C, shaking, for 24 or 72 h depending on the growth rate in each condition, and monitored to determine the optical density at 600 nm every ten min by a TECAN Infinite M200microplate reader. Finally, results from growth conditions were analyzed using an R package called Growthcurver v0.2.1 [57].
Phenotypic analyses: biofilm formation assay
The capacity to form biofilms was assayed as described previously [30]. Briefly, studied isolates and controls (CBS138, moderate biofilm formation capacity; PEU-382 and PEU-427, high biofilm formation capacity were cultured overnight in YPD medium at 37°C. The optical density was determined at 600 nm (Ultrospec 1000) and adjusted to a value of 2 using sterile NaClphysiol. 50 μL aliquots of the cell suspensions were placed into 96-well polystyrol microtiter plates (Greiner Bio-one) and incubated for 24 h at 37°C. The medium was removed and the attached biofilms washed once with 200 μL distilled water. Cells were stained for 30 min in 100 μL of 0.1% (w/v) crystal violet (CV) solution. Excess CV was removed and the biofilm carefully washed once with 200 μL distilled water. To release CV from the cells, 200 μL 1% (w/v) SDS in 50% (v/v) ethanol were added and the cellular material resuspended by pipetting. CV absorbance was quantified at 490 nm using a microtiter plate reader (MRX TC Revelation). The data shown is the average of three independent biological experiments, each including four technical repeats.
Antifungal drug susceptibility testing
Prior to analysis, the isolates were cultured overnight on Sabouraud (Oxoid) agar plates. Antifungal drug susceptibilities toward Fluconazole, Isavuconazole, Posaconazole, Voriconazole, Micafungin, Caspofungin, 5-Fluorcytosine, and Amphotericin B were determined according to EUCAST EDef 7.1 method [63]. The MIC values of each isolates were calculated according to EUCAST guidelines (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Clinical_breakpoints/Antifungal_breakpoints_v_8.0_November_2015.xlsx, accessed Nov 16th 2016)
Pulsed-field gel electrophoresis
Intact chromosomes were separated using pulsed-field gel electrophoresis (PFGE) as described before [25]. To better visualize differences between small chromosomes (size range CBS 138 ChrA-K) conditions were modified to using 1.2% agarose at 17°C and pulse times from 40-100 s. Large chromosomes (size range CBS138 ChrK-M) were resolved with pulse times form 60-140 s.
Quantification and Statistical Analysis
All statistical details (statistical test used, number of samples, and p values) for each experiment can be found in the text and in the Figure labels.
Fisher tests were computed using R and plots were obtained using the ggplot2 package for R [58]. Multiple Correspondence Analysis (MCA) was performed using ade4 package for R to establish the main relationships between all sequenced strains and the reference [59]. MCA is a technique similar to principal component analysis (PCA) but specific for nominal categorical data, which is used to detect and represent underlying structures in datasets. Principal Component Analysis (PCA) was used to understand the relationship between the analysis from the growth curves and the distribution of our genomes using stats package for R.
Data and Software Availability
Sequence data produced for this project has been deposited at short read archive under the accession PRJNA361477.
Acknowledgments
The T.G. group acknowledges the support of the Spanish Ministry of Economy and Competitiveness grants “Centro de Excelencia Severo Ochoa 2013–2017” SEV-2012-0208 and BFU2015-67107 cofunded by the European Regional Development Fund (ERDF); European Union and ERC Seventh Framework Programme (FP7/2007–2013) under grant agreement ERC-2012-StG-310325; Catalan Research Agency (AGAUR) SGR857; CERCA Programme/Generalitat de Catalunya; and a grant from the European Union’s Horizon 2020 research and innovation programme under Marie Sklodowska-Curie grant agreement H2020-MSCA-ITN-2014-642095. C.F.’s and T.G.’s groups acknowledge support from the GDRI “iGenolevures” of the French CNRS for travel and meeting funds. T.G., O.B., and E.G.-M. acknowledge funding from the European Union under grant agreement FP7-PEOPLE-2013-ITN-606786 “ImresFun.” All authors acknowledge the technical support of the UPF-CRG FACS facility and the CRG Genomics facility.
