ABSTRACT
Interactions between photoautotrophic and heterotrophic microorganisms are central to the marine microbial ecosystem. Lab cultures of one of the dominant marine photoautotrophs, Synechococcus, have historically been difficult to render axenic, presumably because these bacteria depend upon other organisms to grow under these conditions. These tight associations between Synechococcus and heterotrophic bacteria represent a good relevant system to study interspecies interactions. Ten individual Synechococcus strains, isolated from eutrophic and oligotrophic waters, were chosen for investigation. Four to six dominant associated heterotrophic bacteria were detected in the liquid cultures of each Synechococcus isolate, comprising members of the Cytophaga-Flavobacteria-Bacteroides (CFB) group (mainly from Flavobacteriales and Cytophagales), Alphaproteobacteria (mainly from the Roseobacter clade), Gammaproteobacteria (mainly from the Alteromonadales and Pseudomonadales), and Actinobacteria. The presence of the CFB group, Gammaproteobacteria, and Actinobacteria showed clear geographic patterns related to the isolation environments of the Synechococcus bacteria. An investigation of the population dynamics within a growing culture (XM-24) of one of the isolates, including an evaluation of the proportions of cells that were free-living versus aggregated/attached, revealed interesting patterns for different bacterial groups. In Synechococcus sp. strain XM-24 culture, flavobacteria, which was the most abundant group throughout the culture period, tended to be aggregated or attached to the Synechococcus cells, whereas the actinobacteria demonstrated a free-living lifestyle, and roseobacters displayed different patterns depending on the culture growth phase. Factors contributing to these succession patterns for the heterotrophs likely include interactions among the culture community members, their relative abilities to utilize different compounds produced by Synechococcus cells and changes in the compounds released as culture growth proceeds, and their responses to other changes in the environmental conditions throughout the culture period.
IMPORTANCE Marine microbes exist within an interactive ecological network, and studying their interactions is an important part of understanding their roles in global biogeochemical cycling and the determinants of microbial diversity. In this study, the dynamic relationships between Synechococcus spp. and their associated heterotrophic bacteria were investigated. Synechococcus-associated heterotrophic bacteria had similar geographic distribution patterns as their “host” and displayed different lifestyles (free-living versus attached/aggregated) according to the Synechococcus culture growth phases. Combined organic carbon composition and bacterial lifestyle data indicated a potential for succession in carbon utilization patterns by the dominant associated heterotrophic bacteria. Comprehending the interactions between photoautotrophs and heterotrophs and the patterns of organic carbon excretion and utilization is critical to understanding their roles in oceanic biogeochemical cycling.
KEYWORDS: Flavobacteria, Roseobacter, Synechococcus, heterotrophic bacteria, metabolic interaction
INTRODUCTION
Microbes play important roles in biogeochemical cycling and productivity in the ocean (1–3). Marine microbes are part of an interacting ecological ecosystem, and comprehending their inter- and intraspecies interactions is critical to understanding their community compositions and distribution patterns in the environment (4–8). Microbes can affect each other through a variety of synergistic interactions (such as cooperation and symbiosis) and antagonistic interactions (such as competition) that influence their function and behavior (5, 6, 9).
Interactions between photoautotrophic and heterotrophic microorganisms are fundamental in the marine food web (1, 10). Heterotrophs utilize photosynthetically fixed organic carbon and other nutrients supplied by photoautotrophs. Free-living heterotrophic bacteria mainly take up labile dissolved organic carbon (DOC) released by photoautotrophs to the surrounding water, while some heterotrophic bacteria attach to photoautotrophs or photoautotroph-derived detrital particles and metabolize particulate organic carbon (POC) as the main sources of organic carbon (10–12). In return, heterotrophic bacteria benefit photoautotrophs by providing essential micronutrients, such as vitamins, amino acids, and bioavailable trace metals (13–17), and reducing the levels of toxic reactive oxygen species, such as hydrogen peroxide (18, 19). Cultures of marine unicellular cyanobacteria, including Synechococcus and Prochlorococcus, frequently contain associated heterotrophic bacteria, likely due in part to this intimate codependency (20–24). Associations between Synechococcus and heterotrophic bacteria were also detected in the environment (25, 26), and geological evidence for their interactions dates to 440 million years ago (mya) (27).
Synechococcus, an ancient and genetically diverse clade, is ubiquitous in the global ocean and is abundant in both estuarine and coastal waters (28, 29). Marine Synechococcus members have been classified into three major subclusters (5.1, 5.2, and 5.3), and each of these contains dozens of genotypes (28–30). There are clear geographic distribution patterns for diverse Synechococcus clades (29, 30). Those belonging to clades I and IV (subcluster 5.1) are dominant in coastal and higher-latitude regions (31–34). Clade III members (subcluster 5.1) are largely distributed in global oligotrophic waters, similar to clade II (subcluster 5.1) in subtropical/tropical open-ocean waters (29, 32–34). Subcluster 5.2 is typically an estuarine group, with diverse representation in estuaries, such as the Chesapeake Bay (35, 36), whereas subcluster 5.3 members are widely found in the open ocean (e.g., East China, Mediterranean, Sargasso, and South China Seas) (28, 30, 37, 38).
Many different bacteria associate and form close relationships with eukaryotic hosts (e.g., diatoms, corals, and sponges) (10, 13, 39, 40). Compared with eukaryotic entities, how do unicellular Synechococcus spp. act as “microbial habitats” and interact with surrounding heterotrophic bacteria? Some heterotrophic bacteria contribute to aggregate formation and particle sinking in diatom cultures (41). During phytoplankton blooms, although the bulk of the bacterial biomass is free-living, the proportion of bacteria attached to algae or particles increases to up to 20% of the total prokaryotes (1). Therefore, investigation of the bacterial community compositions in free-living and attached/aggregated fractions in Synechococcus cultures might supply clues for understanding the interactions among these community members and with surrounding environments in natural systems.
The aims of this study were to (i) identify the heterotrophic bacteria that occur in coculture with Synechococcus isolates from different environments, (ii) determine whether the heterotrophic bacteria that were found in coculture with Synechococcus isolates have similar geographic distribution patterns as their “host,” (iii) characterize the patterns and dynamics in the bacterial community corresponding to different lifestyles (free-living versus attached/aggregated) in a Synechococcus culture, and (iv) evaluate the potential for succession in carbon utilization patterns by the dominant associated heterotrophic bacteria in these cultures.
RESULTS AND DISCUSSION
Synechococcus isolates.
Ten Synechococcus isolates were selected for an investigation of the community structure of their associated heterotrophic bacteria. Five isolates were isolated from coastal/estuary eutrophic waters, and five isolates were isolated from oligotrophic water in the South China Sea (see Table S1 in the supplemental material). Four Synechococcus strains (XM-5, XM-11, XM-24, and XM-13) were isolated from the Xiamen coastal/estuary region, and one strain (Cy04) was isolated from the coastal Yellow Sea. Three of the Xiamen Synechococcus strains (XM-5, XM-11, and XM-24) belonged to clade CB5 in subcluster 5.2, and the fourth strain (XM-13) was classified into clade IX in subcluster 5.1. Strain Cy04 was grouped into clade VIII in subcluster 5.1 (Table S1). The five oligotrophic Synechococcus strains were isolated from the South China Sea. Two strains (YX-A3-2 and ZS02-2) were classified into clade II in subcluster 5.1, and another two strains (YX02-3 and YX04-3) were grouped into clade III in subcluster 5.1. The fifth strain (ZS01-1) belonged to clade V in subcluster 5.1 (Table S1).
According to the classifications of Synechococcus in previous studies (28–30, 35), three of the Synechococcus strains (XM-5, XM-11, and XM-24) were typical eutrophic estuary genotypes, and four strains (YX-A3-2, ZS02-2, YX02-3, and YX04-3) were oligotrophic water genotypes.
Community composition of Synechococcus-associated heterotrophic bacteria.
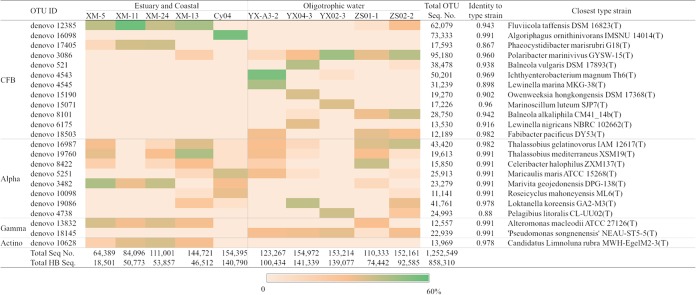
To explore the heterotrophic bacterial community composition associated with Synechococcus isolates from different environments, a total of 1,252,549 16S rRNA gene sequences with paired-end 250-bp reads, ranging from 64,389 to 154,972 per sample, with an average length of 225 bp, were obtained from the 10 Synechococcus culture samples after passing quality filtering and chimera checks, and these were grouped into 939 operational taxonomic units (OTUs). Bacteroidetes (here Cytophaga-Flavobacteria-Bacteroides [CFB] group) and Alphaproteobacteria and Gammaproteobacteria dominated in all samples, and Actinobacteria were found in all five eutrophic Synechococcus cultures. The top 23 OTU among all samples made up approximately 83% of the total heterotrophic bacteria. These 23 OTU were mostly classified into the CFB group (mainly Flavobacteriales and Cytophagales), Alphaproteobacteria (mainly the Roseobacter clade), Gammaproteobacteria (mainly Alteromonadales and Pseudomonadales), and Actinobacteria (Fig. 1 and Table S2).
