ABSTRACT
Chemotaxis can provide bacteria with competitive advantages for survival in complex environments. The CheZ chemotaxis protein is a phosphatase, affecting the flagellar motor in Escherichia coli by dephosphorylating the response regulator phosphorylated CheY protein (CheY∼P) responsible for clockwise rotation. A cheZ gene has been found in Azorhizobium caulinodans ORS571, in contrast to other rhizobial species studied so far. The CheZ protein in strain ORS571 has a conserved motif similar to that corresponding to the phosphatase active site in E. coli. The construction of a cheZ deletion mutant strain and of cheZ mutant strains carrying a mutation in residues of the putative phosphatase active site showed that strain ORS571 participates in chemotaxis and motility, causing a hyperreversal behavior. In addition, the properties of the cheZ deletion mutant revealed that ORS571 CheZ is involved in other physiological processes, since it displayed increased flocculation, biofilm formation, exopolysaccharide (EPS) production, and host root colonization. In particular, it was observed that the expression of several exp genes, involved in EPS synthesis, was upregulated in the cheZ mutant compared to that in the wild type, suggesting that CheZ negatively controls exp gene expression through an unknown mechanism. It is proposed that CheZ influences the Azorhizobium-plant association by negatively regulating early colonization via the regulation of EPS production. This report established that CheZ in A. caulinodans plays roles in chemotaxis and the symbiotic association with the host plant.
IMPORTANCE Chemotaxis allows bacteria to swim toward plant roots and is beneficial to the establishment of various plant-microbe associations. The level of CheY phosphorylation (CheY∼P) is central to the chemotaxis signal transduction. The mechanism of the signal termination of CheY∼P remains poorly characterized among Alphaproteobacteria, except for Sinorhizobium meliloti, which does not contain CheZ but which controls CheY∼P dephosphorylation through a phosphate sink mechanism. Azorhizobium caulinodans ORS571, a microsymbiont of Sesbania rostrata, has an orphan cheZ gene besides two cheY genes similar to those in S. meliloti. In addition to controlling the chemotaxis response, the CheZ-like protein in strain ORS571 is playing a role by decreasing bacterial adhesion to the host plant, in contrast to the general situation where chemotaxis-associated proteins promote adhesion. In this study, we identified a CheZ-like protein among Alphaproteobacteria functioning in chemotaxis and the A. caulinodans-S. rostrata symbiosis.
KEYWORDS: Azorhizobium caulinodans, CheZ, Sesbania rostrata, chemotaxis, colonization
INTRODUCTION
The chemotactic response is an important mechanism that motile prokaryotes have developed to colonize appropriate niches, conferring a selective advantage to improve viability in harsh environments (1–3). The response to external stimuli, detected by specialized chemoreceptors known as methyl-accepting chemotaxis proteins (MCPs), is mediated through a signal transduction regulatory cascade controlling the flagellar motor, leading to movement toward attractants or away from repellents (2, 4, 5).
The chemotaxis signal transduction system, best studied in Escherichia coli, involves a histidine kinase CheA and a response regulator CheY (2, 4). MCP binding of a signal molecule leads to a conformational change affecting the activity of CheA, which can be autophosphorylated by ATP. The phosphorylated CheA (CheA∼P) then phosphorylates the response regulator CheY, and CheY∼P binds to the flagellar motor protein enabling flagellar clockwise rotation. The spontaneous dephosphorylation of CheY∼P is accelerated by the phosphatase CheZ, modulating the cell's response to external signal stimulation.
CheZ dephosphorylation of CheY∼P is essential to control flagellar rotation and to improve chemotaxis efficiency (4). CheZ, first described in Gammaproteobacteria and Betaproteobacteria (2), is also present in Campylobacter and Helicobacter belonging to the epsilon subclass (5). However, in bacteria that do not possess a cheZ gene, other mechanisms of CheY∼P dephosphorylation are known (2, 6). For example, CheC and FliY, belonging to another class of phosphatases, are found in Bacillus subtilis, and CheX is found in spirochetes (7). The situation in Sinorhizobium meliloti, a member of Alphaproteobacteria, shows another level of complexity, because this bacterium possesses two genes encoding the CheY proteins, cheY1 and cheY2 (8, 9). CheY2 is the actual response regulator, which once phosphorylated by CheA∼P, interacts with the flagellar motor. Modulation of CheY2 phosphorylation is also achieved by CheA, which can retransfer the phosphoryl group to CheY1. Thus, CheY1 functions only as a sink for phosphoryl groups, and CheA together with another protein, CheS, can achieve dephosphorylation of CheY1 (8, 9). In addition, other proteins such as CheV and hybrid CheA kinases, which have additional REC domains, may also play a role in CheY∼P dephosphorylation (10). Helicobacter pylori possesses a CheZ phosphatase (11) that is distant from that of E. coli CheZ, but it also contains a hybrid CheAY protein, functioning as a phosphate sink (12). Moreover, Wuichet and Zhulin (6), by performing an extensive bioinformatics analysis of genomes, revealed the presence of CheZ protein families in all subclasses of Proteobacteria, showing that CheZs from Epsilonproteobacteria and Alphaproteobacteria structurally differ from those in the beta and gamma subgroups. To date, there is no experimental evidence that CheZ proteins from the alpha subclass have a function in chemotaxis.
Chemotaxis is known to play important roles in many processes, such as plant and animal tissue colonization by pathogenic bacteria (13–15) and the colonization of soil and plant root systems (16, 17). A significant correlation between chemotaxis and attachment or adhesion in the process of colonization of plant roots or seeds has been established (18–20). In the symbiotic association of rhizobia with legumes, the attachment to the root surface of the host plant is considered an important prerequisite for successful nodulation (17). Indeed, motility and chemotaxis toward root exudates play roles in the interaction of rhizobia and the host system (16, 21), and it was reported long ago that chemotactic mutations can decrease the efficiency or competitiveness of nodulation (22). Recently, we reported that a soluble chemotactic receptor of Azorhizobium caulinodans ORS571 was required for the nodulation of the host plant (23).
A. caulinodans ORS571 is a nitrogen-fixing bacterium belonging to the Alphaproteobacteria subclass (24, 25), forming nodules on the roots and stems of the host Sesbania rostrata (26, 27). Strain ORS571 can sense amino acid, sugar, and flavonoid substances secreted by host plants and move toward plant roots (28). The complete nucleotide sequence of the genome of strain ORS571 was determined (29), and this led us to search for gene products possibly involved in chemotaxis in this bacterium. A. caulinodans contains two copies of the gene encoding CheY, similar to the situation found in S. meliloti, and a copy of the gene encoding CheZ (25), in agreement with the former suggestion that CheZ was present in the Alphaproteobacteria subclass (30).
The present work reports a functional analysis of the cheZ gene product of A. caulinodans ORS571 based on the construction of mutant strains. We found that CheZ of strain ORS571 inhibited bacterial adhesion to the host plant, in contrast to the general situation where the chemotaxis-associated proteins promote adhesion (31). According to data obtained on the flocculation and biofilm formation of cheZ mutant strains, it is proposed that the strong adhesion of the cheZ mutant to the host is the result of multiple effects, including chemotaxis and exopolysaccharide (EPS) production.
RESULTS
Presence of an orphan cheZ gene in the A. caulinodans ORS571 genome.