Author Contributions
T.G., C.F., and O.B. supervised the research. T.G. and L.C. designed the research. L.C., C.P., E.K., D.L., E.G.-M., E.S., and S.I.-G. performed experiments and analyzed the data. T.G., L.C., and C.P. wrote the first draft of the paper.
Published: December 14, 2017
Footnotes
Supplemental Information includes seven figures, five tables, and two data files and can be found with this article online at https://doi.org/10.1016/j.cub.2017.11.027.
Supplemental Information
Columns indicate, in this order: strain name or ID; host; isolation site; country of isolation; mating type (if any); experiment name in short read archive (SRA); number of SRA run.
The first column indicates the gene functional category (based on their function in S. cerevisiae). The following columns indicate gene ID, gene name (only for S. cerevisiae), and πN/πS for C. glabrata, C. albicans, and S. cerevisiae genes.
Columns indicate, in this order: condition used for the analysis; sample; clade; carrying capacity (K); population size at time 0 (n0); growth rate (r); time when the population density reaches ½K (t_mid); fastest possible generation time (t_gen); area under the logistic curve measured by taking the integral of the logistic equation (auc_l); empirical area under the curve (auc_e); measure of the goodness of fit of the parameters for the logistic equation (a small value indicates a better fit of the curve) (sigma).
Range of sensitivity levels (triplicate experiments) to amphotericin B (MIC90), 5-fluorocytosine, fluconazole, voriconazole, posaconazole, isavuconazole, micafungin, and caspofungin (all MIC50) and, where available (http://www.eucast.org/clinical_breakpoints), classification according to clinical breakpoints. S, sensitive; I, intermediate; R, resistant. Red and boldfaced names and values indicate isolates with minimal inhibitory concentration (MIC) values measured deviating from the wild-type distribution. There are no species-specific clinical breakpoints available for 5-fluorocytosine (5FC), voriconazole, posaconazole, isavuconazole, or caspofungin. The measured MIC range in most isolates (three technical repeats) encompasses both “susceptible” and “resistant” interpretations according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoint definition: S ≤ 0.032, R > 0.032 for micafungin.
Columns indicate, in this order: strain name; clade; non-synonymous variant; Saccharomycotina species presenting the same non-synonymous mutations (five-letter codes correspond to species mnemonic based on the UniProt taxonomy database [33]); reduced sensitivity of the strain to any antifungal tested; genome-wide πN; genome-wide πS; genome-wide πN/πS.
Columns indicate, in this order: duplication or deletion number in this study (corresponds to Figure 2); gene ID; gene name; name of S. cerevisiae one-to-one ortholog (if any); description.
Columns of private non-synonymous SNPs indicate, in this order: strain ID; chromosome and position affected by the SNP; gene ID; amino acid substitution; gene name; description. The first column based on CNV indicates the strain analyzed. The following columns indicate the gene affected and description of gene affected.
References
- 1.Brown G.D., Denning D.W., Gow N.A.R., Levitz S.M., Netea M.G., White T.C. Hidden killers: human fungal infections. Sci. Transl. Med. 2012;4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Diekema D., Arbefeville S., Boyken L., Kroeger J., Pfaller M. The changing epidemiology of healthcare-associated candidemia over three decades. Diagn. Microbiol. Infect. Dis. 2012;73:45–48. doi: 10.1016/j.diagmicrobio.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Gabaldón T., Martin T., Marcet-Houben M., Durrens P., Bolotin-Fukuhara M., Lespinet O., Arnaise S., Boisnard S., Aguileta G., Atanasova R. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics. 2013;14:623. doi: 10.1186/1471-2164-14-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabaldón T., Carreté L. The birth of a deadly yeast: tracing the evolutionary emergence of virulence traits in Candida glabrata. FEMS Yeast Res. 2016;16:fov110. doi: 10.1093/femsyr/fov110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabaldón T., Naranjo-Ortíz M.A., Marcet-Houben M. Evolutionary genomics of yeast pathogens in the Saccharomycotina. FEMS Yeast Res. 2016;16:fow064. doi: 10.1093/femsyr/fow064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dodgson A.R., Pujol C., Pfaller M.A., Denning D.W., Soll D.R. Evidence for recombination in Candida glabrata. Fungal Genet. Biol. 2005;42:233–243. doi: 10.1016/j.fgb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Dodgson A.R., Pujol C., Denning D.W., Soll D.R., Fox A.J. