ABSTRACT
Vibrio parahaemolyticus and Vibrio vulnificus are naturally occurring estuarine bacteria and are the leading causes of seafood-associated infections and mortality in the United States. Though multiple-antibiotic-resistant V. parahaemolyticus and V. vulnificus strains have been reported, resistance patterns in vibrios are not as well documented as those of other foodborne bacterial pathogens. Salinity relaying (SR) is a postharvest processing (PHP) treatment to reduce the abundances of these pathogens in shellfish harvested during the warmer months. The purpose of this study was to evaluate the antimicrobial susceptibility (AMS), pathogenicity, and genetic profiles of V. parahaemolyticus and V. vulnificus recovered from oysters during an oyster relay study. Isolates (V. parahaemolyticus [n = 296] and V. vulnificus [n = 94]) were recovered from oysters before and during the 21-day relaying study to detect virulence genes (tdh and trh) and genes correlated with virulence (vcgC) using multiplex quantitative PCR (qPCR). AMS to 20 different antibiotics was investigated using microbroth dilution, and pulsed-field gel electrophoresis (PFGE) was used to study the genetic profiles of the isolates. Twenty percent of V. vulnificus isolates were vcgC+, while 1 and 2% of V. parahaemolyticus were tdh+ and trh+, respectively. More than 77% of the V. vulnificus isolates and 30% of the V. parahaemolyticus isolates were resistant to at least one antimicrobial. Forty-eight percent of V. vulnificus and 8% of V. parahaemolyticus isolates were resistant to two or more antimicrobials. All isolates demonstrated a high genetic diversity, even among those isolated from the same site and having a similar AMS profile. No significant effects of the relaying process on AMS, virulence genes, or PFGE profiles of V. vulnificus and V. parahaemolyticus were observed.
IMPORTANCE Analysis of the antibiotic resistance profiles of V. vulnificus and V. parahaemolyticus isolated from oysters during this study indicated that more than 48% of V. vulnificus isolates were resistant to two or more antimicrobials, including those recommended by the CDC for treating Vibrio infections. Also, the V. parahaemolyticus isolates showed high MICs for some of the Vibrio infection treatment antibiotics. Monitoring of AMS profiles of this bacterium is important to ensure optimal treatment of infections and improve food safety. Our study showed no significant differences in the AMS profiles of V. vulnificus (P = 0.26) and V. parahaemolyticus (P = 0.23) isolated from the oysters collected before versus after relaying. This suggests that the salinity of the relaying sites did not affect the AMS profiles of the Vibrio isolates, although it did reduce the numbers of these bacteria in oysters (S. Parveen et al., J Food Sci 82:484–491, 2017, https://doi.org/10.1111/1750-3841.13584).
KEYWORDS: oysters, relay, Vibrio parahaemolyticus, Vibrio vulnificus
INTRODUCTION
Vibrio parahaemolyticus and Vibrio vulnificus, naturally occurring estuarine bacteria, are the leading causes of seafood-borne mortality and illness in the United States (1–3). V. parahaemolyticus causes the highest number of seafood-associated bacterial gastroenteritis in the United States and in Asian countries (4–6). The primary classification of V. parahaemolyticus is a serotyping scheme, which depends mainly on the antigenic properties of the somatic (O) and capsular (K) antigens (7). V. parahaemolyticus is a multiserogroup pathogen, with at least 13 O serogroups and 71 K serotypes detected (8, 9).
Thermostable direct hemolysin (tdh) and TDH-related hemolysin (trh) are two major virulence factors associated with V. parahaemolyticus which are closely related to its pathogenicity (10). Epidemiological investigations indicated that tdh is one of the major pathogenic factors in V. parahaemolyticus and is prevalent in almost all (95%) clinical isolates (11, 12).