The presence of deduced protein products corresponding to CheZ in some members of Alphaproteobacteria was reported (6), but this analysis did not contain A. caulinodans, whose genome nucleotide sequencing was reported later (29). Using the MiST2.2 database (see Materials and Methods), a search for genes possibly involved in chemotaxis in the genome of A. caulinodans ORS571 (GenBank accession no. AP009384.1) was performed (25), and the organization of the che gene clusters was compared to that of other members of Alphaproteobacteria and to that of E. coli and Salmonella enterica serovar Typhimurium (Fig. 1; see also Fig. S1 in the supplemental material). In the species exemplified by E. coli, cheZ is located in the conserved che cluster together with cheA, cheW, cheR, and cheB (Fig. 1). The conservation of gene organization in operons may reflect physical interaction between the encoded proteins within the same cluster (32). However, in Alphaproteobacteria genomes, the organization of the che clusters and the presence of a cheZ gene revealed major differences. Orphan cheZ genes were located outside the che cluster and often were adjacent to a gene encoding an orphan response regulator, CheY (Fig. S1). This is the case in A. caulinodans, which contains a single copy of cheZ adjacent to a cheY gene (Fig. 1), and also in Methylobacterium radiotolerans JCM 2831 (GenBank accession no. NC_010505.1), Nitrobacter hamburgensis X14 (GenBank accession no. NC_007964.1), and Rhodospirillum centenum SW (GenBank accession no. CP000613.2) (Fig. S1). This organization in Alphaproteobacteria is quite unique, and it raises the question as to whether ORS571 CheZ plays a role in chemotaxis in A. caulinodans. Indeed, the presence of CheZ does not necessarily mean its involvement in chemotaxis as reported in some members of the gamma subclass, such as Vibrio cholerae (33). In contrast, no cheZ is present in S. meliloti (Fig. 1) (34), and none was found in Rhizobium leguminosarum, Rhizobium etli, or Bradyrhizobium japonicum (25, 34) (Fig. S1). Thus, A. caulinodans differs from other rhizobia.
FIG 1.
Comparison of the organization of the chemotaxis gene clusters in the chromosomes of S. meliloti, A. caulinodans, and E. coli. The genes encoding CheY and CheZ are indicated in black and red, respectively. In A. caulinodans, cheY and cheZ, which are distant from che cluster, are orphan genes.
The CheZ polypeptide of 26,122 Da with 236 residues is similar in size to the CheZ found in Gammaproteobacteria, and it shows significant similarity (44.1% and 43.9%) and identity (25.6% and 26.6%) with those in E. coli and Salmonella Typhimurium, respectively (Fig. 2). Within the Alphaproteobacteria (Fig. S1, Table S1), the similarity of CheZ with A. caulinodans CheZ is approximately 50% and the identity is approximately 36%. The highest values recorded are with M. radiotolerans JCM 2831, where CheZ displays 58.6% and 45.6% similarity and identity, respectively (Table S1). An alignment of ORS571 CheZ with those in E. coli and S. Typhimurium showed a significant conservation of the motif responsible for the phosphatase activity in E. coli (DXXXQ) (Fig. 2). In particular, Asp 165 and Gln 169 residues are conserved, but there is a substitution, an Ile (at position 166 in the A. caulinodans sequence) instead of Lys, in the active motif of ORS571 CheZ (see Fig. S2). These results suggest that CheZ in strain ORS571 may have a similar function as those in E. coli and S. Typhimurium.
FIG 2.
Amino acid sequence alignment of CheZ proteins from A. caulinodans ORS571, E. coli, and S. Typhimurium. The numbers on the left and right show the positions of the residues in E. coli, S. Typhimurium, and A. caulinodans. Gaps indicated by dashes are introduced to maximize the alignments. The similarity between the homologous proteins is highlighted by different shading: black, all amino acids in a column are identical; gray, the amino acids in a column belong to a weak similarity group. The conserved phosphatase active-site motif in E. coli is boxed, and the two key amino residues in the motif, D and Q, are marked with asterisks.
ORS571 CheZ plays a role in chemotaxis.
To determine if strain ORS571 plays a role in chemotaxis, we constructed a mutant strain, AC601, in which the cheZ gene is disrupted. The growth rate of the mutant strain was the same as that of the wild type (data not shown). The chemotactic behavior of the cheZ mutant was analyzed in the semisoft agar assay. In this assay, cells can sense and migrate toward different attractant gradients, resulting in a characteristic ring (7). With succinate as the sole carbon source, the wild type (ORS571) can form a sharp and large-diameter chemotactic ring, while the cheZ mutant (AC601) cannot (Fig. 3A), suggesting its chemotactic response was abolished. Similar data were obtained when malate was the carbon source (Fig. 3A). Bacteria were picked up from the edge of the chemotactic ring and observed under a microscope. Both the mutant and wild-type strains were able to move rapidly, suggesting that the defect in ring formation was not due to a defect of motility in the mutant strain. Interestingly, compared to those in strain ORS571, bacterial cells of the mutant strain were located preferentially on the edge of the microscope coverslip (see Movies S1 and S2). Furthermore, chemotactic behavior was restored when strain AC601 was complemented with a broad-host-range vector expressing the wild-type cheZ gene (strain AC602) (Fig. 3B). By measuring the relative expression level of cheZ gene using reverse transcriptase quantitative PCR (RT-qPCR), we can see the expression level of cheZ in strain AC602 is 1.25 times higher than that in the wild type, and there is no cheZ mRNA detected in strain AC601 (see Fig. S3). So, the complementation strain AC602 does not fully restore the wild-type chemotaxis phenotype. This clearly indicates that the defect of chemotactic behavior of strain AC601 is caused by the inactivation of the cheZ gene.
FIG 3.
A. caulinodans chemotaxis behavior. (A) Comparison of the chemotactic responses of the wild-type ORS571 and the cheZ deletion mutant, AC601. The chemotactic ring diameters were measured after 48 h for each strain in the presence of the desired chemoeffector, succinate or malate, in a soft agar plate assay with or without NH4Cl as a nitrogen source. (B) Comparison of the chemotactic responses of ORS571(pBBR1MCS-2), AC601(pBBR1MCS-2), and AC602(pBBR2CheZ) complemented mutants and AC603(pBBR2CheZD165A) and AC604(pBBR2CheZQ169A) mutant strains obtained after site-directed mutagenesis of the putative phosphatase active site. Examples of the chemotaxis rings observed after 48 h in the presence of succinate and NH4Cl by each counterpart strain are shown in panels A and B. (C) Competitive chemotactic responses of A. caulinodans strains using the quantitative capillary assay. Strains were mixed in equal ratios; the capillary contained either 10 mM succinate or PBS only as a control. (D) Chemotaxis ring formation of the E. coli wild type (RP437), the cheZ mutant (UU2685), and strain UU2685CheZ containing the ORS571 cheZ gene. Error bars in panels A, B, and C show standard deviations of the means from triplicates. The scale bars in panels A and B represent 10 mm.
To quantitatively compare the chemotactic behaviors of the wild type and the cheZ mutant, a competitive capillary assay was performed using 10 mM succinate as an attractant and phosphate-buffered saline (PBS) as a control. These capillaries were immersed in a bacterial suspension with a 1:1 mixture of strains ORS571 and AC601. As can be seen in Fig. 3C, the total number of bacterial cells from the suspension entering the capillaries containing succinate as the chemoattractant was approximately three times more than that entering the capillary containing only PBS. The ratio of the wild-type and AC601 strains was close to 4:1 in the presence of the chemoeffector, while it remained 1:1 when using only PBS, confirming that the chemotactic ability of strain AC601 was impaired (Fig. 3C). The chemotactic ability of strain AC602 is similar to that of the wild-type strain (Fig. 3C). Thus, it is clear that CheZ plays an important role in the chemotaxis to organic acid in strain ORS571.
CheZ controls the swimming motility bias.
In E. coli, CheY∼P can interact with the flagellar motor proteins, especially FliM, to change the rotational bias of the motor (35), resulting in an increase in the reorientation frequency (36). CheZ controls the reorientation by accelerating the dephosphorylation of CheY∼P. The inactivation of cheZ in E. coli mutant strains decreases CheY∼P dephosphorylation, causing an increase in the reorientation frequency (37).
To examine the swimming behavior of the A. caulinodans cheZ mutant, the swimming pattern was recorded and analyzed. Strains ORS571 and AC602 tended to swim smoothly, and their trajectories were predominantly linear. Strain AC601 tended to swim in circles with constant changes in direction (Fig. 4A). We calculated the average velocity of the bacteria, and there were no significant differences between wild-type and cheZ mutant strains (data not shown). To further recognize the differences in swimming behavior, the motility bias was quantified by measuring the frequencies of direction changes. The cheZ mutant displayed a reorientation frequency of 1.58 per second, almost 2-fold that recorded for the wild type (0.77 per second) (Fig. 4B), while the reorientation frequency was 0.34 per second in strain AC602 (Fig. 4B), whose cheZ gene was overexpressed (Fig. S3). These results suggested that the hyperreversal behavior was associated with the loss of function of CheZ in strain ORS571, as also reported for E. coli.