FIG 1.
The relative abundances of the dominant heterotrophic bacterial OTUs in each 16S rRNA-sequenced Synechococcus culture sample (after removing Synechococcus sequences). Total Seq. No., the total number of sequences including Synechococcus and heterotrophic bacteria in each sample; Total HB No., total number of heterotrophic bacterial sequences (without Synechococcus) in each sample; Total OTU Seq. No., the sum of sequences for each OTU in the 10 samples. Alpha, Alphaproteobacteria; Gamma, Gammaproteobacteria; Actino, Actinobacteria.
In each Synechococcus culture, 1 to 4 dominant OTU (>1% relative abundance) belonging to the CFB group and 1 to 3 dominant alphaproteobacterial OTU were usually found (Fig. 1). At least one dominant gammaproteobacterial OTU was present in all Synechococcus cultures, except for strain Cy04. In the strain ZS01-1 culture, one gammaproteobacterial OTU (denovo14255, with 4,289 sequences) accounted for 5.7% of the total heterotrophic bacteria. As mentioned above, Actinobacteria were found in all eutrophic Synechococcus cultures, with the same actinobacterial OTU found at relatively high abundance in the cultures of the four Synechococcus strains isolated from the coastal Xiamen location (Fig. 1). The Cy04 culture also included an OTU (denovo9467, with 4,567 sequences) belonging to the Actinobacteria, which accounted for 3.2% of its total heterotrophic bacteria. An evaluation of a coastal Xiamen environmental microbial community assembly showed that the dominant Synechococcus-associated heterotrophic bacteria were not abundant in the environmental samples (Table S3), with the exception of denovo10628, which is a member of the Actinobacteria, with corresponding 16S sequences widely distributed in the Xiamen coastal area (Table S3). Overall, it appears that the core heterotrophic bacterium assemblages comprise at least one species from each of the CFB group, Alphaproteobacteria, and Gammaproteobacteria in the coculture system, because at least one of each of these was present in every culture (Fig. 1), and Actinobacteria regularly associate with eutrophic Synechococcus bacteria.
The representative sequences of the dominant OTUs belonging to the CFB group showed much lower identities with type strains, with an average of 94.2% (Fig. 1), in comparison to the representative sequences of dominant OTUs belonging to Alphaproteobacteria and Gammaproteobacteria, which showed 97.4% and 99.1% average identities to type strains, respectively (Fig. 1). The distant relationships of the dominant CFB sequences to known strains indicate that novel representatives were prevalent in the Synechococcus cultures. One possible explanation is that the media used in previous isolation studies could not support the growth of these strains. Another is that these bacteria have been difficult to isolate independently because of their dependency upon Synechococcus and/or other heterotrophs.
The CFB OTU denovo12385 was dominant and common in the four Xiamen coastal Synechococcus cultures, and it shared 94.3% 16S rRNA gene sequence identity with Fluviicola taffensis DSM 16823. Fluviicola-related bacteria have frequently been found attached on particles and during phytoplankton blooms (42–44).
The OTU denovo3086 that was dominant and common among the oligotrophic Synechococcus strains shared 95.3% 16S rRNA gene identity with Polaribacter sp. strain MED152, which is a representative of one of the major genera of Flavobacteria within the CFB group (45). Members of the genus Polaribacter are more abundant in oceanic regions with higher phytoplankton abundance and show significant correlations with chlorophyll a fluorescence (46–48). Genomic analysis showed that strain MED152 possessed a substantial number of genes related to attachment to surfaces or particles and to polymer degradation (45). Flavobacteria are thought to be the “first responders” to phytoplankton blooms and can degrade diverse complex high-molecular-weight organic matter by direct attachment and exoenzymatic attack of algal cells and algae-derived detrital particles (11, 49, 50).
OTUs belonging to Alphaproteobacteria (mainly in the Roseobacter clade) had no clear distribution pattern related to the geographic origins of the Synechococcus (Fig. 1), which likely results from their widespread distributions (51–54). Two OTUs (denovo16987 and denovo19760) with high identity to members of the genus Thalassobius were found in both eutrophic and oligotrophic Synechococcus cultures (Fig. 1). The OTU denovo3482 was dominant in three Synechococcus cultures isolated from the Xiamen coastal site, and it was also detected in the estuary regions in Xiamen coastal environmental samples collected in the winter (Table S3). Its known relatives, Marivita spp., were also found in coastal areas (e.g., Chesapeake Bay and coastal sediments) (55, 56) and phytoplankton cultures, such as those of Pyrodinium bahamense, Coccolithus braarudii, and Emiliania huxleyi (57–59).
The gammaproteobacterial OTU denovo13825, which had high sequence identity to members of the genus Alteromonas, was abundant in four Xiamen coastal Synechococcus cultures, and it was also frequently found in summer samples from the Xiamen coastal area (Table S3). In contrast, the OTU denovo18145, which was common in the oligotrophic Synechococcus cultures, was not identified in the Xiamen coastal environmental samples (Table S3). Members of the genus Alteromonas were reported to contribute to the total bacterial biomass at levels similar to those of Roseobacter and CFB bacteria in phytoplankton blooms (9, 60, 61).
The actinobacterial OTU denovo10626 was classified into the genus Aquiluna and was prevalent in Xiamen coastal microbial communities, especially in the estuary station (Table S3), which indicates they prefer low-salinity eutrophic environments. The presence of Actinobacteria accompanying cyanobacterial blooms was frequently detected in diverse aquatic environments, with their relative abundance changing significantly with bloom development (62–64).
In addition to the Actinobacteria, the CFB group members and Gammaproteobacteria also displayed clear geographic distribution patterns with their host Synechococcus isolation environments (Table S1 and Fig. 1). The OTUs denovo10626 (Actinomycetales), denovo12385 (Flavobacteriales), denovo17405 (Cytophagales), and denovo13832 (Alteromonadales) were mainly obtained from (at least three) eutrophic Synechococcus isolates, whereas OTUs denovo3086 (Flavobacteriales), denovo18503 (Cytophagales), and denovo18145 (Pseudomonadales) were mainly detected from (at least three) oligotrophic isolates.
After examining the community structure of associated heterotrophic bacteria in the Synechococcus cultures isolated from different environments, we next sought to investigate the temporal interactions between heterotrophic bacteria and the “host” Synechococcus cells. A specific eutrophic Synechococcus culture (strain XM-24) was chosen for further analysis. Although we recognize that this study of only one strain limits our ability to know how ubiquitous the observed phenomena are, we believe it is still valuable for probing more deeply into these interactions.
Morphologies of Synechococcus and associated heterotrophic bacteria in the Synechococcus sp. XM-24 culture.
To investigate possible interactions of the Synechococcus cells and associated heterotrophic bacteria in the Synechococcus sp. XM-24 culture, transmission electron microscopy (TEM) was used to visualize cells from the cocultures. The Synechococcus cells were short rods or coccoid-shaped (0.6 ± 0.2 μm by 0.8 ± 0.2 μm) with relatively strong electron density (Fig. S1A). Aggregated cells with different shapes were frequently detected from samples after late-exponential phase (Fig. S1B to D). In addition to the short rods or coccoid-shaped cells, long rod-shaped cells (0.3 ± 0.1 μm by 2 ± 0.5 μm) were also abundant in the cultures. Aggregations of cells in irregular clusters have frequently been observed for many marine bacteria, including cyanobacteria, and these might be caused by changes in the growth conditions and surrounding environments (65). Conjoined Synechococcus and heterotrophic bacterial cells were also observed in the marine environment by atomic force microscopy (25, 26).
Dynamic relationship between Synechococcus sp. XM-24 and its cocultured heterotrophic bacteria.
To characterize the dynamics in the Synechococcus culture, the variations in the abundances of Synechococcus strain XM-24 and heterotrophic bacteria were determined by flow cytometry. The similar dynamic patterns were observed in the different original inoculum ratios of Synechococcus to heterotrophic bacteria (1.77 × 107 to 2.52 × 107, 1.54 × 107 to 6.01 × 105 cells · ml−1, and 3.69 × 107 to 1.77 × 107 cells · ml−1, respectively) (Fig. 2A and S2). Therefore, the Synechococcus culture described in Fig. 2A was chosen to be representative of the following analyses about dynamic patterns, organic carbon concentrations, and community compositions. The Synechococcus strain was inoculated at 1.77 × 107 cells · ml−1 and reached a peak density of 2.24 × 109 cells · ml−1. The heterotrophic bacterial abundance ranged from 2.52 × 107 to 1.51 × 109 cells · ml−1 over the incubation period. Four phases (lag, exponential, stationary, and decline) were visible from the Synechococcus growth curve (Fig. 2A and S2). Interestingly, a sharp decline in heterotrophic bacterial abundance was seen during the Synechococcus exponential phase. The heterotrophic bacterial abundance slowly increased when Synechococcus began to decline and exceeded Synechococcus numbers over the second half of the decline phase. Our preliminary data showed this dynamic relationship was universal among eutrophic Synechococcus cultures, whereas a different pattern was found in the oligotrophic Synechococcus cultures.