Vibrio vulnificus is responsible for more than 95% of seafood-related deaths in the United States (13), especially among high-risk consumers with immunocompromised conditions or liver disease (14, 15). Despite the high number of at-risk consumers and the fact that in some regions, oysters during the summer months typically contain 103 to 104 CFU/g of V. vulnificus, the incidence of disease is relatively low (16–19), leading to the hypothesis that not all strains of V. vulnificus are equally virulent. Several biomarkers, e.g., the virulence-correlated gene (vcg), 16S rRNA, and the capsular polysaccharide operon (CPS), have been used to differentiate virulent- from nonvirulent-type V. vulnificus strains (20). There are, however, no available molecular markers with sufficient resolving power to categorize with absolute certainty the pathogenicity of V. vulnificus strains (17). Rosche et al. (21) reported that 90% of the C-type strains were clinical isolates (vcgC positive), while 93% of environmental isolates were classified as E-type (vcgC negative). Therefore, vcgC may serve as a reliable biomarker to screen for potentially virulent V. vulnificus strains.
According to the U.S. Centers for Disease Control and Prevention (CDC), the incidence of Vibrio infections has increased since 2001 (almost tripling during the period between 1996 and 2010), while all other foodborne infection rates have remained the same or decreased (13). Also, in 2013, the FoodNet found that the incidence of Vibrio infections was significantly (32%) higher than that during 2010 to 2012 (22). In the severe form of V. vulnificus infection (septicemia), the fatality rates for patients significantly increased with the delays between the onset of illness and initiation of antibiotic treatment (14); the awareness of antimicrobial resistance of these two pathogens is not as well documented as those for other foodborne bacterial pathogens. Vibrio spp. are susceptible to most antimicrobial agents of veterinary and human significance (3). Recent studies indicated that V. parahaemolyticus and V. vulnificus have developed multiple antimicrobial resistances, which may be due to the discharging of wastewater containing pathogenic bacteria with antimicrobial resistance genes (23). Among individuals consuming raw or undercooked contaminated seafood and seafood products, this can lead to serious public health issues (24).
Because of health concerns associated with the consumption of raw product, the National Shellfish Sanitation Program (NSSP) provides dealers the option to use an approved and validated postharvest processing (PHP) method to reduce Vibrio levels and make safety-added labeling claims. The few studies performed on the efficacy of relaying and depuration as PHP methods for reducing the abundance of Vibrio bacteria, especially V. vulnificus, in oysters have shown promising results. Interestingly, flowthrough depuration was successful in reducing V. vulnificus in artificially contaminated oysters from >104 most probable number (MPN)/g to <30 MPN/g when incoming water salinity was higher than (30 ppt) after 6 days (25). Recently, we reported that relaying of oysters to high-salinity field sites (29 to 34 ppt) or transfer to high-salinity recirculating aquaculture systems (RAS) (32 to 34 ppt) can reduce average levels of these bacteria in oysters by 2 to 5 logs after 21 to 28 days. These methods were more effective in reducing V. vulnificus than V. parahaemolyticus. Oyster mortality rates averaged 4% or less and did not exceed 7% (26).
It has been reported that salinity can affect the growth and survival of V. parahaemolyticus in aquatic environments (26, 27). Whitaker et al. (28) investigated the growth of V. parahaemolyticus at different salt concentrations and observed that salt can affect its response to pH and temperature. The same study also reported that the cytotoxic effects of V. parahaemolyticus in human intestinal cells were greater when this bacterium was grown in a medium containing 1% salt than with 3% salt. However, it is undetermined whether or not high-salinity relaying has any appreciable effect on the prevalence of pathogenic strains (virulence genes), antibiotic resistance, and genetic profiles of V. parahaemolyticus and V. vulnificus.
In this study, we evaluated the phenotypic and genotypic characteristics of PCR-confirmed Vibrio isolates recovered from oyster samples before and during the 21-day relaying process. The objectives of this study were to (i) investigate the antimicrobial susceptibility profiles of the PCR-confirmed V. parahaemolyticus and V. vulnificus isolates, as well as the predominant O serogroups of V. parahaemolyticus recovered during the relaying process; (ii) study the virulence properties of the V. parahaemolyticus and V. vulnificus strains isolated during the relaying trials; and (iii) investigate the genetic relationships of these isolates based on their susceptibility profiles using the pulsed-field gel electrophoresis (PFGE) technique.