FIG 4.
Analysis of swimming behaviors of A. caulinodans wild-type ORS571, cheZ mutant AC601, and complemented strain AC602. (A) Sample trajectories. Individual motile cells were tracked over time and their motion was compiled into trajectories that trace their movement. To track motile cells, we used high frequency (10-ms interval) phase contrast imaging. Some swimming direction changes are labeled with arrows as examples. (B) Flagellar rotation frequency. Up to 50 cells were examined for each strain; the average frequency of direction changes for each strain is indicated by black solid lines. Dashed line indicates the baseline for smooth swimming in the wild-type strain, which was observed immediately after adding an attractant. Each strain was tested by using at least three biological replicates.
Key amino acid residues of phosphatase activity for CheZ are essential.
The CheZ protein contributes to chemotaxis by regulating the swimming bias in strain ORS571, and the CheZ motif involved in catalysis in E. coli is relatively well conserved in strain ORS571, at residues 165 to 169 (Fig. 2). In particular, two key amino acid residues, D143 and Q147, which are playing a key role in the catalysis of E. coli CheZ, are also conserved in strain ORS571, at locations 165 and 169, respectively (Fig. 2). Therefore, we constructed mutant strains by site-directed mutagenesis, in which residues D165 and Q169 were replaced by alanine residues (CheZD165A and CheZQ169A, respectively), named AC603 and AC604, respectively. The chemotactic behaviors of these two mutants were assayed. As can be seen in Fig. 3B, strains AC603 and AC604 do not form normal chemotaxis rings, indicating that CheZ in strain ORS571 plays an essential role in chemotaxis, which could possibly be linked through phosphatase activity, as in E. coli.
However, when a broad-host-range vector expressing the ORS571 cheZ wild-type gene was transferred into an E. coli cheZ mutant strain, UU2685 (38) (Table 1), the loss of chemotactic behavior of strain UU2685 could not be complemented on a soft agar plate (Fig. 3D). The lack of complementation may be due to poor expression of the complementing plasmid in E. coli or more likely, to the fact that CheZ lacks the CheA-binding region (30) critical for the interaction between CheA and CheZ in E. coli. This implies that the mechanisms of CheZ function in the N terminus or in other regions, except phosphatase active sites, differ between strain ORS571 and E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− supE44 lacA-U169 ϕ80lacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Transgen |
| RP437 | thr(Am)-1 leuB6 his-4 metF(Am)159 eda-50 rpsL136 [thi-1 ara-14 lacY1 mtl-1 xyl-5 tonA31 tsx-78]b | 37 |
| UU2685 | (cheZ)Δ4211 thr(Am)-1 leuB6 his-4 metF(Am)159 rpsL136 [thi-1 ara-14 lacY1 mtl-1 xyl-5 tonA31 tsx-78] | 38 |
| UU2685CheZ | UU2685 derivative, contains pBBRCheZ; Kmr | This study |
| Azorhizobium caulinodans | ||
| ORS571 | Type strain; Ampr, Nalr | 24 |
| AC601 | ORS571 derivative, ΔcheZ; Ampr, Nalr, Gmr | This study |
| AC602 | AC601 derivative, contains pBBRCheZ; Kmr, Ampr, Nalr, Gmr | This study |
| AC603 | AC601 derivative, contains the mutated plasmid pBBRCheZD150A; Ampr, Nalr, Gmr, Kmr | This study |
| AC604 | AC601 derivative, contains the mutated plasmid pBBRCheZQ154A; Ampr, Nalr, Gmr, Kmr | This study |
| Plasmids | ||
| pCM351 | Allelic exchange vector; Gmr, Tcr | 61 |
| pRK2013 | Helper plasmid, ColE1 replicon; Tra+, Kmr | 62 |
| pBBR1MCS-2 | Broad-host-range plasmid; Kmr | 63 |
| pBBRCheZ | pBBR1MCS-2 with cheZ open reading frame and 406-bp upstream promoter region; Kmr | This study |
| pBBRCheZD150A | pBBRCheZ carrying D150A substitution | This study |
| pBBRCheZQ154A | pBBRCheZ carrying Q154A substitution | This study |
Ampr, ampicillin resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance; Nalr, nalidixic acid resistance; Tcr, tetracycline resistance.
thr(Am)-1 and metF(Am)159 are amber (UAG) nonsense mutations in genes that cause auxotrophic requirements for the amino acids threonine and methionine, respectively. Other genetic markers are listed in the brackets.
CheZ impairs EPS production and flocculation properties of strain ORS571.
Previous observations revealed that the AC601 mutant had an increased tendency to adhere to coverslips. The efficiency of bacterial attachment can be facilitated by polysaccharides, proteins, and nucleic acids (39). Thus, we hypothesized that the adhesion phenotype of the cheZ mutant may result from an increased EPS production. Indeed, the cheZ mutant on Congo red plates containing succinate formed colonies more viscous than those of the wild type (Fig. 5A). Quantitative data also showed that the mutant produced a significant excess of polysaccharides, which were increased by 42.5% compared to that in the wide type (Fig. 5B). To further assay the relationship between CheZ and extracellular polysaccharide production, we performed RT-qPCR analyses of exp genes, responsible for the synthesis of EPS (40, 41). Three exp genes (AZC_1833, AZC_1834, and AZC_3326) (42) distributed in two gene clusters were selected. It was found that the relative expression levels of the genes in strain AC601 were approximately 2-fold higher than those in strain ORS571, while the relative expression levels of the genes in strain AC602 were approximately 6-fold lower than those in strain ORS571 (Fig. 5C). These results indicate that CheZ negatively regulates the expression of the exp genes in strain ORS571 through an unknown mechanism.
FIG 5.
EPS staining and production and relative expression of exp genes in A. caulinodans. (A) Wild-type ORS571 (left) and cheZ deletion mutant AC601 (right) colony morphologies on Congo red plates. Photographs were taken after 3 days of growth. (B) EPS production by the wild type and the cheZ mutant. The EPS was extracted and quantified as described in Materials and Methods. Error bars show the standard deviations from the means. *, P < 0.05 versus the wild-type strain. (C) RT-qPCR analyses of exp genes AZC_1833, AZC_1834, and AZC_3326. The expression levels were assessed by normalization to the 16S rRNA level.
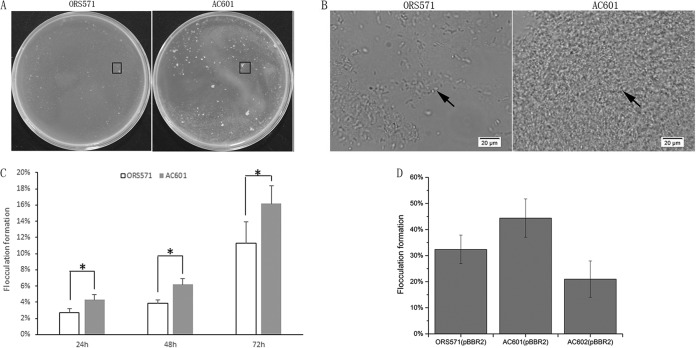
In addition, a positive correlation between chemotactic behavior and flocculation was reported in Azospirillum brasilense (43). Therefore, we analyzed the differences of flocculation between strains ORS571 and AC601. The measurements of flocculation formation after 24 h, 48 h, and 72 h revealed a significant increase in the total amount of flocculation by both strains (Fig. 6), but the flocculation rate was significantly higher with the AC601 cheZ mutant than with strain ORS571. Microscopic examination showed larger, denser, and thicker flocs formed by strain AC601 than by strain ORS571 (Fig. 6A and B; see also Fig. S4), suggesting that the mutant tended to form bigger clumps. In addition, the kinetics of floc formation differed; big clumps were formed earlier in strain AC601 (Movie S2).