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J. Clin. Microbiol. 2003;41:5709–5717. doi: 10.1128/JCM.41.12.5709-5717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brisse S., Pannier C., Angoulvant A., de Meeus T., Diancourt L., Faure O., Muller H., Peman J., Viviani M.A., Grillot R. Uneven distribution of mating types among genotypes of Candida glabrata isolates from clinical samples. Eukaryot. Cell. 2009;8:287–295. doi: 10.1128/EC.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fundyga R.E., Lott T.J., Arnold J. Population structure of Candida albicans, a member of the human flora, as determined by microsatellite loci. Infect. Genet. Evol. 2002;2:57–68. doi: 10.1016/s1567-1348(02)00088-6. [DOI] [PubMed] [Google Scholar]
- 10.Hirakawa M.P., Martinez D.A., Sakthikumar S., Anderson M.Z., Berlin A., Gujja S., Zeng Q., Zisson E., Wang J.M., Greenberg J.M. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 2015;25:413–425. doi: 10.1101/gr.174623.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tavanti A., Davidson A.D., Fordyce M.J., Gow N.A.R., Maiden M.C.J., Odds F.C. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J. Clin. Microbiol. 2005;43:5601–5613. doi: 10.1128/JCM.43.11.5601-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cromie G.A., Hyma K.E., Ludlow C.L., Garmendia-Torres C., Gilbert T.L., May P., Huang A.A., Dudley A.M., Fay J.C. Genomic sequence diversity and population structure of Saccharomyces cerevisiae assessed by RAD-seq. G3 (Bethesda) 2013;3:2163–2171. doi: 10.1534/g3.113.007492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liti G., Carter D.M., Moses A.M., Warringer J., Parts L., James S.A., Davey R.P., Roberts I.N., Burt A., Koufopanou V. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabre E., Muller H., Therizols P., Lafontaine I., Dujon B., Fairhead C. Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing, and subtelomeres. Mol. Biol. Evol. 2005;22:856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- 15.Enache-Angoulvant A., Bourget M., Brisse S., Stockman-Pannier C., Diancourt L., François N., Rimek D., Fairhead C., Poulain D., Hennequin C. Multilocus microsatellite markers for molecular typing of Candida glabrata: application to analysis of genetic relationships between bloodstream and digestive system isolates. J. Clin. Microbiol. 2010;48:4028–4034. doi: 10.1128/JCM.02140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cormack B.P., Falkow S. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics. 1999;151:979–987. doi: 10.1093/genetics/151.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller H., Thierry A., Coppée J.-Y., Gouyette C., Hennequin C., Sismeiro O., Talla E., Dujon B., Fairhead C. Genomic polymorphism in the population of Candida glabrata: gene copy-number variation and chromosomal translocations. Fungal Genet. Biol. 2009;46:264–276. doi: 10.1016/j.fgb.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Lin C.Y., Chen Y.C., Lo H.J., Chen K.W., Li S.Y. Assessment of Candida glabrata strain relatedness by pulsed-field gel electrophoresis and multilocus sequence typing. J. Clin. Microbiol. 2007;45:2452–2459. doi: 10.1128/JCM.00699-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., De Montigny J., Marck C., Neuvéglise C., Talla E. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 20.Xu N., Ye C., Chen X., Liu J., Liu L., Chen J. Genome sequencing of the pyruvate-producing strain Candida glabrata CCTCC M202019 and genomic comparison with strain CBS138. Sci. Rep. 2016;6:34893. doi: 10.1038/srep34893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biswas C., Chen S.C.-A., Halliday C., Kennedy K., Playford E.G., Marriott D.J., Slavin M.A., Sorrell T.C., Sintchenko V. Identification of genetic markers of resistance to echinocandins, azoles and 5-fluorocytosine in Candida glabrata by next-generation sequencing: a feasibility study. Clin. Microbiol. Infect. 2017;23:676.e7–676.e10. doi: 10.1016/j.cmi.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Håvelsrud O.E., Gaustad P. Draft genome sequences of Candida glabrata isolates 1A, 1B, 2A, 2B, 3A, and 3B. Genome Announc. 2017;5:e00328-16. doi: 10.1128/genomeA.00328-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L.-M., Li Y., Li H.-Z., Chen J. Manipulating the pyruvate dehydrogenase bypass of a multi-vitamin auxotrophic yeast Torulopsis glabrata enhanced pyruvate production. Lett. Appl. Microbiol. 2004;39:199–206. doi: 10.1111/j.1472-765X.2004.01563.x. [DOI] [PubMed] [Google Scholar]