FIG 2.
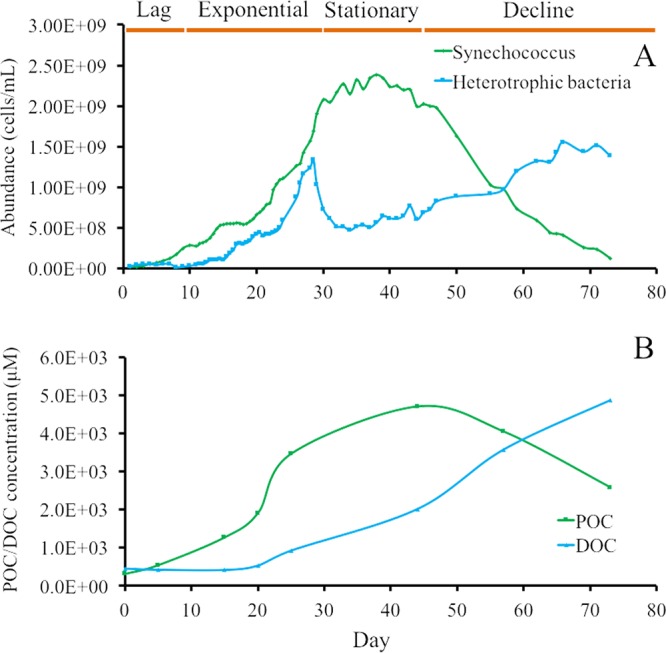
The dynamic relationships between Synechococcus and heterotrophic bacteria (A) and POC and DOC (B) in Synechococcus sp. XM-24 culture over a 72-day growth period. The abundances of Synechococcus and heterotrophic bacteria were measured by flow cytometry. The original abundances of Synechococcus and heterotrophic bacteria were 1.76 × 107 and 2.52 × 107 cells · ml−1, respectively. A similar dynamic pattern with different initial inocula of Synechococcus sp. XM-24 liquid cultures was observed (Fig. S2 in the supplemental material).
The concentration of POC showed a pattern similar to that of Synechococcus abundance, with a rapid increase during the exponential phase and peaking during stationary phase (Fig. 2B). The DOC concentration rose slowly throughout the culture incubation beginning in exponential phase and then surpassed POC in the decline phase.
Although it is not visible in Fig. 2, the abundance of heterotrophic bacteria was slightly higher than that of Synechococcus in the early lag phase (2.52 × 107 versus 1.77 × 107 cells · ml−1, respectively). The abundance of Synechococcus cells exceeded the heterotrophic bacteria on the fifth day, and this trend continued to the late-decline phase. However, the highest concentration of heterotrophic bacteria was lower than the peak Synechococcus number. Based on the correlations of the cell abundances and POC/DOC concentrations, Synechococcus was the major source of organic matter in this coculture system, and the labile organic matter consisted of dissolved organic matter (mainly in the exponential and stationary phases) directly released by Synechococcus cells and Synechococcus-derived detrital particles (mainly in the decline phase).
During the lag phase, Synechococcus and the heterotrophic bacteria tended to aggregate, as most of the cells were in the larger-size fraction (Fig. S3). After the lag phase, the Synechococcus bacteria grew exponentially as free-living cells (Fig. S3). In the late-exponential phase, the heterotrophic bacteria tended to attach to Synechococcus again, possibly because of biopolymers (such as polysaccharides) secreted by Synechococcus (66, 67), which accounted for the sudden decline in heterotrophic bacterial abundance (Fig. 2A). Since the heterotrophic bacterial number was calculated as total bacteria minus Synechococcus bacteria, it would be underestimated as result of the attachment or aggregation. After the stationary phase, in the decline period, the Synechococcus cells were likely degraded by exoenzymatic attack from certain heterotrophic bacteria (11, 50). Tight associations between Synechococcus and the heterotrophic bacteria appeared to be important for the decomposition of organic matter and nutrient cycling in this system (Fig. 2).
Free-living versus attached lifestyles of Synechococcus sp. XM-24 over the culture period.
To access the different lifestyles (free-living versus attached/aggregated) in the Synechococcus culture, sequencing of 16S rRNA gene amplicons from 58 size-fractionated samples collected over the culture growth generated a total of 1,083,985 sequences (530,451 sequences from the 0.22- to 3-μm size fraction and 553,534 sequences from the >3-μm size fraction) with paired-end 450-bp reads that passed the quality control pipeline, with an average length of 392.5 bp.
The average relative abundances of Synechococcus in the total 16S rRNA sequences for each size fraction represented 64.3% and 36.9% of total bacteria in the 0.22- to 3-μm and >3-μm size fractions, respectively, over the entire culture growth. Except during the first day, the relative abundance of Synechococcus increased rapidly in both size fractions in the lag and early exponential phases and decreased in the decline phase (Fig. 3). After the first day, Synechococcus maintained a high relative abundance (>58%) over most of the incubation period, until the decline phase, where it was reduced to <30% of the total bacteria in the 0.22- to 3-μm fraction samples (Fig. 3). In the middle of the exponential phase (from the 13th to the 29th day), Synechococcus maintained a high relative abundance (average, 47.7%) in the >3-μm fraction samples. The relative abundance of Synechococcus decreased from the late-exponential phase (from the 31st day) and represented only 32.4% of the total bacteria from the 33rd to the 43rd day (during the stationary phase) in the >3-μm fraction samples. A similar trend could be observed in the flow cytometry data, which showed that more than one heterotrophic bacterial cell aggregated or attached to a Synechococcus cell at the late-exponential phase, leading to an increased relative abundance of free-living Synechococcus and a reduced relative abundance of attached cells (Fig. 3, S3, and S4).
FIG 3.
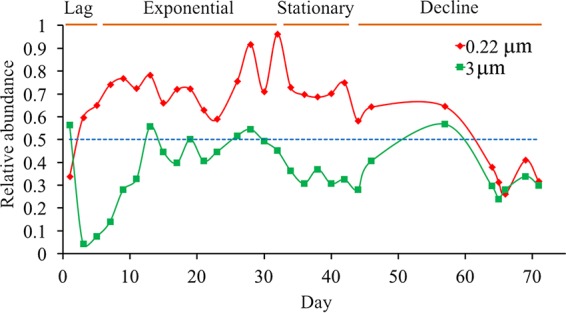
The relative abundances of Synechococcus in the total 16S rRNA sequences for each time point from the 0.22- to 3-μm (red) and >3-μm (green) size fractions during the Synechococcus sp. XM-24 culture growth. The blue line shows where the relative abundance equals 50%.
Free-living versus attached lifestyles of the heterotrophic bacteria in the Synechococcus sp. XM-24 culture.
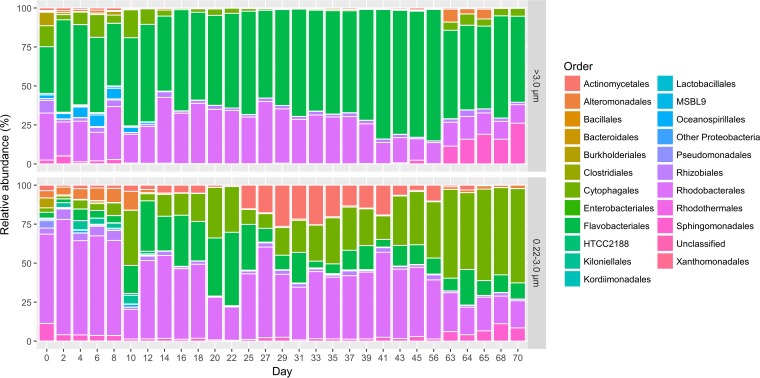
A total of 193,630 and 351,644 sequences were obtained from the 0.22- to 3-μm size fractions, respectively, after removing Synechococcus reads. These sequences were grouped into 380 OTU, which mostly were classified in seven orders: Flavobacteriales (44.2%), Rhodobacterales (28.8%), Cytophagales (13.1%), Sphingomonadales (4.1%), Actinomycetales (2.4%), Rhizobiales (2.3%), and Alteromonadales (1.5%) (Fig. 4). The most numerous OTU in each of these seven orders comprised 66.8 to 97.3% of the total sequences within each respective order, and each of the orders contained 1 to 3 different dominant OTUs (Table S4). Each of the most abundant OTUs contained approximately 3 to 5 dominant genetic types that showed 1 to 3 single-nucleotide polymorphisms, suggesting that each dominant OTU represented a specific strain. As different primer sets were used to investigate microbial assemblages in the 10 cultures versus the XM-24 culture time course, the two sequence data sets could not be put together for analysis. However, the OTUs present in the Synechococcus sp. XM-24 culture experiment were only a subset of those identified in the 10 Synechococcus cultures in the above-described investigation.
FIG 4.