FIG 6.
Comparative flocculation of wild-type ORS571 and cheZ mutant AC601 in L3 minimal medium containing low nitrogen. (A) Observation of floc formation in plate assays at 48 h. (B) Enlargements of boxes in panel A for flocs formed after 48 h. (C) Quantification of flocculation formation. There are obvious differences in the total flocculation formation between the wild type and strain AC601 after 24 h, 48 h, and 72 h. (D) Complementation assays of the cheZ mutant. Flocculation formation of strain AC602 was quantified after 24 h. Strains ORS571 and AC601 harboring the empty plasmid pBBR1MCS-2 were used as controls. *, P < 0.05 versus the wild-type strain.
CheZ negatively regulates colonization of strain ORS571 on Sesbania roots.
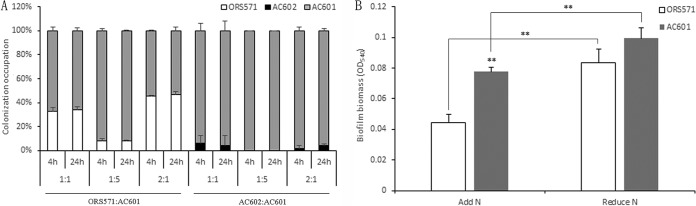
Chemotaxis plays an essential role in microbial colonization and symbiosis with plant hosts (13, 44, 45), and numerous studies have shown that chemotaxis affects Rhizobium-host interactions (16). The colonization of Rhizobium on the roots of the host was seen as an essential step for successful nodulation (17). To further assess the biological significance of ORS571 CheZ in the symbiotic relationship with the host plant, we performed competitive colonization experiments with Sesbania roots. The incubation of roots with equal proportions of wild-type and mutant strains revealed that the cheZ mutant had a competitive advantage over the wild type, since the estimation of the relative number of bacteria of the mutant strain AC601 colonized on the roots was approximately 2 times of that of the wild type (Fig. 7A). When strains AC602 and AC601 were mixed equally to inoculate the roots of Sesbania, the competitive advantage of strain AC601 was more noticeable (Fig. 7A). This suggested that CheZ plays a negative role in the colonization ability at the root surface. To confirm these results, the wild type and the mutant were further mixed in the proportions of 2:1 and 1:5, respectively, and the mixtures of bacteria were used to conduct competitive root colonization tests. The results showed that the cheZ mutant had an increased ability to colonize the root surface compared to that of the wild type, regardless of the inoculation ratios and time (Fig. 7A). When strains AC602 and AC601 were mixed in the proportions of 2:1 and 1:5, respectively, strain AC602 remained less competitive than the mutant AC601 and barely colonized the roots (Fig. 7A).
FIG 7.
Surface adhesion properties of the A. caulinodans wild type and the cheZ mutant. (A) Competitive adhesion to plant roots of S. rostrata between strain ORS571 and the cheZ deletion mutant AC601 and between the complemented strains AC602 and AC601. The pattern of competitive colonization of the strains at different ratios is expressed on the x axis and the ratios of the strains (ORS571 to AC601 or AC602 to AC601) are indicated below each column. (B) Biofilm formation of the cheZ mutant compared to the wild type. Cells were grown for 72 h in L3 medium with or without a nitrogen source. The biofilm is stained with CV, and the amount of CV staining was quantified as described in Materials and Methods. Error bars show standard errors of the means from at least three replicates.
It was reported that adhesion is critical for the formation of biofilms (46). To investigate whether the ability of biofilm formation was affected, a biofilm formation experiment with the wild type and the mutant on abiotic surfaces was performed. Glass was used as the adhesion medium, and the culture was carried out for 3 days. The results showed that the biofilm formation ability of strain AC601 was stronger than that of the wild type under both N-free and N-sufficient conditions (43% and 16%, respectively) (Fig. 7B), and this was consistent with the results of competitive colonization experiments. Biofilm formation of both ORS571 and AC601 strains under nitrogen-limiting conditions was more pronounced than that under N-sufficient conditions. This is consistent with the ability of bacteria to form biofilms to improve their stress-resistant abilities in unfavorable environments (47, 48).
DISCUSSION
The present work establishes unambiguously that the orphan cheZ gene found in A. caulinodans ORS571 encodes a functional protein playing an essential role in the chemotaxis process affecting the swimming behavior of the bacteria (Fig. 3 and 4). However, the chemotactic response of the cheZ mutant strain (AC601) was not fully complemented when the wild-type gene was introduced in the mutant strain (AC602). This may be due to the high expression in strain AC602 of cheZ, which is approximately 1.25-fold higher than in strain ORS571 (see Fig. S3).
The CheZ product showed extensive similarity with E. coli CheZ (Fig. 2), suggesting it may also be a phosphatase. Indeed, the motif responsible for the phosphatase active site identified in E. coli (49–51), DXXXQ (Fig. 2), is conserved in ORS571 CheZ. In addition, this motif is well conserved in the 200 CheZ proteins aligned from the gamma and alpha subclasses of Proteobacteria. Moreover, the substitution of alanine for the key amino acids D165 and Q169 in the ORS571 CheZ protein resulted in mutant strains (AC603 and AC604) incapable of chemotaxis (Fig. 3B). As ORS571 cheZ is adjacent to a cheY gene (Fig. 1), it is hypothesized that CheZ is playing a role in the chemotactic process by hydrolyzing CheY∼P, similar to that established in E. coli.
The phosphate sink model, involving two CheY proteins (encoded by different cheY genes), was regarded as the most common mechanism for dephosphorylating CheY∼P in other rhizobia, such as S. meliloti, not containing cheZ genes (52). As two cheY genes are also present in strain ORS571 (Fig. 1), this raises the question as to whether a phosphate sink is also functioning in A. caulinodans. CheY1 (whose gene is located next to cheZ) shares 57.7% amino acid sequence identity with CheY in E. coli and 33.9% identity with CheY2 in S. meliloti. CheY2 shares 38.3% amino acid sequence identity with E. coli CheY and 36.8% identity with S. meliloti CheY1, shown to act as a phosphate sink (52). Further experiments are required to establish if the two CheY∼P dephosphorylation patterns, CheZ, and the phosphate sink are functional in strain ORS571. However, as the chemotactic response of strain ORS571 was completely lost when the cheZ gene was deleted, it can be concluded that the CheY∼P dephosphorylation mechanisms differ between A. caulinodans and S. meliloti. In addition, the conservation of the che operon and cheZ gene in a number of species from Alphaproteobacteria (see Fig. S1 and Table S1 in the supplemental material) suggests that the control of chemotaxis through CheZ phosphatase may be more common among Alphaproteobacteria than initially thought.
In addition to a role in the control of the chemotactic response, A. caulinodans CheZ appears to be involved in other cellular processes, such as the formation of cellular aggregates (referred to as flocculation), the production of polysaccharides, and adhesion to biotic (root) and abiotic surfaces (Fig. 5, 6, and 7).
In a hostile environment, bacteria can regulate transient cell flocculation to adapt to changes in signal perception before forming biofilms. In Azospirillum brasilense, flocculation and biofilm formation are related to the production of extracellular polysaccharides (53). During biofilm formation, EPS production was seen as the result of the stimulation of mechanosensing signals caused by flagellar rotation (54), and the accumulation of EPS can promote the attachment of cells. In addition, chemotactic responses can disrupt the formation of flocs in Azospirillum by regulating the direction of motility (43). Interestingly, the flocculation under adverse conditions observed with the cheZ mutant, strain AC601, was higher than that of the wild-type ORS571 (Fig. 6), suggesting that CheZ might be involved in controlling floc formation by mechanisms not directly related to the control of chemotaxis. The increase in flocculation was correlated with an increase in polysaccharide production and an increased expression of exp genes by the cheZ mutant compared to those by the wild type (Fig. 5). Therefore, it is likely that the increased flocculation and EPS production resulted from the increased expression of exp genes.