- 24.de Groot P.W.J., Bader O., de Boer A.D., Weig M., Chauhan N. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot. Cell. 2013;12:470–481. doi: 10.1128/EC.00364-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bader O., Schwarz A., Kraneveld E.A., Tangwattanachuleeporn M., Schmidt P., Jacobsen M.D., Gross U., De Groot P.W., Weig M. Gross karyotypic and phenotypic alterations among different progenies of the Candida glabrata CBS138/ATCC2001 reference strain. PLoS ONE. 2012;7:e52218. doi: 10.1371/journal.pone.0052218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richard G.-F., Kerrest A., Lafontaine I., Dujon B. Comparative genomics of hemiascomycete yeasts: genes involved in DNA replication, repair, and recombination. Mol. Biol. Evol. 2005;22:1011–1023. doi: 10.1093/molbev/msi083. [DOI] [PubMed] [Google Scholar]
- 27.Andrulis E.D., Zappulla D.C., Ansari A., Perrod S., Laiosa C.V., Gartenberg M.R., Sternglanz R. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol. Cell. Biol. 2002;22:8292–8301. doi: 10.1128/MCB.22.23.8292-8301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller H., Hennequin C., Gallaud J., Dujon B., Fairhead C. The asexual yeast Candida glabrata maintains distinct a and alpha haploid mating types. Eukaryot. Cell. 2008;7:848–858. doi: 10.1128/EC.00456-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boisnard S., Zhou Li Y., Arnaise S., Sequeira G., Raffoux X., Enache-Angoulvant A., Bolotin-Fukuhara M., Fairhead C. Efficient mating-type switching in Candida glabrata induces cell death. PLoS ONE. 2015;10:e0140990. doi: 10.1371/journal.pone.0140990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gómez-Molero E., de Boer A.D., Dekker H.L., Moreno-Martínez A., Kraneveld E.A., Ichsan, Chauhan N., Weig M., de Soet J.J., de Koster C.G. Proteomic analysis of hyperadhesive Candida glabrata clinical isolates reveals a core wall proteome and differential incorporation of adhesins. FEMS Yeast Res. 2015;15:fov098. doi: 10.1093/femsyr/fov098. [DOI] [PubMed] [Google Scholar]
- 31.Tsai H.-F., Krol A.A., Sarti K.E., Bennett J.E. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 2006;50:1384–1392. doi: 10.1128/AAC.50.4.1384-1392.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Healey K.R., Zhao Y., Perez W.B., Lockhart S.R., Sobel J.D., Farmakiotis D., Kontoyiannis D.P., Sanglard D., Taj-Aldeen S.J., Alexander B.D. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat. Commun. 2016;7:11128. doi: 10.1038/ncomms11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pundir S., Magrane M., Martin M.J., O’Donovan C., UniProt Consortium Searching and navigating UniProt databases. Curr. Protoc. Bioinformatics. 2015;50:1.27.1–1.27.10. doi: 10.1002/0471250953.bi0127s50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nurk S., Bankevich A., Antipov D., Gurevich A.A., Korobeynikov A., Lapidus A., Prjibelski A.D., Pyshkin A., Sirotkin A., Sirotkin Y. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer S., Brunk B.P., Chen F., Gao X., Harb O.S., Iodice J.B., Shanmugam D., Roos D.S., Stoeckert C.J., Jr. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinformatics. 2011;35:6.12.1–6.12.19. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auton A., McVean G. Recombination rate estimation in the presence of hotspots. Genome Res. 2007;17:1219–1227. doi: 10.1101/gr.6386707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mostowy R., Croucher N.J., Andam C.P., Corander J., Hanage W.P., Marttinen P. Efficient inference of recent and ancestral recombination within bacterial populations. Mol. Biol. Evol. 2017;34:1167–1182. doi: 10.1093/molbev/msx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin D., Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 48.Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 50.Darling A.C.E., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Angiuoli S.V., Salzberg S.L. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics. 2011;27:334–342. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamatakis A., Ludwig T., Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 55.Hubisz M.J., Falush D., Stephens M., Pritchard J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfeifer B., Wittelsbürger U., Ramos-Onsins S.E., Lercher M.J. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 2014;31:1929–1936. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sprouffske K., Wagner A. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics. 2016;17:172. doi: 10.1186/s12859-016-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wickham, H. (2009). ggplot2. http://link.springer.com/10.1007/978-0-387-98141-3.