The heterotrophic bacterial community structure from the 0.22- to 3-μm (top) and >3-μm (bottom) size fractions. The relative abundances of different bacterial orders in the total heterotrophic bacterial 16S rRNA sequences (after removing Synechococcus sequences) of each time point from the size-fractionated samples collected at different time points are shown.
The top 10 OTU made up 89.4% of the total heterotrophic bacterial sequences. Some patterns for lifestyle preference (free-living versus attached) and responses according to the Synechococcus growth phases could be discerned (Table S4 and Fig. 5). Two OTUs (OTU1 and OTU4) belonging to the Flavobacteria were most abundant in the attached/aggregated fraction (Table S4 and Fig. 5). OTU1 (corresponding to denovo12385 in Fig. 1), which is classified into the order Flavobacteriales and genus Fluviicola, was the most dominant heterotrophic bacterial OTU. Its relative abundance from the >3-μm fraction was much higher than from the 0.22- to 3-μm fraction over the entire incubation (Fig. 5). Its relative abundance in the 0.22- to 3-μm fraction did increase during the exponential phase, with lower abundance in the other three phases. However, its relative abundance in the >3-μm fraction increased over the entire incubation until the decline phase, when it decreased quickly.
FIG 5.
Comparison of relative abundances of the top 10 heterotrophic bacterial OTUs from the 0.22- to 3-μm and >3-μm size fractions over the Synechococcus sp. XM-24 culture growth period. OTUs 1 and 4, order Flavobacteriales; OTUs 2, 8, and 9, order Rhodobacterales; OTU 3, order Cytophagales; OTU 5, Sphingomonadales; OTU6, order Actinomycetales; OTU7, order Rhizobiales; OTU10, order Alteromonadales.
The second most dominant OTU (OTU2, corresponding to denovo3482 in Fig. 1), which belongs to the genus Marivita in the Roseobacter clade, shifted between free-living and attached over the culture growth period (Table S4 and Fig. 5). In the lag and decline phases, it was more abundant in the free-living fraction, while its distribution was more even during the exponential and stationary phases. This pattern is consistent with the known abilities of related bacteria to metabolize biopolymers generated by marine phytoplankton using hydrolytic enzymes (17) and to directly import low-molecular-weight DOC (53, 68).
The third most dominant OTU (OTU3, corresponding to denovo17405 in Fig. 1), which belonged to the genus Roseivirga in the order Cytophagales, was most abundant in the free-living fraction (Table S4 and Fig. 5). Its relative abundance from the 0.22- to 3-μm fraction surpassed that from >3-μm fraction in the early exponential phase and increased greatly in the decline phase, and it had relatively low relative abundance in the >3-μm fraction in all four growth phases. The lifestyles of the three most dominant OTUs were also proven in another two incubation systems of Synechococcus sp. XM-24 cultures (Fig. S4).
Both OTU4 and OTU5, belonging to the orders Flavobacteriales and Sphingomonadales, respectively, had a clear response in the decline phase (Table S4 and Fig. 5), and the response in the >3-μm fraction was much stronger than in the 0.22- to 3-μm size fraction. This indicates that these bacteria grow quickly by direct attachment to other microbial cells or derived detrital particles. Members of the Sphingomonadales are prevalent within phytoplankton blooms and are abundant in particle-attached samples (69–72).
OTU6 (corresponding to denovo10628 in Fig. 1), which was classified into the genus Aquiluna in the order Actinomycetales, was mainly found in the 0.22- to 3-μm size fraction and had higher relative abundance in the late-exponential and stationary phases (Table S4 and Fig. 5). This corresponds to when the Synechococcus cells began to release more DOC (Fig. 2). Although there was a much higher DOC concentration in the decline phase, most of this represents by-products from Synechococcus and the degradation of other bacterial cells. This suggests that this actinobacterium might prefer fresh DOC released by Synechococcus. Previous studies showed that Actinobacteria could be found at high abundance in both free-living and particle-attached lifestyles, especially in temperate freshwater ecosystems (73, 74), whereas others have suggested that actinobacterial abundance is significantly higher in free-living than in particle-attached states (37, 75). The high diversity within Actinobacteria might account for different lifestyles (76). However, there was only one group of Actinobacteria found in these Synechococcus cultures, and they were most similar to the known “Candidatus Aquiluna” sp. strain IMCC13023 that possesses a relatively small genome size (1.36 Mb) (77).
Both OTU7 and OTU8, which belonged to the orders Rhizobiales and Rhodobacterales, respectively, showed no differences in their relative abundances between the two size fractions, although both were in relatively high abundance in the lag phase. OTU9, belonging to the Rhodobacterales, showed higher relative abundance in the early growth stage in the free-living fraction. OTU10, corresponding to denovo13832 in Table S4 and classified in the order Alteromonadales, was abundant in the free-living fraction at the early growth stage and then in the attached fraction late in the decline phase (Table S4 and Fig. 5).
Some of the heterotrophic bacteria had obvious preferences for a free-living (e.g., OTU3 and OTU6) or attached/aggregated (e.g., OTU1) lifestyle. Others, such as OTU2, shifted between the two different lifestyles. Similarly, some environmental microbes have been found to occur in different size fractions (78, 79). Therefore, these lifestyles are not only determined by the bacterial genetic features but also by changes in the surrounding environment.
Origin of Synechococcus-associated heterotrophic bacteria.
In this work, some of the Synechococcus strains were isolated using dilution methods on plates, which have a low efficiency of isolation success, and continuous dilution streaking of the Synechococcus onto plates failed to produce axenic isolates. It was previously found that axenic cultures of some cyanobacteria, obtained by antibiotic treatment, grew efficiently only if the inoculum was large (14, 18, 19). This would help explain the difficulty in obtaining Synechococcus colonies using dilution methods, where the inoculum is much lower. However, previous studies showed that colonies of cyanobacteria formed easily when they were cocultured with heterotrophic bacteria that helped the cyanobacteria reduce oxidative stress (14, 18). In addition, Synechococcus bacteria were observed as being conjoined with heterotrophic bacteria in the environment (25, 26). In line with this logic and going through the isolation process used here, at the early stage of dilution and spreading of the water samples on the plates, the Synechococcus bacteria interacting with heterotrophic bacteria would be able to grow (Fig. S5). A few heterotrophic bacteria might be able to utilize agar components and form small colonies on the plate (Fig. S5), but most marine heterotrophic bacteria will not grow on the SN medium (95) employed unless they are associated with growing Synechococcus cells. After the Synechococcus colonies begin growing, organic material is released around the colonies, and the associated heterotrophic bacteria in the plate continue to survive and grow (Fig. S5). The heterotrophic bacteria form close associations with and attach to the Synechococcus cells. In order to obtain additional fresh organic matter, the heterotrophic bacteria that were utilizing agar components or surviving through other means might also attach to the Synechococcus colonies (Fig. S5). These original heterotrophic bacteria that facilitate Synechococcus colony formation and that attach and grow with the colony represent the Synechococcus-associated heterotrophic bacteria that were present in the subsequent liquid cultures.
It is possible that the agar itself is a potential carbon source for some marine heterotrophic bacteria, and the agar concentration used for the Synechococcus isolations might affect heterotrophic bacterial growth and composition. In addition, the different capacities with respect to motility and chemotaxis of heterotrophic bacteria on the agar plates might impact the formation of the heterotrophic bacterial pool.
In the liquid culture, the bacteria responded to multiple selection pressures, and some became abundant at different growth phases. The tight associations between Synechococcus and the heterotrophic bacteria reflect the complementation of physiological or biochemical functions, and the resultant heterotrophic bacterial community structure presumably allows for optimal metabolic performance and enhanced productivity.
Community succession during utilization of organic matter from Synechococcus cells.
In order to easily understand and visualize the interactions between Synechococcus and associated dominant heterotrophic bacteria, a conceptual model was built based on the different lifestyles employed over the culture growth and on carbon availability and utilization changes in the cultures (Fig. 6). It displays a successional utilization relationship related to the organic matter availability. This agrees with previous publications (10, 50). Analysis of the associated heterotrophic bacterial assemblages indicates that at least one species from each of the Flavobacteria, Alphaproteobacteria, and Gammaproteobacteria consists of the core and abundant components in the Synechococcus coculture system.
FIG 6.
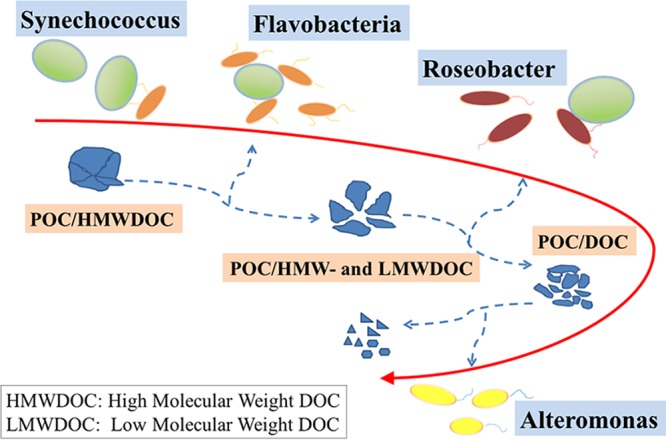
Schematic representation of the patterns of utilization of the organic matter produced by Synechococcus by different heterotrophic bacteria. HMWDOC, high-molecular-weight DOC; LMWDOC, low-molecular-weight DOC. Flavobacteria prefer an attached lifestyle in the Synechococcus culture and can degrade POC and complex HMWDOC. Roseobacters shift between free-living and attached lifestyles, and they not only utilize POC/HMWDOC but also uptake labile LMWDOC, including by-products from the growth of Synechococcus and Flavobacteria. Alteromonas demonstrates broad substrate preferences and may use different complementary growth strategies relative to the other two bacterial groups.