In the particular case of the Dif system, one of the chemotaxis pathways of myxobacteria, it was found that DifE (CheA-like protein) regulates the production of polysaccharides by phosphorylating EpsW, a specific response regulator responsible for EPS biosynthesis. DifD (CheY-like) and DifG (CheZ-like) proteins in the pathway can indirectly affect polysaccharide production by diverting the phosphorylation of DifE (55). Hence, in strain ORS571, a similar mechanism may account for the regulatory role of CheZ in eps gene expression and the inhibition of the production of polysaccharides. To date, no gene homologous to myxobacterial epsW has been found in the ORS571 genome. Moreover, among Proteobacteria, the amino acid sequences of the C-terminal regions of CheZ proteins are more conserved than those of the highly variable N-terminal regions. In particular, CheZ proteins in Alphaproteobacteria lack the CheA-binding regions presented in those in Betaproteobacteria, Gammaproteobacteria, and Epsilonproteobacteria (30). These structural differences may account for the differences in the regulatory circuity of CheZ phosphatases/response regulators controlling other physiological processes, including polysaccharide synthesis.
Chemotaxis was reported to enhance the interactions between rhizobia and their host plants (16). It is generally believed that chemotaxis promotes Rhizobium adherence to the host root hair surfaces (17). In the particular case of A. caulinodans, the isolation of a deletion mutant strain of the main che operon led to a strain that displayed less adhesion to the host roots (W. Liu, Y. Sun, X. Dang, X. Liu, F. Sui, Y. Li, Z. Zhang, G. Alexandres. C. Elmerich, and Z. Xie, unpublished data). Surprisingly, compared with strain ORS571, strain AC601 displays an increased colonization of the host root surface (Fig. 7A). This phenomenon is in contradiction to the belief that chemotaxis could improve the ability of the bacteria to colonize the plants (56), suggesting CheZ of strain ORS571 may have an unusual function, not like known CheZs in other organisms. Generally, EPS secreted by bacteria can promote the process of infection (57, 58). In the case of A. caulinodans, it was previously reported that a strain with a mutation affecting the production of extracellular polysaccharides had impaired nodulation properties (59). The improvement of colonization for strain AC601 is in agreement with the increased EPS production and biofilm formation (Fig. 7B). Thus, CheZ in the wild type, by downregulating EPS production, may have a negative regulatory role in the adhesion process.
It was observed that the motility process was strongly impaired in the cheZ mutant, which showed a hyperreversal phenotype (Fig. 4). This phenotype may prevent bacteria from moving specifically toward the root system. Recently, Zhou and Nan (60) reported that EPS can inhibit cellular reversal in Myxococcus xanthus in social motility (S motility). Though S motility is not found in strain ORS571, EPS may play a role, to some extent, in increasing the colonization of the cheZ mutant by inhibiting the hyperreversal behavior.
Taken together, the results from this study expand the horizon with regard to chemotaxis in Alphaproteobacteria. However, the specific mechanism of regulating EPS production through CheZ is not elucidated, and the functions of CheZ in the processes of crack entry and nodulation formation need to be further studied.
MATERIALS AND METHODS
Media and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. A. caulinodans ORS571 (24) and its derivatives were grown at 37°C in tryptone-yeast extract (TY) medium or in L3 minimal medium containing 10 mg/ml dl-sodium lactate (23, 40), which was or was not supplemented with 10 mM NH4Cl (designated L3+N or L3−N medium, respectively). In L3 medium, sodium lactate can be replaced with other carbon sources as the sole carbon source functioning as the attractant. Antibiotics at the following final concentrations were added: ampicillin (Amp), 100 μg/ml; nalidixic acid, 25 μg/ml; gentamicin, 50 μg/ml; and kanamycin, 50 μg/ml.
Plasmid and strain construction.
To construct the cheZ mutant, a 785-bp upstream fragment (UF) and a 725-bp downstream fragment (DF) of the cheZ gene were amplified by PCR by using two primer pairs, cheZUF-cheZUR and cheZDF-cheZDR (Table 2). The amplicons were digested with appropriate restriction enzymes (for UF, KpnI and NdeI; for DF, AgeI and SacI) before linking them together to generate a KpnI-SacI fragment. The DNA fragment obtained was inserted into an allelic exchange vector, pCM351 (61), digested with KpnI-SacI. This resulting construct was introduced into the wild-type strain by triparental conjugation for allelic exchange (using the helper pRK2013) (Table 1), as described previously (62).
TABLE 2.
PCR primers used in this study
| Primers | Sequences (5′→3′)a | Source or reference |
|---|---|---|
| cheZUF KpnI | GGGGTACCTGCCCTTCAGCGTCTGC | This study |
| cheZUR NdeI | GGAATTCCATATGGCGTCCTGGATCGTCTC | This study |
| cheZDF AgeI | GACCGGTCGACCGACGAGAATGTGG | This study |
| cheZDR SacI | CGAGCTCGCTGCCGATCCTCTATGC | This study |
| cheZcomF KpnI | GGGGTACCGAAATCACGAGGCCGTAC | This study |
| cheZcomR BamHI | CGGGATCCCATCCATCAAGCCAGCAA | This study |
| SDMDF | TTCCAGGCCATCACCGGCCA | This study |
| SDMDR | TGGCCGGTGATGGCCTGGAA | This study |
| SDMQF | CATCACCGGCGCGCGCATC | This study |
| SDMQR | GATGCGCGCGCCGGTGATG | This study |
| RT-cheZ-F | AACCATCGACAATTCCATCG | This study |
| RT-cheZ-R | TGAAGACCATCAGCTTCACG | This study |
| RT-1833-F | GCGGTTGTCGAGATACCAGT | This study |
| RT-1833-R | ACCTCATCACCTTCGTGACC | This study |
| RT-1834-F | TAGATGAAGGGGCTGTCGAG | This study |
| RT-1834-R | GCGTGCTCTACAAGGTGGAC | This study |
| RT-3326-F | GGTTCCACGACGTGATCTTC | 40 |
| RT-3326-R | CGTCTCCTGAGTTTCCGAAC | 40 |
| RT-16S-F | ACGGATTTCTTCCAGCAATG | 40 |
| RT-16S-R | ACCGGCAGTCCCTTTAGAGT | 40 |
Engineered restriction sites are underlined.
Homologous recombinants lacking the cheZ gene were selected by antibiotics on TY plates. The cheZ mutant can grow on TY plates containing ampicillin, nalidixic acid, and gentamicin (final concentrations, 100 μg/ml) but not on TY plates containing tetracycline (final concentration, 100 μg/ml), and correct recombination was verified by PCR. One resulting mutant strain was named AC601 (Table 1) and was used in subsequent experiments.
To complement the cheZ mutant strain AC601, a fragment encompassing the 406-bp region upstream of the promoter of the cheZ gene and the intact open reading frame for cheZ was amplified by PCR using the primer pair cheZcomF-cheZcomR (Table 2). The amplified fragment was cloned into the KpnI and BamHI sites of the broad-host-range vector pBBR1MCS-2 (63), and the DNA sequence was verified by sequencing. Then, the resulting plasmid was introduced into strain AC601 via triparental mating and selecting for kanamycin resistance. The resulting strain was named AC602 (Table 1).
Swim plate and competitive capillary assays for chemotaxis.
The soft agar plate and capillary assays for chemotaxis in A. caulinodans ORS571 were performed essentially as previously described (64, 65), with some modifications. For the soft agar assay, 5 μl of cells suspended in chemotaxis buffer (10 mM K2HPO4, 10 mM KH2PO4, 0.1 mM EDTA [pH 7.0]) at an optical density at 600 nm (OD600) of 0.6 was inoculated at the center of L3 minimal soft agar plates solidified with 0.3% agar and containing different carbon sources added to a final concentration of 10 mM. Photographs were taken after 48 h of incubation at 37°C. E. coli cells were inoculated into VBC minimal medium (66) with 0.3% agar for 24 h at 37°C. Experiments were performed at least three times, with a minimum of six replicates per sample.