RESULTS
Virulence genes of V. vulnificus and V. parahaemolyticus isolates.
In this study, the vcgC gene of V. vulnificus isolated from oyster samples was detected. Fifty-five of 263 (20.9%) PCR-confirmed V. vulnificus isolates were vcgC+; specifically, 27 of 100 (27%) tested isolates from the moderate-salinity site and 25 of 127 (19.7%) tested isolates from the high-salinity site were vcgC+ (Table 1). There was no significant difference between the frequencies of detection of the virulence gene between isolates from the moderate- and the high-salinity sites.
TABLE 1.
vcgC-positive Vibrio vulnificus isolated during the high- and moderate-salinity relaying process
| Day/sitea | No. of isolates | % vcgC+ |
|---|---|---|
| 0 | 36 | 8.3 |
| 7/S1 | 36 | 16.6 |
| 14/S1 | 37 | 27.0 |
| 21/S1 | 27 | 40.7 |
| 7/S2 | 47 | 17.0 |
| 14/S2 | 43 | 18.6 |
| 21/S2 | 37 | 24.3 |
S1, moderate-salinity site, 12 to 18.7 ppt; S2, high-salinity site, 28 to 32.5 ppt.
The V. parahaemolyticus multiplex PCR assay was used to detect the presence of tdh+ and/or trh+ genes on PCR-confirmed V. parahaemolyticus isolated from oyster samples during the relaying trials. Out of 278 tested isolates, only two of them were tdh+ (0.72%), and six isolates were trh+ (2.2%) (Table 2). Similar to V. vulnificus, we observed no difference between the frequencies of detection of the pathogenicity markers of V. parahaemolyticus isolates at relaying sites of differing salinity.
TABLE 2.
tdh- and trh-positive V. parahaemolyticus isolated during the high- and moderate-salinity relaying process
| Day/sitea | No. of isolates | % tdh+ | % trh+ |
|---|---|---|---|
| 0 | 37 | 0 | 8.1 |
| 7/S1 | 34 | 0 | 5.9 |
| 14/S1 | 35 | 2.9 | 0 |
| 21/S1 | 36 | 0 | 0 |
| 7/S2 | 47 | 0 | 2.1 |
| 14/S2 | 43 | 0 | 0 |
| 21/S2 | 46 | 2.2 | 0 |
S1, moderate-salinity site, 12 to 18.7 ppt; S2, high-salinity site, 28 to 32.5 ppt.
Identification of the O serogroups of V. parahaemolyticus isolates.
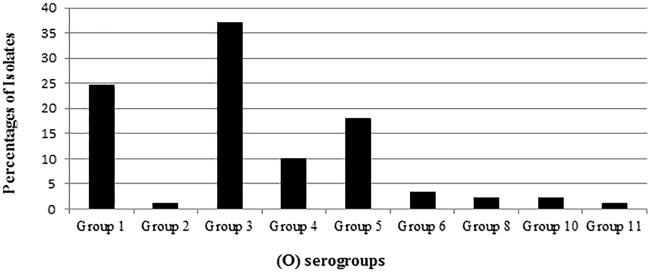
Among the 92 V. parahaemolyticus isolates obtained from the oysters during the relaying study, 3 were untypeable. The most predominant O serogroups among the 89 typeable V. parahaemolyticus isolates were 3 and 1, as they represent 37% and 25% of the isolates, respectively, while none of the isolates belonged to serogroups 7, 9, 12, and 13 (Fig. 1).
FIG 1.
Somatic (O) serogroups of Vibrio parahaemolyticus isolated during an oyster relay study.
Prevalence of antimicrobial resistance in V. vulnificus.