Flavobacteria, which were the most abundant group and preferred an attached lifestyle in the Synechococcus cultures (Fig. 1 and 5), seem to be specialized in the degradation of complex high-molecular-weight organic matter (Fig. 6) (45, 50, 80, 81). Members of the Roseobacter lineage, the second-most dominant group in the cultures, shifted between free-living and attached lifestyles. As ecological generalists, roseobacters not only utilize biopolymers with hydrolytic enzymes (17) but also uptake labile low-molecular-weight DOC, including by-products from the growth of Synechococcus and Flavobacteria (Fig. 6) (50, 53, 68). Marine Gammaproteobacteria, specifically members of the Alteromonadales, demonstrate broad substrate preferences (82, 83) and have obvious responses during phytoplankton blooms (9, 60, 61). According to their behavior in the Synechococcus cultures in this study, with the largest responses in the early growth stage and decline phase, they cycled opposite of the Flavobacteria and roseobacters, suggesting they may use different complementary growth strategies relative to the other two dominant bacterial groups.
Cyanobacteria contribute approximately 40% of the global ocean primary production (84–87), and organic matter excretion by these primary producers drives carbon cycling in the upper ocean (88, 89). The utilization and metabolism of the released organic carbon are then mainly carried out by heterotrophic microbes. Marine heterotrophic bacteria can incorporate organic carbon into biomass, respire it into CO2, or degrade the labile fractions into refractory compounds by successive utilization (3). Synechococcus has also frequently been detected in deep-sea sediment traps and was estimated to contribute significant amounts of the total sediment trap POC flux (90–93). Aggregates of Synechococcus and heterotrophic bacteria might facilitate the sinking process. Understanding the processes of organic carbon excretion and utilization is complicated by the interactions between phototrophs and heterotrophs and how this affects global biogeochemical cycling.
In the global ocean, Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes are cosmopolitan and abundant bacterioplankton populations (49, 94). In the Synechococcus cultures, the core and dominant microbial assemblies also contained these three bacterial clades. Is this a coincidence as a result of their abundance, or do they have some complementary metabolic characteristics and synergistic effects to sustain the ecosystem? Further metagenomic and metatranscriptomic data would shed new light on understanding the interactions between photoautotrophs and associated heterotrophic bacteria and community assembly principles relevant to natural microbial communities.
MATERIALS AND METHODS
Synechococcus strain isolations.
Water samples were collected from the coastal region near Xiamen (∼24°N, ∼118°E) and near Dalian (∼38°N, ∼121°E) and from the South China Sea near Yongxing Island (∼17°N, ∼112° E) in April 2014. The coastal area near Xiamen is a typical tropical or subtropical eutrophic oceanic region (Table S5), whereas the water near Yongxing Island is oligotrophic (Table S5). Seawater samples from the subsurface (1 m) were filtered through 3.0-μm-pore-size polycarbonate filters by gravity and the filtrates used for isolations.
Synechococcus was isolated using SN medium (95) with reduced salinity (25 ppt) for the coastal Xiamen samples. The top agar overlay method was applied to improve the efficiency of isolation (96). The bottom layer agar concentration was 1.0% (wt/vol), and the top layer concentration was 0.5%. The culture plates were incubated at 25°C under constant cool white fluorescent light (Philips, Somerset, NJ, USA) at 10 μmol photons · m−2 · s−1. Synechococcus colonies usually appeared after 1 to 2 months of incubation. Colonies with different appearances or morphologies were picked and inoculated to fresh plates for further purification. Single colonies (after 2 to 3 rounds of purification) were then inoculated into 96-well microtiter plates (Corning, NY, USA), with each well containing 200 μl of SN medium. The cultures were then scaled up in steps (10 ml, 100 ml, and 500 ml). Synechococcus strain Cy04 was isolated independently from the coastal region near Dalian with the same method described above. Synechococcus isolations from the South China Sea samples used the same incubation conditions as described above but were done in PRO2 liquid medium (21), with the water samples diluted until a single Synechococcus internal transcribed spacer (ITS) sequence (after ∼4 to 7-rounds of dilutions) was obtained.
The scaled-up Synechococcus sp. XM-24 culture was incubated at 25°C at 10 μmol photons · m−2 · s−1 without shaking (to reduce the risk of external contamination). However, the culture was mixed well in the laminar flow work area before each sampling. Three replicates that were inoculated by different growth phases of Synechococcus liquid cultures were performed to detect the dynamic patterns of Synechococcus and associated heterotrophic bacteria with growth.
Electron microscopy.
Cell morphologies were examined by transmission electron microscopy (JEM-1230; JEOL), using negative staining with phosphotungstic acid on cells collected in the stationary phase of growth.
Cell collections, DNA extractions, and PCR amplification of Synechococcus ITS regions.
For extractions of total culture DNA from the 10 different environmental isolates, 6 to 8 ml of liquid Synechococcus culture (in the stationary or decline phase) was collected and centrifuged. For collection of size-fractionated samples, 6 to 8 ml of the Synechococcus liquid culture was sequentially filtered through 3- and 0.22-μm-pore-size polycarbonate filters (47-mm diameter; Millipore) at a pressure of <0.03 MPa. To remove free-living cells from the 3-μm filters, a second clean 3-μm filter was placed on top of the first one with cells, and the filters were turned over. Autoclaved 0.22-μm-filtered seawater was then used to wash the cells located between the two filters and the two filters were retained to represent the >3.0-μm fraction. Cell pellets and filters were flash-frozen in liquid nitrogen and then stored at −80°C until DNA extraction.
Nucleic acids were extracted with hot sodium dodecyl sulfate, phenol, chloroform, and isoamyl alcohol (97). The quality and quantity of DNA were assessed using a NanoDrop device (ND-2000; Thermo Fisher). The DNA was stored at −80°C until use.
Synechococcus ITS DNA fragments were amplified using the 16S-1247f and 23S-241r primer set (98). The PCR mixtures (50 μl total volume) contained 5 μl of 10× Taq polymerase buffer, 0.2 mM each dinucleoside triphosphate (dNTP), 0.5 μM each primer, 1.25 U of Taq DNA polymerase (TaKaRa, Japan), and 20 to 200 ng of template DNA.
TOC and DOC sampling and measurement.
For total organic carbon (TOC) sampling, 20-ml samples were collected directly into 40-ml glass vials (CNW, Germany) and immediately stored at −20°C for further analysis. For DOC sampling, 20-ml samples were filtered through precombusted (450°C for 4 h) 0.7-μm-pore-size GF/F filters (47-mm diameter; Whatman, Maidstone, UK) into 40-ml glass vials and stored at −20°C after sampling. To prevent carbon contamination, all glass materials used for sample collection and storage were acid-washed, rinsed with Milli-Q (ion- and nuclease-free) water, and precombusted, as described above. TOC and DOC concentrations were measured by high-temperature catalytic oxidation (HTCO) using a Shimadzu TOC-VCPH analyzer. Before analysis, samples were melted and acidified to a pH of ≈2 with phosphoric acid. POC concentrations were obtained by subtracting the DOC concentrations from the TOC concentrations. The Synechococcus culture with original inoculum ratio of Synechococcus to heterotrophic bacteria (1.77 × 107 to 2.52 × 107 cells · ml−1, described in Fig. 2A) was chosen to be representative of the analyses of organic carbon concentrations.
Counts of Synechococcus and heterotrophic bacterial cells.
Synechococcus and heterotrophic bacteria cell numbers were measured using flow cytometry, as described previously (99), in Synechococcus sp. XM-24 culture. Briefly, an Epics Altra II flow cytometer (Beckman Coulter, Inc., Brea, CA, USA) equipped with an external quantitative sample injector (Harvard Apparatus PHD 2000; Instech Laboratories, Inc.) was used to quantify Synechococcus and the total bacterial cells. For Synechococcus, they were identified from plots of side scatter versus red fluorescence and orange fluorescence versus red fluorescence (100). Total bacteria were enumerated according to the method of Marie et al. (101) in plots of red fluorescence versus green fluorescence and side scatter versus green fluorescence after staining with SYBR green I. Subtracting the Synechococcus count from the total bacteria count gave the number of heterotrophic bacteria. All flow cytometry data were analyzed using the Expo 32 Multi-COMP software (Beckman Coulter, Inc.).
Sequence generation and processing.