For the quantitative capillary assay, cells were grown in TY medium and centrifuged and washed 3 to 5 times with PBS, and the suspension was adjusted to an OD600 of 0.01. The wild-type and mutant strains were mixed at 1:1 ratios, and 200 μl of the mixed bacterial solution was added to the 96-well plate. The capillary is open at one end and sealed by flame at the other end. The capillary is gently swept over the flame and then quickly inserted into the PBS or 10 mM succinate solution for 3 to 5 min, so that the capillary liquid height stabilized at about 1 cm. Capillary tubes inserted into both kinds of solutions were moved to 96-well plates containing the mixed bacteria solution. The bacteria were cultured at 37°C for 1 h. The liquid in the capillaries was then pipetted into 1 ml PBS and plated on TY solid medium containing nalidixic acid and ampicillin. The numbers of colonies on the plates were counted, and PCR amplification was carried out using specific primers to determine the ratios of wild-type and mutant strains in the bacterial solutions entering the capillaries.
Analyses of swimming behavior.
A. caulinodans strains were cultured overnight with shaking in TY medium. Five microliters of culture was added to a microscope slide, and the swimming behavior of strains was recorded using cellSens Dimension 1.7 imaging software (Olympus Inc.) and an Olympus DP73 digital camera on an Olympus BX53 system microscope at ×40 and ×100 magnifications. Every culture selected from more than 3 independent biological replicates was recorded at least 3 times. For each A. caulinodans strain, the swimming paths from at least 50 cells were manually tracked on video recordings with ICY software (67). Each cell was tracked until it was not visible for more than two frames. The reorientation frequency of each strain was determined by counting the number of changes in swimming direction in 5 s.
Flocculation formation assay.
Flocculation was estimated using the method described in reference 68, with the following modifications reported in reference 23. Bacterial cells in 50-ml conical centrifuge tubes containing L3 minimal medium with 5 mM NH4Cl were incubated at 37°C and 200 rpm for 24, 48, and 72 h and allowed to stand at room temperature for half an hour so that flocs settled to the bottoms of the tubes. The OD600 values of the nonflocculated cells remained in suspension (ODs), and the total turbidity (ODt) was determined after the cultures were dispersed by treatment in a tissue homogenizer for 1 min. The percentage of flocculation was estimated as follows: % flocculation = ([ODt − ODs] × 100)/ODt. The experiment was carried out at least three times, with three replicates per sample.
Site-directed mutagenesis.
Site-specific mutations were generated by overlapping PCR (69). Strain AC603 was constructed by expressing the mutated cheZD165A gene from its native promoter in plasmid pBBR1MCS-2 (pBBRCheZD165A) (63). The primers (SDMDF/SDMDR) that replaced A with C in the GAC encoding aspartic acid were designed. The other pair of primers was the same as the upstream and downstream primers of cheZcomF/cheZcomR (containing sequences for two endonucleases, KpnI and BamHI, respectively). First, two pairs of primers, cheZcomF/SDMDR and SDMDF/cheZcomR, were used for amplification. Then, PCR products were directly mixed as the template, and the PCR was carried out using cheZcomF/cheZcomR as primers. Amplification products were ligated into the pEASY plasmid, pEASYCheZD165A. The pEASYCheZD165A and pBBR1MCS-2 plasmids were digested separately with KpnI and BamHI endonucleases. The two digested fragments were then ligated with T4 ligase to construct the pBBRCheZD165A plasmid. The plasmid was transformed into strain AC601 by triparental mating. The CheZQ169A site-specific mutant strain, AC604, was constructed using a similar method. All constructed plasmids were verified by sequencing.
Plant growth and colonization of Sesbania roots.
Sesbania seeds were surface sterilized by treatment with concentrated sulfuric acid for 20 min, followed by three washes with sterile water. All seeds were germinated in sterile petri dishes in the dark at 37°C for 48 to 72 h. A. caulinodans and AC601 cells were grown overnight in TY liquid medium to an OD600 of 0.8 to 1.0. Germinated seeds were soaked in a mixture of different proportions of the wild-type and mutant strains for 4 or 24 h. The surfaces of roots were washed 4 times with sterile water and then were mashed with sterile pipette tips sealed by flame. Bacteria were reisolated from root surfaces by using serial dilutions plated on TY agar plates containing ampicillin and nalidixic acid. After 24 to 36 h of growth at 37°C, single colonies were identified by PCR amplification of the cheZ gene using the primers cheZcomF and cheZcomR. For each competition experiment, at least 100 colonies were PCR amplified. In addition, since strain AC601 is Gmr, the growth of the colonies was also assayed on TY agar plates with gentamicin.
Biofilm formation measurement and quantification of EPS.
The quantification of biofilm formation was performed using the crystal violet (CV) staining method (70) according to a protocol previously described (23). The OD540 was determined using a microplate reader (Tecan Infinite M200) to quantify the amount of biofilm. The quantification of EPS was performed according to reference 40, with modifications previously reported (23). The EPS content was estimated using d-glucose as the standard. For qualitative evaluation of EPS production, bacteria grown on L3+N plates containing Congo red (40 μg/ml) were observed and photographed after 3 days.
Expression analyses of EPS production-related genes.
Total RNA was isolated from 25 ml of A. caulinodans bacterial cells grown in TY medium, using the TransZol Up Plus RNA kit (Transgen Biotech, China) according to the instructions. The concentration and quality of RNA were determined by a Nanodrop 2000. One thousand nanograms of cDNA was synthesized with a One-Step gDNA removal kit (Transgen Biotech, China). The synthesized cDNA was diluted 10-fold and 105-fold with RNA-free water and then was used in the subsequent RT-qPCR. The comparative CT method (71) was used to analyze the expression levels of EPS genes.
Bioinformatics analysis.
CheZ amino acid sequences were selected from the Mist2.2 database (http://mistdb.com/proteins/slice?repcon_id=1804&class=chemotaxis) (72). The selected amino acid sequences of CheZ were aligned using MEGA5. The evolutionary tree was inferred using the neighbor-joining method. The analysis involved 13 16S RNA sequences and was conducted in MEGA5. The alignment of CheZ sequences from A. caulinodans, E. coli, and S. Typhimurium was performed using Bioedit (73). Chemotaxis genes and proteins present in the bacterial genome were searched in the Mist2.2 database, and the information on the distribution and organization of chemotaxis genes was also collected from Mist2.2. The protein sequences selected randomly were aligned using MEGA5, and the alignment file was put into Jalview to develop a graphical representation, which showed the level of amino acid conservation (74). An amino acid region near the phosphatase active site was the most conserved and was selected and put into WebLogo (75).
Statistical analysis.
Statistical analyses for behavioral assays, expression assays, and competitive colonization experiments were performed using SPSS. Student t tests assuming equal variances (P < 0.05) were used to determine significant differences between conditions. Chi-square tests were used to determine differences between inoculation and recovery ratios (P < 0.001 and P < 0.05 were tested).
Supplementary Material
ACKNOWLEDGMENTS
We thank John Parkinson, University of Utah, for E. coli mutants and Gladys Alexandre and John Parkinson for helpful discussions and insightful comments on the manuscript.
This study was financed by the National Natural Science Foundation of China (31370108 and 31570063), the One Hundred-Talent Plan of the Chinese Academy of Sciences, the High-Tech Industrialization Cooperation Funds of Jilin province and the Chinese Academy of Sciences (2017SYHZ0007), and Shandong Key Research and Development Program (2016CYJS05A01-1). This study was conducted with the support of the Institut Pasteur, Paris, France.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01827-17.