- 59.Tenenhaus M., Young F.W. An analysis and synthesis of multiple correspondence analysis, optimal scaling, dual scaling, homogeneity analysis and other methods for quantifying categorical multivariate data. Psychometrika. 1985;50:91–119. [Google Scholar]
- 60.Boeva V., Zinovyev A., Bleakley K., Vert J.-P., Janoueix-Lerosey I., Delattre O., Barillot E. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics. 2011;27:268–269. doi: 10.1093/bioinformatics/btq635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McVean G.A.T., Myers S.R., Hunt S., Deloukas P., Bentley D.R., Donnelly P. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 62.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arendrup M.C., Cuenca-Estrella M., Lass-Flörl C., Hope W., EUCAST-AFST EUCAST technical note on the EUCAST definitive document EDef 7.2: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST) Clin. Microbiol. Infect. 2012;18:E246–E247. doi: 10.1111/j.1469-0691.2012.03880.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Columns indicate, in this order: strain name or ID; host; isolation site; country of isolation; mating type (if any); experiment name in short read archive (SRA); number of SRA run.
The first column indicates the gene functional category (based on their function in S. cerevisiae). The following columns indicate gene ID, gene name (only for S. cerevisiae), and πN/πS for C. glabrata, C. albicans, and S. cerevisiae genes.
Columns indicate, in this order: condition used for the analysis; sample; clade; carrying capacity (K); population size at time 0 (n0); growth rate (r); time when the population density reaches ½K (t_mid); fastest possible generation time (t_gen); area under the logistic curve measured by taking the integral of the logistic equation (auc_l); empirical area under the curve (auc_e); measure of the goodness of fit of the parameters for the logistic equation (a small value indicates a better fit of the curve) (sigma).
Range of sensitivity levels (triplicate experiments) to amphotericin B (MIC90), 5-fluorocytosine, fluconazole, voriconazole, posaconazole, isavuconazole, micafungin, and caspofungin (all MIC50) and, where available (http://www.eucast.org/clinical_breakpoints), classification according to clinical breakpoints. S, sensitive; I, intermediate; R, resistant. Red and boldfaced names and values indicate isolates with minimal inhibitory concentration (MIC) values measured deviating from the wild-type distribution. There are no species-specific clinical breakpoints available for 5-fluorocytosine (5FC), voriconazole, posaconazole, isavuconazole, or caspofungin. The measured MIC range in most isolates (three technical repeats) encompasses both “susceptible” and “resistant” interpretations according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoint definition: S ≤ 0.032, R > 0.032 for micafungin.
Columns indicate, in this order: strain name; clade; non-synonymous variant; Saccharomycotina species presenting the same non-synonymous mutations (five-letter codes correspond to species mnemonic based on the UniProt taxonomy database [33]); reduced sensitivity of the strain to any antifungal tested; genome-wide πN; genome-wide πS; genome-wide πN/πS.
Columns indicate, in this order: duplication or deletion number in this study (corresponds to Figure 2); gene ID; gene name; name of S. cerevisiae one-to-one ortholog (if any); description.
Columns of private non-synonymous SNPs indicate, in this order: strain ID; chromosome and position affected by the SNP; gene ID; amino acid substitution; gene name; description. The first column based on CNV indicates the strain analyzed. The following columns indicate the gene affected and description of gene affected.