DNA extractions from Synechococcus culture samples, without size fractionation, were used for amplification of the bacterial hypervariable V4 region of 16S rRNA gene using the primers 520F (5′-AYTGGGYDTAAAGNG-3′) and 802R (5′-TACNVGGGTATCTAATCC-3′) (102). Sequencing libraries were constructed using the NEBNext Ultra DNA library prep kit for Illumina (New England BioLabs, USA), according to the manufacturer's recommendations in terms of NEBNext end prep, adaptor ligation, size selection, or cleanup of adaptor-ligated DNA, PCR enrichment of adaptor-ligated DNA, and cleanup of PCR amplification. The library quality was assessed with a Qubit 2.0 fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. The library was sequenced on the Illumina MiSeq (Illumina, San Diego, CA, USA) platform. The low-quality reads (k-mer 5 < Q20, length < 150 bp, or with ambiguous base calls) were removed, and the paired-end 250-bp reads were then combined using the FLASH software version 1.2.7 (103). Questionable reads, with homopolymer length of >8 bases or a 5′ primer mismatch of >1, were removed from the total read pool by QIIME version 1.8.0 (104). Chimeras were removed by USEARCH version 5.2.236 (105), and UCLUST was employed to cluster the remaining high-quality reads to generate operational taxonomic units (OTUs) with an identity cutoff of 0.97 (106). The OTUs were taxonomically classified based on the Greengenes release 13.8 (107) within the QIIME pipeline, with default parameters (104). OTUs with a relative abundance of <0.001% were removed to avoid inflated estimates of diversity (108).
For the 58 size-fractionated samples from Synechococcus strain XM-24 cultures, the sequence manipulations were similar to those described above, but different primers, targeting the bacterial V3-V4 region (515F, 5′-GTGCCAGCMGCCGCGGTAA-3′; and 907R, 5′-CCGTCAATTCMTTTRAGTTT-3′) of the 16S rRNA gene (109, 110), were used. With changes in sequencing technology, the read length (with paired-end 450-bp reads) that could be sequenced by Illumina greatly increased. Therefore, the 16S rRNA primers covering a longer region were used for the subsequent 58 samples.
Accession number(s).
The OTU sequence data were deposited in the NCBI Sequence Read Archive under BioProject no. PRJNA378717. The sequences of the size-fractionated samples were deposited in the NCBI database with BioProject no. PRJNA362761. The ITS sequences of 10 Synechococcus strains have been submitted to the GenBank databases with accession numbers KY930773 to KY930776 and KY933643 to KY933648.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhao Zhao, Wei Yan, and Xilin Xiao for supplying the Synechococcus strains and Yuyu Liao for collecting the samples.
This work was supported by the National Key Research Programs (grants 2016YFA0601400 and 2013CB955700), a National Natural Science Foundation of China (NSFC) project (41776145), the State Oceanic Administration (SOA) project (GASI-03-01-02-05), the China National Offshore Oil Corp (CNOOC) grants CNOOC-KJ125FZDXM00TJ001-2014 and CNOOC-KJ125FZDXM00ZJ001-2014, and the China National Marine Science Talent Training Base project (2015Z01). J.S. is supported by a National Natural Science Foundation of China (NSFC) project (41676112).
We declare no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01517-17.
REFERENCES
- 1.Azam F, Fenchel T, Field J, Gray J, Meyer-Reil L, Thingstad F. 1983. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 10:257–263. doi: 10.3354/meps010257. [DOI] [Google Scholar]
- 2.Cotner JB, Biddanda BA. 2002. Small players, large role: microbial influence on biogeochemical processes in pelagic aquatic ecosystems. Ecosystems 5:105–121. doi: 10.1007/s10021-001-0059-3. [DOI] [Google Scholar]
- 3.Jiao N, Herndl GJ, Hansell DA, Benner R, Kattner G, Wilhelm SW, Kirchman DL, Weinbauer MG, Luo T, Chen F, Azam F. 2010. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat Rev Microbiol 8:593–599. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- 4.Bassler BL, Losick R. 2006. Bacterially speaking. Cell 125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nat Rev Microbiol 5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 6.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stocker R. 2012. Marine microbes see a sea of gradients. Science 338:628–633. doi: 10.1126/science.1208929. [DOI] [PubMed] [Google Scholar]
- 8.Brussaard CP, Bidle KD, Pedrós-Alió C, Legrand C. 2016. The interactive microbial ocean. Nat Microbiol 2:16255. doi: 10.1038/nmicrobiol.2016.255. [DOI] [PubMed] [Google Scholar]
- 9.Sarmento H, Gasol JM. 2012. Use of phytoplankton-derived dissolved organic carbon by different types of bacterioplankton. Environ Microbiol 14:2348–2360. doi: 10.1111/j.1462-2920.2012.02787.x. [DOI] [PubMed] [Google Scholar]
- 10.Buchan A, Lecleir GR, Gulvik CA, González JM. 2014. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol 12:686–698. doi: 10.1038/nrmicro3326. [DOI] [PubMed] [Google Scholar]
- 11.Gómez-Pereira PR, Schüler M, Fuchs BM, Bennke C, Teeling H, Waldmann J, Richter M, Barbe V, Bataille E, Glöckner FO, Amann R. 2012. Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean. Environ Microbiol 14:52–66. doi: 10.1111/j.1462-2920.2011.02555.x. [DOI] [PubMed] [Google Scholar]
- 12.Arandia-Gorostidi N, Weber PK, Alonso-Sáez L, Morán XAG, Mayali X. 2017. Elevated temperature increases carbon and nitrogen fluxes between phytoplankton and heterotrophic bacteria through physical attachment. ISME J 11:641–650. doi: 10.1038/ismej.2016.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amin SA, Green DH, Hart MC, Küpper FC, Sunda WG, Carrano CJ. 2009. Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc Natl Acad Sci U S A 106:17071–17076. doi: 10.1073/pnas.0905512106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi S, Itoh K, Suyama K. 2011. Growth of the cyanobacterium Synechococcus leopoliensis CCAP1405/1 on agar media in the presence of heterotrophic bacteria. Microbes Environ 26:120–127. doi: 10.1264/jsme2.ME10193. [DOI] [PubMed] [Google Scholar]
- 15.Kazamia E, Czesnick H, Nguyen TTV, Croft MT, Sherwood E, Sasso S, Hodson SJ, Warren MJ, Smith AG. 2012. Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ Microbiol 14:1466–1476. doi: 10.1111/j.1462-2920.2012.02733.x. [DOI] [PubMed] [Google Scholar]
- 16.Xie B, Bishop S, Stessman D, Wright D, Spalding MH, Halverson LJ. 2013. Chlamydomonas reinhardtii thermal tolerance enhancement mediated by a mutualistic interaction with vitamin B12-producing bacteria. ISME J 7:1544–1555. doi: 10.1038/ismej.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christie-Oleza JA, Scanlan DJ, Armengaud J. 2015. “You produce while I clean up,” a strategy revealed by exoproteomics during Synechococcus-Roseobacter interactions. Proteomics 15:3454–3462. doi: 10.1002/pmic.201400562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. 2008. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol 74:4530–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. 2011. Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean's surface. PLoS One 6:e16805. doi: 10.1371/journal.pone.0016805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chisholm SW, Frankel SL, Goericke R, Olson RJ, Palenik B, Waterbury JB, West-Johnsrud L, Zettler ER. 1992. Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch Microbiol 157:297–300. doi: 10.1007/BF00245165. [DOI] [Google Scholar]
- 21.Moore LR, Coe A, Zinser ER, Saito MA, Sullivan MB, Lindell D, Frois-Moniz K, Waterbury J, Chisholm SW. 2007. Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr Methods 5:353–362. doi: 10.4319/lom.2007.5.353. [DOI] [Google Scholar]
- 22.Cole JK, Hutchison JR, Renslow RS, Kim Y-M, Chrisler WB, Engelmann HE, Dohnalkova AC, Hu D, Metz TO, Fredrickson JK, Lindemann SR. 2014. Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: model systems for the study of autotroph-heterotroph interactions. Front Microbiol 5:109. doi: 10.3389/fmicb.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biller SJ, Coe A, Chisholm SW. 2016. Torn apart and reunited: impact of a heterotroph on the transcriptome of Prochlorococcus. ISME J 10:2831–2843. doi: 10.1038/ismej.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aharonovich D, Sher D. 2016. Transcriptional response of Prochlorococcus to co-culture with a marine Alteromonas: differences between strains and the involvement of putative infochemicals. ISME J 10:2892–2906. doi: 10.1038/ismej.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malfatti F, Azam F. 2009. Atomic force microscopy reveals microscale networks and possible symbioses among pelagic marine bacteria. Aquatic Microb Ecol 58:1–14. doi: 10.3354/ame01355. [DOI] [Google Scholar]
- 26.Malfatti F, Samo TJ, Azam F. 2010. High-resolution imaging of pelagic bacteria by atomic force microscopy and implications for carbon cycling. ISME J 4:427–439. doi: 10.1038/ismej.2009.116. [DOI] [PubMed] [Google Scholar]