REFERENCES
- 1.Neumann S, Grosse K, Sourjik V. 2012. Chemotactic signaling via carbohydrate phosphotransferase systems in Escherichia coli. Proc Natl Acad Sci U S A 109:12159–12164. doi: 10.1073/pnas.1205307109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szurmant H, Ordal GW. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB. 2016. Cache domains that are homologous to, but different from pas domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol 12:e1004862. doi: 10.1371/journal.pcbi.1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisenbach M. 1996. Control of bacterial chemotaxis. Mol Microbiol 20:903–910. doi: 10.1111/j.1365-2958.1996.tb02531.x. [DOI] [PubMed] [Google Scholar]
- 5.Lertsethtakarn P, Ottemann KM, Hendrixson DR. 2011. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szurmant H, Muff TJ, Ordal GW. 2004. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J Biol Chem 279:21787–21792. doi: 10.1074/jbc.M311497200. [DOI] [PubMed] [Google Scholar]
- 8.Dogra G, Purschke FG, Wagner V, Haslbeck M, Kriehuber T, Hughes JG, Van Tassell ML, Gilbert C, Niemeyer M, Ray WK, Helm RF, Scharf BE. 2012. Sinorhizobium meliloti CheA complexed with CheS exhibits enhanced binding to CheY1, resulting in accelerated CheY1 dephosphorylation. J Bacteriol 194:1075–1087. doi: 10.1128/JB.06505-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riepl H, Maurer T, Kalbitzer HR, Meier VM, Haslbeck M, Schmitt R, Scharf B. 2008. Interaction of CheY2 and CheY2-P with the cognate CheA kinase in the chemosensory-signalling chain of Sinorhizobium meliloti. Mol Microbiol 69:1373–1384. doi: 10.1111/j.1365-2958.2008.06342.x. [DOI] [PubMed] [Google Scholar]
- 10.Alexander RP, Lowenthal AC, Harshey RM, Ottemann KM. 2010. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol 18:494–503. doi: 10.1016/j.tim.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lertsethtakarn P, Ottemann KM. 2010. A remote CheZ orthologue retains phosphatase function. Mol Microbiol 77:225–235. doi: 10.1111/j.1365-2958.2010.07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiménez-Pearson MA, Delany I, Scarlato V, Beier D. 2005. Phosphate flow in the chemotactic response system of Helicobacter pylori. Microbiology 151:3299–3311. doi: 10.1099/mic.0.28217-0. [DOI] [PubMed] [Google Scholar]
- 13.Aihara E, Closson C, Matthis AL, Schumacher MA, Engevik AC, Zavros Y, Ottemann KM, Montrose MH. 2014. Motility and chemotaxis mediate the preferential colonization of gastric injury sites by Helicobacter pylori. PLoS Pathog 10:e1004275. doi: 10.1371/journal.ppat.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antúnez-Lamas M, Cabrera-Ordonez E, Lopez-Solanilla E, Raposo R, Trelles-Salazar O, Rodriguez-Moreno A, Rodriguez-Palenzuela P. 2009. Role of motility and chemotaxis in the pathogenesis of Dickeya dadantii 3937 (ex Erwinia chrysanthemi 3937). Microbiology 155:434–442. doi: 10.1099/mic.0.022244-0. [DOI] [PubMed] [Google Scholar]
- 15.Novak EA, Sekar P, Xu H, Moon KH, Manne A, Wooten RM, Motaleb MA. 2016. The Borrelia burgdorferi CheY3 response regulator is essential for chemotaxis and completion of its natural infection cycle. Cell Microbiol 18:1782–1799. doi: 10.1111/cmi.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scharf BE, Hynes MF, Alexandre GM. 2016. Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol Biol 90:549–559. doi: 10.1007/s11103-016-0432-4. [DOI] [PubMed] [Google Scholar]
- 17.Vande Broek A, Vanderleyden J. 1995. The role of bacterial motility, chemotaxis, and attachment in bacteria plant interactions. Mol Plant Microbe Interact 8:800–810. doi: 10.1094/MPMI-8-0800. [DOI] [Google Scholar]
- 18.Burdman S, Okon Y, Jurkevitch E. 2000. Surface characteristics of Azospirillum brasilense in relation to cell aggregation and attachment to plant roots. Crit Rev Microbiol 26:91–110. doi: 10.1080/10408410091154200. [DOI] [PubMed] [Google Scholar]
- 19.Dupuy LX, Silk WK. 2016. Mechanisms of early microbial establishment on growing root surfaces. Vadose Zone J 15:13. doi: 10.2136/vzj2015.06.0094. [DOI] [Google Scholar]
- 20.Espinosa-Urgel M, Salido A, Ramos JL. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J Bacteriol 182:2363–2369. doi: 10.1128/JB.182.9.2363-2369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webb BA, Hildreth S, Helm RF, Scharf BE. 2014. Sinorhizobium meliloti chemoreceptor McpU mediates chemotaxis toward host plant exudates through direct proline sensing. Appl Environ Microbiol 80:3404–3415. doi: 10.1128/AEM.00115-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caetano-Anollés G, Wall LG, De Micheli AT, Macchi EM, Bauer WD, Favelukes G. 1988. Role of motility and chemotaxis in efficiency of nodulation by Rhizobium meliloti. Plant Physiol 86:1228–1235. doi: 10.1104/pp.86.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang N, Liu W, Li Y, Wu H, Zhang Z, Alexandre G, Elmerich C, Xie Z. 2016. A chemotaxis receptor modulates nodulation during the Azorhizobium caulinodans-Sesbania rostrata symbiosis. Appl Environ Microbiol 82:3174–3184. doi: 10.1128/AEM.00230-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dreyfus B, Garcia JL, Gillis M. 1988. Characterization of Azorhizobium-caulinodans gen. nov., sp. nov., a stem-nodulating nitrogen-fixing bacterium isolated from Sesbania rostrata. Int J Syst Bacteriol 38:89–98. doi: 10.1099/00207713-38-1-89. [DOI] [Google Scholar]
- 25.Jiang N, Liu W, Li Y, Xie Z. 2016. Comparative genomic and protein sequence analyses of the chemotaxis system of Azorhizobium caulinodans. Wei Sheng Wu Xue Bao 56:1256–1265. [PubMed] [Google Scholar]
- 26.Dreyfus BL, Dommergues YR. 1981. Nitrogen-fixing nodules induced by Rhizobium on the stem of the tropical legume Sesbania rostrata. FEMS Microbiol Lett 10:313–317. doi: 10.1111/j.1574-6968.1981.tb06262.x. [DOI] [Google Scholar]
- 27.Dreyfus BL, Elmerich C, Dommergues YR. 1983. Free-living Rhizobium strain able to grow on N2 as the sole nitrogen-source. Appl Environ Microbiol 45:711–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stone PJ, O'Callaghan KJ, Davey MR, Cocking EC. 2001. Azorhizobium caulinodans ORS571 colonizes the xylem of Arabidopsis thaliana. Mol Plant Microbe Interact 14:93–97. doi: 10.1094/MPMI.2001.14.1.93. [DOI] [PubMed] [Google Scholar]
- 29.Lee KB, De Backer P, Aono T, Liu CT, Suzuki S, Suzuki T, Kaneko T, Yamada M, Tabata S, Kupfer DM, Najar FZ, Wiley GB, Roe B, Binnewies TT, Ussery DW, D'Haeze W, Herder JD, Gevers D, Vereecke D, Holsters M, Oyaizu H. 2008. The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9:271. doi: 10.1186/1471-2164-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wuichet K, Alexander RP, Zhulin IB. 2007. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol 422:1–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Weert S, Vermeiren H, Mulders IH, Kuiper I, Hendrickx N, Bloemberg GV, Vanderleyden J, De Mot R, Lugtenberg BJ. 2002. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol Plant Microbe Interact 15:1173–1180. doi: 10.1094/MPMI.2002.15.11.1173. [DOI] [PubMed] [Google Scholar]
- 32.Dandekar T, Snel B, Huynen M, Bork P. 1998. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci 23:324–328. doi: 10.1016/S0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 33.Butler SM, Camilli A. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci U S A 101:5018–5023. doi: 10.1073/pnas.0308052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller LD, Yost CK, Hynes MF, Alexandre G. 2007. The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol Microbiol 63:348–362. doi: 10.1111/j.1365-2958.2006.05515.x. [DOI] [PubMed] [Google Scholar]
- 35.McEvoy MM, Bren A, Eisenbach M, Dahlquist FW. 1999. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein fliM. J Mol Biol 289:1423–1433. doi: 10.1006/jmbi.1999.2830. [DOI] [PubMed] [Google Scholar]