All tested V. vulnificus isolates were uniformly susceptible to four of the 20 tested antibiotics, two of which were recommended by the CDC for treating Vibrio infections (ciprofloxacin, levofloxacin, piperacillin, and piperacillin-tazobactam) (Table 3). More than 77% of V. vulnificus isolates were resistant to at least one antimicrobial, and more than 48% were resistant to two or more antimicrobials (see Appendix SA-1 in the supplemental material). Ten percent of tested isolates were found to be completely susceptible to all antimicrobials tested.
TABLE 3.
Antimicrobial resistance patterns among V. vulnificus strains isolated during the oyster relaying process
| Antibiotic | % with profilea: |
||
|---|---|---|---|
| S | I | R | |
| Amikacinb | 88 | 12 | 0 |
| Amoxicillin-clavulanic acid | 66 | 9 | 26 |
| Ampicillin | 72 | 2 | 26 |
| Ampicillin-sulbactam | 93 | 3 | 3 |
| Cefepime | 93 | 5 | 2 |
| Cefotaximeb | 91 | 2 | 7 |
| Cefoxitin | 36 | 47 | 17 |
| Ceftazidimeb | 72 | 16 | 12 |
| Ceftriaxoneb | 78 | 10 | 12 |
| Cephalothin | 31 | 2 | 67 |
| Chloramphenicol | 86 | 12 | 2 |
| Ciprofloxacinb | 100 | 0 | 0 |
| Doxycyclineb | 71 | 9 | 21 |
| Imipenem | 76 | 9 | 16 |
| Levofloxacinb | 100 | 0 | 0 |
| Meropenem | 95 | 5 | 0 |
| Piperacillin | 100 | 0 | 0 |
| Piperacillin-tazobactam | 100 | 0 | 0 |
| Tetracyclineb | 64 | 7 | 29 |
| Trimethoprim-sulfamethoxazoleb | 98 | 0 | 2 |
S, susceptible; I, intermediate; R, resistant.
CDC-recommended antibiotic for Vibrio infections.
Overall, the most common resistances were those to cephalothin (67%), tetracycline (29%), amoxicillin-clavulanic acid, and ampicillin (26%). With regard to the recommended antibiotics for the treatment of V. vulnificus infections, V. vulnificus tested isolates exhibited resistance against ceftriaxone (12%), ceftazidime (12%), and cefotaxime (7%), a third-generation cephalosporin. Within the tetracycline class of antibiotics, isolates exhibited high resistance to tetracycline (29%) and doxycycline (21%). Also, 2% resistance was observed against trimethoprim-sulfamethoxazole. Intermediate resistance was expressed against ceftazidime (16%), amikacin (12%), ceftriaxone (10%), doxycycline, and tetracycline (7%). Quinolones (ciprofloxacin and levofloxacin) were the only recommended class of drug to which all V. vulnificus isolates were completely susceptible. V. vulnificus displayed the highest percentage of intermediate resistance (47%) to cefoxitin (Table 3).
Of the 11 isolates positive for vcgC, 8 isolates displayed resistance to one or more of the tested antibiotics, and 7 isolates expressed resistance to more than 4 tested antibiotics (Appendix SA-1).
Prevalence of antimicrobial resistance in V. parahaemolyticus.
All tested isolates were susceptible to five of the 20 tested antibiotics, including four of those recommended by the CDC for treating Vibrio infections (ciprofloxacin, levofloxacin, piperacillin-tazobactam, and trimethoprim-sulfamethoxazole) (Table 4). Only about 8% of the isolates showed resistance to two or more tested antibiotics (Appendix SA-2).
TABLE 4.