- 27.Tomescu AM, Honegger R, Rothwell GW. 2008. Earliest fossil record of bacterial-cyanobacterial mat consortia: the early Silurian Passage Creek biota (440 Ma, Virginia, USA). Geobiology 6:120–124. doi: 10.1111/j.1472-4669.2007.00143.x. [DOI] [PubMed] [Google Scholar]
- 28.Dufresne A, Ostrowski M, Scanlan DJ, Garczarek L, Mazard S, Palenik BP, Paulsen IT, de Marsac NT, Wincker P, Dossat C, Ferreira S, Johnson J, Post AF, Hess WR, Partensky F. 2008. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol 9:R90. doi: 10.1186/gb-2008-9-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. 2009. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev 73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang S, Wilhelm SW, Harvey HR, Taylor K, Jiao N, Chen F. 2012. Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J 6:285–297. doi: 10.1038/ismej.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferris MJ, Palenik B. 1998. Niche adaptation in ocean cyanobacteria. Nature 396:226. doi: 10.1038/24297. [DOI] [Google Scholar]
- 32.Toledo G, Palenik B. 2003. A Synechococcus serotype is found preferentially in surface marine waters. Limnol Oceanogr 48:1744–1755. doi: 10.4319/lo.2003.48.5.1744. [DOI] [Google Scholar]
- 33.Zwirglmaier K, Heywood JL, Chamberlain K, Woodward EMS, Zubkov MV, Scanlan DJ. 2007. Basin-scale distribution patterns of picocyanobacterial lineages in the Atlantic Ocean. Environ Microbiol 9:1278–1290. doi: 10.1111/j.1462-2920.2007.01246.x. [DOI] [PubMed] [Google Scholar]
- 34.Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, Not F, Massana R, Ulloa O, Scanlan DJ. 2008. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol 10:147–161. [DOI] [PubMed] [Google Scholar]
- 35.Chen F, Wang K, Kan J, Suzuki MT, Wommack KE. 2006. Diverse and unique picocyanobacteria in Chesapeake Bay, revealed by 16S-23S rRNA internal transcribed spacer sequences. Appl Environ Microbiol 72:2239–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai H, Wang K, Huang S, Jiao N, Chen F. 2010. Distinct patterns of picocyanobacterial communities in winter and summer in the Chesapeake Bay. Appl Environ Microbiol 76:2955–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahlgren NA, Rocap G. 2006. Culture isolation and culture-independent clone libraries reveal new marine Synechococcus ecotypes with distinctive light and N physiologies. Appl Environ Microbiol 72:7193–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi DH, Noh JH. 2009. Phylogenetic diversity of Synechococcus strains isolated from the East China Sea and the East Sea. FEMS Microbiol Ecol 69:439–448. doi: 10.1111/j.1574-6941.2009.00729.x. [DOI] [PubMed] [Google Scholar]
- 39.Rohwer F, Seguritan V, Azam F, Knowlton N. 2002. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10. doi: 10.3354/meps243001. [DOI] [Google Scholar]
- 40.Taylor MW, Schupp PJ, Dahllöf I, Kjelleberg S, Steinberg PD. 2004. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ Microbiol 6:121–130. doi: 10.1046/j.1462-2920.2003.00545.x. [DOI] [PubMed] [Google Scholar]
- 41.Gärdes A, Iversen MH, Grossart H-P, Passow U, Ullrich MS. 2011. Diatom-associated bacteria are required for aggregation of Thalassiosira weissflogii. ISME J 5:436–445. doi: 10.1038/ismej.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bižić-Ionescu M, Zeder M, Ionescu D, Orlić S, Fuchs BM, Grossart HP, Amann R. 2015. Comparison of bacterial communities on limnic versus coastal marine particles reveals profound differences in colonization. Environ Microbiol 17:3500–3514. doi: 10.1111/1462-2920.12466. [DOI] [PubMed] [Google Scholar]
- 43.Rieck A, Herlemann DP, Jürgens K, Grossart H-P. 2015. Particle-associated differ from free-living bacteria in surface waters of the Baltic Sea. Front Microbiol 6:1297. doi: 10.3389/fmicb.2015.01297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delmont TO, Eren AM, Vineis JH, Post AF. 2015. Genome reconstructions indicate the partitioning of ecological functions inside a phytoplankton bloom in the Amundsen Sea, Antarctica. Front Microbiol 6:1090. doi: 10.3389/fmicb.2015.01090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.González JM, Fernández-Gómez B, Fernàndez-Guerra A, Gómez-Consarnau L, Sánchez O, Coll-Lladó M, Campo JD, Escudero L, Rodríguez-Martínez R, Alonso-Sáez L, Latasa M, Paulsen I, Nedashkovskaya O, Lekunberri I, Pinhassi J, Pedros-Alio C. 2008. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria). Proc Natl Acad Sci U S A 105:8724–8729. doi: 10.1073/pnas.0712027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gómez-Pereira PR, Fuchs BM, Alonso C, Oliver MJ, van Beusekom JE, Amann R. 2010. Distinct flavobacterial communities in contrasting water masses of the North Atlantic Ocean. ISME J 4:472–487. doi: 10.1038/ismej.2009.142. [DOI] [PubMed] [Google Scholar]
- 47.Williams TJ, Wilkins D, Long E, Evans F, Demaere MZ, Raftery MJ, Cavicchioli R. 2013. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ Microbiol 15:1302–1317. doi: 10.1111/1462-2920.12017. [DOI] [PubMed] [Google Scholar]
- 48.El-Swais H, Dunn KA, Bielawski JP, Li WKW, Walsh DA. 2015. Seasonal assemblages and short-lived blooms in coastal north-west Atlantic Ocean bacterioplankton. Environ Microbiol 17:3642–3661. doi: 10.1111/1462-2920.12629. [DOI] [PubMed] [Google Scholar]
- 49.Kirchman DL. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol 39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 50.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang S, Mann AJ, Waldmann J, Weber M, Klindworth A, Otto A, Lange J, Bernhardt J, Reinsch C, Hecker M, Peplies J, Bockelmann FD, Callies U, Gerdts G, Wichels A, Wiltshire KH, Glockner FO, Schweder T, Amann R. 2012. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 51.Buchan A, González JM, Moran MA. 2005. Overview of the marine Roseobacter lineage. Appl Environ Microbiol 71:5665–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner-Döbler I, Biebl H. 2006. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol 60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- 53.Mou X, Sun S, Edwards RA, Hodson RE, Moran MA. 2008. Bacterial carbon processing by generalist species in the coastal ocean. Nature 451:708–711. doi: 10.1038/nature06513. [DOI] [PubMed] [Google Scholar]
- 54.Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, Howard EC, King E, Oakley CA, Reisch CR, Rinta-Kanto JM, Sharma S, Sun S, Varaljay V, Vila-Costa M, Westrich JR, Moran MA. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J 4:784–798. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- 55.Budinoff CR, Dunlap JR, Hadden M, Buchan A. 2011. Marivita roseacus sp. nov., of the family Rhodobacteraceae, isolated from a temperate estuary and an emended description of the genus Marivita. J Gen Appl Microbiol 57:259–267. [DOI] [PubMed] [Google Scholar]
- 56.Thongphrom C, Kim JH, Yoon JH, Bora N, Kim W. 2016. Marimonas arenosa gen. nov., sp. nov., isolated from sea sand. Int J Syst Evol Microbiol 67:121–126. [DOI] [PubMed] [Google Scholar]
- 57.Onda DF, Azanza RV, Lluisma AO. 2015. Potential DMSP-degrading Roseobacter clade dominates endosymbiotic microflora of Pyrodinium bahamense var. compressum (Dinophyceae) in vitro. Arch Microbiol 197:965–971. doi: 10.1007/s00203-015-1133-0. [DOI] [PubMed] [Google Scholar]
- 58.Green DH, Echavarribravo V, Brennan D, Hart MC. 2015. Bacterial diversity associated with the coccolithophorid algae Emiliania huxleyi and Coccolithus pelagicus f. braarudii. BioMed Res Int 2015:194540. doi: 10.1155/2015/194540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cruz-López R, Maske H. 2016. The vitamin B1 and B12 required by the marine dinoflagellate Lingulodinium polyedrum can be provided by its associated bacterial community in culture. Front Microbiol 7:560. doi: 10.3389/fmicb.2016.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tada Y, Taniguchi A, Nagao I, Miki T, Uematsu M, Tsuda A, Hamasaki K. 2011. Differing growth responses of major phylogenetic groups of marine bacteria to natural phytoplankton blooms in the western North Pacific Ocean. Appl Environ Microbiol 77:4055–4065. doi: 10.1128/AEM.02952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romera-Castillo C, Sarmento H, Gasol JM, Marrasé C. 2011. Net production and consumption of fluorescent colored dissolved organic matter by natural bacterial assemblages growing on marine phytoplankton exudates. Appl Environ Microbiol 77:7490–7498. doi: 10.1128/AEM.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eiler A, Bertilsson S. 2004. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ Microbiol 6:1228–1243. doi: 10.1111/j.1462-2920.2004.00657.x. [DOI] [PubMed] [Google Scholar]
- 63.Zeder M, Peter S, Shabarova T, Pernthaler J. 2009. A small population of planktonic Flavobacteria with disproportionally high growth during the spring phytoplankton bloom in a prealpine lake. Environ Microbiol 11:2676–2686. doi: 10.1111/j.1462-2920.2009.01994.x. [DOI] [PubMed] [Google Scholar]