- 36.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkinson JS. 1978. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol 135:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang IY, Tan MH, Koh E, Ho CL, Poh CL, Chang MW. 2014. Reprogramming microbes to be pathogen-seeking killers. ACS Synth Biol 3:228–237. doi: 10.1021/sb400077j. [DOI] [PubMed] [Google Scholar]
- 39.Fujishige NA, Kapadia NN, Hirsch AM. 2006. A feeling for the micro-organism: structure on a small scale. Biofilms on plant roots. Bot J Linn Soc 150:79–88. doi: 10.1111/j.1095-8339.2006.00492.x. [DOI] [Google Scholar]
- 40.Nakajima A, Aono T, Tsukada S, Siarot L, Ogawa T, Oyaizu H. 2012. Lon protease of Azorhizobium caulinodans ORS571 is required for suppression of reb gene expression. Appl Environ Microbiol 78:6251–6261. doi: 10.1128/AEM.01039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker A, Ruberg S, Kuster H, Roxlau AA, Keller M, Ivashina T, Cheng HP, Walker GC, Puhler A. 1997. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J Bacteriol 179:1375–1384. doi: 10.1128/jb.179.4.1375-1384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato S, Siarot L, Matsuoka J, Aono T, Oyaizu H. 2016. An Azorhizobium caulinodans ORS571 mutant with deletion of a gene encoding a TIGR02302 family protein overproduces exopolysaccharides and is defective in infection into plant host cells. Soil Sci Plant Nutr 62:392–398. doi: 10.1080/00380768.2016.1200954. [DOI] [Google Scholar]
- 43.Alexandre G. 2015. Chemotaxis control of transient cell aggregation. J Bacteriol 197:3230–3237. doi: 10.1128/JB.00121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He L, Wen X, Yan X, Ding L, Cao S, Huang X, Wu R, Wen Y. 2016. Effect of cheY deletion on growth and colonization in a Haemophilus parasuis serovar 13 clinical strain EP3. Gene 577:96–100. doi: 10.1016/j.gene.2015.11.046. [DOI] [PubMed] [Google Scholar]
- 45.Merritt PM, Danhorn T, Fuqua C. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J Bacteriol 189:8005–8014. doi: 10.1128/JB.00566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogino PC, Oliva Mde L, Sorroche FG, Giordano W. 2013. The role of bacterial biofilms and surface components in plant-bacterial associations. Int J Mol Sci 14:15838–15859. doi: 10.3390/ijms140815838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang D, Xu A, Elmerich C, Ma LZ. 2017. Biofilm formation enables free-living nitrogen-fixing rhizobacteria to fix nitrogen under aerobic conditions. ISME J 11:1602–1613. doi: 10.1038/ismej.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yaryura PM, Leon M, Correa OS, Kerber NL, Pucheu NL, Garcia AF. 2008. Assessment of the role of chemotaxis and biofilm formation as requirements for colonization of roots and seeds of soybean plants by Bacillus amyloliquefaciens BNM339. Curr Microbiol 56:625–632. doi: 10.1007/s00284-008-9137-5. [DOI] [PubMed] [Google Scholar]
- 49.Lertsethtakarn P, Howitt MR, Castellon J, Amieva MR, Ottemann KM. 2015. Helicobacter pylori CheZ(HP) and ChePep form a novel chemotaxis-regulatory complex distinct from the core chemotaxis signaling proteins and the flagellar motor. Mol Microbiol 97:1063–1078. doi: 10.1111/mmi.13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silversmith RE. 2010. Auxiliary phosphatases in two-component signal transduction. Curr Opin Microbiol 13:177–183. doi: 10.1016/j.mib.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao R, Collins EJ, Bourret RB, Silversmith RE. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol 9:570–575. doi: 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
- 52.Scharf B, Schmitt R. 2002. Sensory transduction to the flagellar motor of Sinorhizobium meliloti. J Mol Microbiol Biotechnol 4:183–186. [PubMed] [Google Scholar]
- 53.Bahat-Samet E, Castro-Sowinski S, Okon Y. 2004. Arabinose content of extracellular polysaccharide plays a role in cell aggregation of Azospirillum brasilense. FEMS Microbiol Lett 237:195–203. doi: 10.1111/j.1574-6968.2004.tb09696.x . [DOI] [PubMed] [Google Scholar]
- 54.Belas R. 2014. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 55.Black WP, Wang L, Davis MY, Yang Z. 2015. The orphan response regulator EpsW is a substrate of the DifE kinase and it regulates exopolysaccharide in Myxococcus xanthus. Sci Rep 5:17831. doi: 10.1038/srep17831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allard-Massicotte R, Tessier L, Lecuyer F, Lakshmanan V, Lucier JF, Garneau D, Caudwell L, Vlamakis H, Bais HP, Beauregard PB. 2016. Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. mBio 7:e01664-16. doi: 10.1128/mBio.01664-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawaharada Y, Nielsen MW, Kelly S, James EK, Andersen KR, Rasmussen SR, Fuchtbauer W, Madsen LH, Heckmann AB, Radutoiu S, Stougaard J. 2017. Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat Commun 8:14534. doi: 10.1038/ncomms14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszynski A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, Vinther M, Andersen SU, Krusell L, Thirup S, Jensen KJ, Ronson CW, Blaise M, Radutoiu S, Stougaard J. 2015. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523:308–312. doi: 10.1038/nature14611. [DOI] [PubMed] [Google Scholar]
- 59.Gao M, D'Haeze W, De Rycke R, Wolucka B, Holsters M. 2001. Knockout of an azorhizobial dTDP-l-rhamnose synthase affects lipopolysaccharide and extracellular polysaccharide production and disables symbiosis with Sesbania rostrata. Mol Plant Microbe Interact 14:857–866. doi: 10.1094/MPMI.2001.14.7.857. [DOI] [PubMed] [Google Scholar]
- 60.Zhou T, Nan B. 2017. Exopolysaccharides promote Myxococcus xanthus social motility by inhibiting cellular reversals. Mol Microbiol 103:729–743. doi: 10.1111/mmi.13585. [DOI] [PubMed] [Google Scholar]
- 61.Marx CJ, Lidstrom ME. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062–1067. [DOI] [PubMed] [Google Scholar]
- 62.Ditta G, Stanfield S, Corbin D, Helinski DR. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A 77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 64.Miller LD, Russell MH, Alexandre G. 2009. Diversity in bacterial chemotactic responses and niche adaptation. Adv Appl Microbiol 66:53–75. doi: 10.1016/S0065-2164(08)00803-4. [DOI] [PubMed] [Google Scholar]
- 65.Reyes-Darias JA, García V, Rico-Jiménez M, Corral-Lugo A, Krell T. 2016. Identification and characterization of bacterial chemoreceptors using quantitative capillary and gradient plate chemotaxis assays. Bio Protoc 6:e1789. doi: 10.21769/BioProtoc.1789. [DOI] [Google Scholar]
- 66.Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218:97–106. [PubMed] [Google Scholar]
- 67.de Chaumont F, Dallongeville S, Chenouard N, Herve N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Le Montagner Y, Lagache T, Dufour A, Olivo-Marin JC. 2012. Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods 9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- 68.Burdman S, Jurkevitch E, Schwartsburd B, Hampel M, Okon Y. 1998. Aggregation in Azospirillum brasilense: effects of chemical and physical factors and involvement of extracellular components. Microbiology 144(Part 7):1989–1999. [DOI] [PubMed] [Google Scholar]
- 69.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 70.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 71.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 72.Ulrich LE, Zhulin IB. 2010. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res 38:D401–D407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hall T. 2011. BioEdit: an important software for molecular biology. GERF Bull Biosci 2:60–61. [Google Scholar]
- 74.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.