Antimicrobial resistance patterns among Vibrio parahaemolyticus strains isolated during the oyster relaying process
| Antibiotic | % with profilea: |
||
|---|---|---|---|
| S | I | R | |
| Amikacinb | 96 | 3 | 1 |
| Amoxicillin-clavulanic acid | 99 | 0 | 1 |
| Ampicillin | 91 | 5 | 3 |
| Ampicillin-sulbactam | 100 | 0 | 0 |
| Cefepime | 95 | 4 | 1 |
| Cefotaximeb | 97 | 0 | 3 |
| Cefoxitin | 92 | 25 | 13 |
| Ceftazidimeb | 87 | 4 | 7 |
| Ceftriaxoneb | 90 | 4 | 5 |
| Cephalothin | 41 | 48 | 11 |
| Chloramphenicol | 99 | 1 | 0 |
| Ciprofloxacinb | 100 | 0 | 0 |
| Doxycyclineb | 98 | 1 | 1 |
| Imipenem | 97 | 3 | 0 |
| Levofloxacinb | 100 | 0 | 0 |
| Meropenem | 98 | 2 | 0 |
| Piperacillin | 98 | 1 | 1 |
| Piperacillin-tazobactam | 100 | 0 | 0 |
| Tetracyclineb | 99 | 0 | 1 |
| Trimethoprim-sulfamethoxazoleb | 100 | 0 | 0 |
S, susceptible; I, intermediate; R, resistant.
CDC-recommended antibiotic for Vibrio infections.
Overall, all V. parahaemolyticus isolates were susceptible or expressed low resistance and intermediate resistance to CDC-recommended antibiotics for Vibrio treatment; the exception was ceftazidime, as 9% of the isolates displayed resistance to this antibiotic. However, even the low percentage of resistance displayed by V. parahaemolyticus isolates to other cephalosporin antibiotics (3% for cefotaxime and 5% for ceftriaxone) might be cause for concern, as the cephalosporins are considered to be one of the best defenses against severe Vibrio infections (29). The highest resistance (13%) in V. parahaemolyticus isolates was for cefoxitin, followed by cephalothin (11%), ceftazidime (5%), and cefotaxime (3%), while 48% of the isolates were characterized by intermediate resistance to cephalothin, followed by cefoxitin (25%) (Table 4).
Among the tdh+ and/or trh+ V. parahaemolyticus isolates, the tdh+ isolates expressed no resistance to any tested antibiotics and intermediate resistance to cephalothin. Each of the trh+ isolates was resistant to one of the tested antibiotics (ampicillin and piperacillin) and expressed intermediate resistance to more than one antibiotic (Appendix SA-2). In general, none of the four pathogenic isolates showed multiple resistance, and all of them showed intermediate resistance to cephalothin.
Comparison between V. vulnificus and V. parahaemolyticus antimicrobial resistance and intermediate resistance.
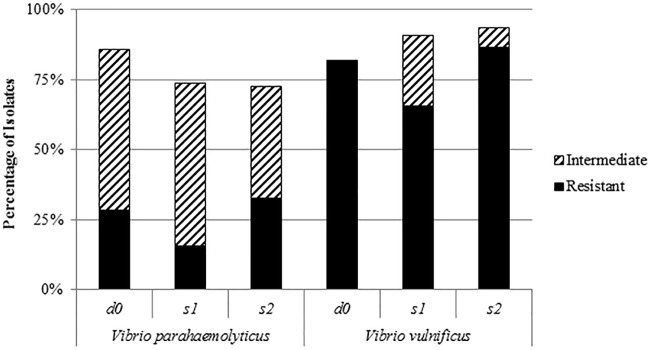
V. vulnificus isolates in this study possessed a higher resistance to all tested antibiotics, including those recommended by the CDC for Vibrio infections (Fig. 2), than did V. parahaemolyticus isolates (Fig. 3). Also, V. vulnificus isolates expressed higher intermediate resistance to all tested antibiotics except cephalothin (Fig. 2). More than 48% of V. vulnificus isolates were resistant to two or more antimicrobials, while only 18% of them were susceptible to all tested antibiotics. On the other hand, only about 8% of V. parahaemolyticus isolates showed resistance to two or more tested antibiotics, and approximately 54% of them were susceptible to all tested antibiotics.
FIG 2.
Percentages of antibiotic resistance and intermediate resistance of Vibrio vulnificus and Vibrio parahaemolyticus isolated from different sites. d0, harvesting day; s1, moderate-salinity site; s2, high-salinity site.