- 64.Berg KA, Lyra C, Sivonen K, Paulin L, Suomalainen S, Tuomi P, Rapala J. 2009. High diversity of cultivable heterotrophic bacteria in association with cyanobacterial water blooms. ISME J 3:314–325. doi: 10.1038/ismej.2008.110. [DOI] [PubMed] [Google Scholar]
- 65.Koblížek M, Komenda J, Masojídek J, Pechar L. 2000. Cell aggregation of the cyanobacterium Synechococcus elongatus: role of the electron transport chain. J Phycol 36:662–668. doi: 10.1046/j.1529-8817.2000.99030.x. [DOI] [PubMed] [Google Scholar]
- 66.Roux JM. 1996. Production of polysaccharide slime by microbial mats in the hypersaline environment of a Western Australian solar saltfield. Int J Salt Lake Res 5:103–130. doi: 10.1007/BF01995826. [DOI] [Google Scholar]
- 67.Deng W, Cruz B, Neuer S. 2016. Effects of nutrient limitation on cell growth, TEP production and aggregate formation of marine Synechococcus. Aquat Microb Ecol 78:39–49. doi: 10.3354/ame01803. [DOI] [Google Scholar]
- 68.Jiao N, Zheng Q. 2011. The microbial carbon pump: from genes to ecosystems. Appl Environ Microbiol 77:7439–7444. doi: 10.1128/AEM.05640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, Xing P, Chen M, Bian Y, Wu QL. 2011. Short-term bacterial community composition dynamics in response to accumulation and breakdown of Microcystis blooms. Water Res 45:1702–1710. doi: 10.1016/j.watres.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 70.Liu M, Dong Y, Zhao Y, Zhang G, Zhang W, Xiao T. 2011. Structures of bacterial communities on the surface of Ulva prolifera and in seawaters in an Ulva blooming region in Jiaozhou Bay, China. World J Microbiol Biotechnol 27:1703–1712. doi: 10.1007/s11274-010-0627-9. [DOI] [Google Scholar]
- 71.Louati I, Pascault N, Debroas D, Bernard C, Humbert J-F, Leloup J. 2015. Structural diversity of bacterial communities associated with bloom-forming freshwater cyanobacteria differs according to the cyanobacterial genus. PLoS One 10:e0140614. doi: 10.1371/journal.pone.0140614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dziallas C, Grossart HP. 2011. Temperature and biotic factors influence bacterial communities associated with the cyanobacterium Microcystis sp. Environ Microbiol 13:1632–1641. doi: 10.1111/j.1462-2920.2011.02479.x. [DOI] [PubMed] [Google Scholar]
- 73.Rösel S, Grossart HP. 2012. Contrasting dynamics in activity and community composition of free-living and particle-associated bacteria in spring. Aquat Microb Ecol 66:169–181. doi: 10.3354/ame01568. [DOI] [Google Scholar]
- 74.Allgaier M, Brückner S, Jaspers E, Grossart HP. 2007. Intra- and inter-lake variability of free-living and particle-associated Actinobacteria communities. Environ Microbiol 9:2728–2741. doi: 10.1111/j.1462-2920.2007.01385.x. [DOI] [PubMed] [Google Scholar]
- 75.Mou X, Jacob J, Lu X, Robbins S, Sun S, Ortiz JD. 2013. Diversity and distribution of free-living and particle-associated bacterioplankton in Sandusky Bay and adjacent waters of Lake Erie western basin. J Great Lakes Res 39:352–357. doi: 10.1016/j.jglr.2013.03.014. [DOI] [Google Scholar]
- 76.Sharma AK, Sommerfeld K, Bullerjahn GS, Matteson AR, Wilhelm SW, Jezbera J, Brandt U, Doolittle WF, Hahn MW. 2009. Actinorhodopsin genes discovered in diverse freshwater habitats and among cultivated freshwater Actinobacteria. ISME J 3:726–737. doi: 10.1038/ismej.2009.13. [DOI] [PubMed] [Google Scholar]
- 77.Kang I, Lee K, Yang SJ, Choi A, Kang D, Lee YK, Cho JC. 2012. Genome sequence of “Candidatus Aquiluna” sp. strain IMCC13023, a marine member of the Actinobacteria isolated from an Arctic fjord. J Bacteriol 194:3550. doi: 10.1128/JB.00586-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeLong EF, Franks DG, Alldredge AL. 1993. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr 38:924–934. doi: 10.4319/lo.1993.38.5.0924. [DOI] [Google Scholar]
- 79.Riemann L, Winding A. 2001. Community dynamics of free-living and particle-associated bacterial assemblages during a freshwater phytoplankton bloom. Microb Ecol 42:274–285. doi: 10.1007/s00248-001-0018-8. [DOI] [PubMed] [Google Scholar]
- 80.Pinhassi J, Azam F, Hemphälä J, Long RA, Martinez J, Zweifel UL, Hagström Å. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat Microb Ecol 17:13–26. doi: 10.3354/ame017013. [DOI] [Google Scholar]
- 81.Cottrell MT, Kirchman DL. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol 66:1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bowman JP, Mccammon SA, Brown MV, Nichols DS, McMeekin TA. 1997. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl Environ Microbiol 63:3068–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Piquet AMT, Bolhuis H, Meredith MP, Buma AGJ. 2011. Shifts in coastal Antarctic marine microbial communities during and after melt water-related surface stratification. FEMS Microbiol Ecol 76:413–427. doi: 10.1111/j.1574-6941.2011.01062.x. [DOI] [PubMed] [Google Scholar]
- 84.Li WKW. 1994. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Oceanogr 39:169–175. [Google Scholar]
- 85.Partensky F, Blanchot J, Vaulot D. 1999. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull Inst Oceanogr Monaco-Numero Special 19:457–476. [Google Scholar]
- 86.Scanlan DJ, West NJ. 2002. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol Ecol 40:1–12. doi: 10.1111/j.1574-6941.2002.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 87.Johnson ZI, Zinser ER, Coe A, Mcnulty NP, Woodward EMS, Chisholm SW. 2006. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 88.Garcia-Pichel F, Belnap J, Neuer S, Schanz F. 2003. Estimates of global cyanobacterial biomass and its distribution. Algol Stud 109:213–227. doi: 10.1127/1864-1318/2003/0109-0213. [DOI] [Google Scholar]
- 89.Bertilsson S, Berglund O, Pullin MJ, Chisholm SW. 2005. Release of dissolved organic matter by Prochlorococcus. Life Environ 55:225–232. [Google Scholar]
- 90.Richardson TL, Jackson GA. 2007. Small phytoplankton and carbon export from the surface ocean. Science 315:838–840. doi: 10.1126/science.1133471. [DOI] [PubMed] [Google Scholar]
- 91.Lomas MW, Moran SB. 2011. Evidence for aggregation and export of cyanobacteria and nano-eukaryotes from the Sargasso Sea euphotic zone. Biogeosciences 8:203–216. doi: 10.5194/bg-8-203-2011. [DOI] [Google Scholar]
- 92.Stukel MR, Décima M, Selph KE, Taniguchi DAA, Landry MR. 2013. The role of Synechococcus in vertical flux in the Costa Rica upwelling dome. Prog Oceanogr 112:49–59. doi: 10.1016/j.pocean.2013.04.003. [DOI] [Google Scholar]
- 93.LeCleir GR, DeBruyn JM, Maas EW, Boyd PW, Wilhelm SW. 2014. Temporal changes in particle-associated microbial communities after interception by nonlethal sediment traps. FEMS Microbiol Ecol 87:153–163. doi: 10.1111/1574-6941.12213. [DOI] [PubMed] [Google Scholar]
- 94.Glöckner FO, Fuchs BM, Amann R. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol 65:3721–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Waterbury JB, Watson SW, Valois FW, Franks DG. 1986. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can Bull Fish Aquat Sci 214:71–120. [Google Scholar]
- 96.Brahamsha B. 1996. A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl Environ Microbiol 62:1747–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fuhrman JA, Horrigan SG, Capone DG. 1988. Use of 13N as tracer for bacterial and algal uptake of ammonium from seawater. Mar Ecol Prog Ser 45:271–278. [Google Scholar]
- 98.Rocap G, Distel DL, Waterbury JB, Chisholm SW. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol 68:1180–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zheng Q, Liu Y, Steindler L, Jiao N. 2015. Pyrosequencing analysis of aerobic anoxygenic phototrophic bacterial community structure in the oligotrophic western Pacific Ocean. FEMS Microbiol Lett 362:fnv034. doi: 10.1093/femsle/fnv034. [DOI] [PubMed] [Google Scholar]
- 100.Jiao N, Yang Y, Hong N, Ma Y, Harada S, Koshikawa H, Watanabe M. 2005. Dynamics of autotrophic picoplankton and heterotrophic bacteria in the East China Sea. Cont Shelf Res 25:1265–1279. doi: 10.1016/j.csr.2005.01.002. [DOI] [Google Scholar]
- 101.Marie D, Brussaard CP, Thyrhaug R, Bratbak G, Vaulot D. 1999. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl Environ Microbiol 65:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Claesson MJ, O'Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O'Toole PW. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 107.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lane D. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed),Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY. [Google Scholar]
- 110.Stubner S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen detection. J Microbiol Methods 50:155–164. doi: 10.1016/S0167-7012(02)00024-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.