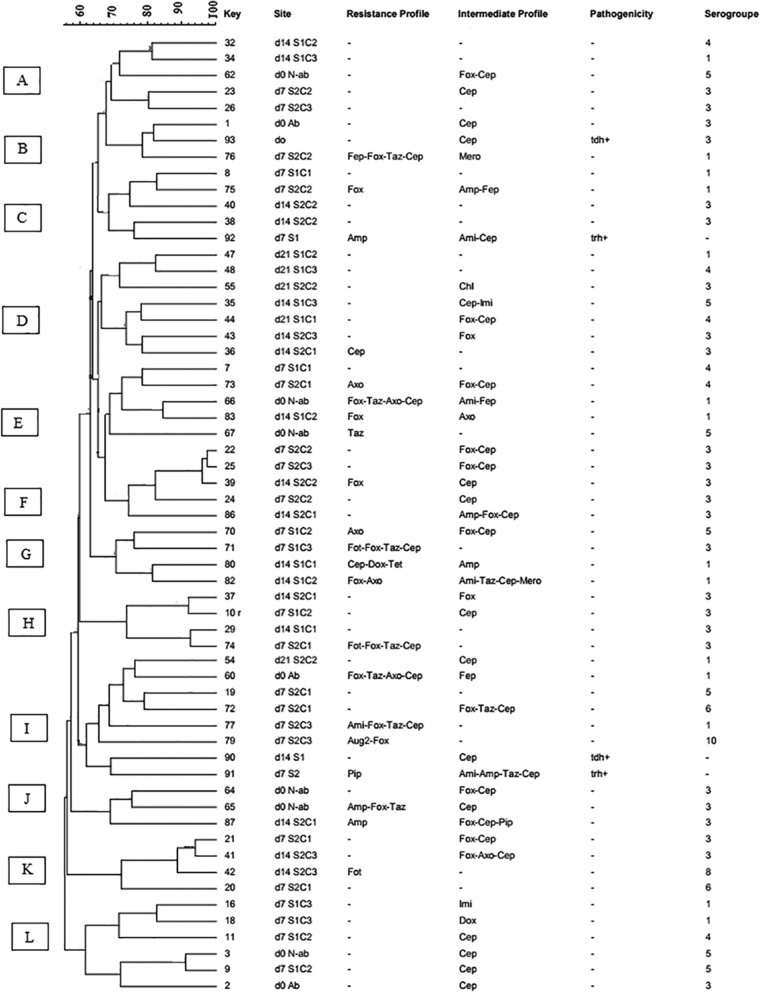
FIG 3.
Dendrogram of PFGE profiles of Vibrio parahaemolyticus isolates. d0, d7, d14, and d21, harvesting days 0, 7, 14, and 21, respectively; S1, moderate-salinity site; S2, high-salinity site; C, oyster composite; tdh, thermostable direct hemolysin; trh, TDH-related hemolysis; Ami, amikacin; Aug2, amoxicillin-clavulanic acid; Amp, ampicillin; Fep, cefepime; Fot, cefotaxime; Fox, cefoxitin; Taz, ceftazidime; Axo, ceftriaxone; Cep, cephalothin; Chl, chloramphenicol; Mero, meropenem; Pip, piperacillin. Minus signs in the resistance profile column indicate susceptibility to all tested antibiotics or untypeable, and those in the pathogenicity column indicate negativity for tdh or trh. Letters A to L on the left represent pulsed-field gel electrophoresis clusters; −. untypeable; Ab, temperature-abused oyster; N-ab, non-abused oyster.
The results of this study showed that there were no significance differences between the antimicrobial susceptibility profiles of V. vulnificus (P = 0.26) and V. parahaemolyticus (P = 0.23) isolated from the oysters before the relaying process (day 0) and after relaying to the two relaying sites, site 1 (moderate salinity) and site 2 (high salinity) (Fig. 4).
FIG 4.
Dendrogram of PFGE profiles of Vibrio vulnificus (vv) isolates. d0, d7, d14, and d21, harvesting days 0, 7, 14, and 21, respectively; S1, moderate-salinity site; S2, high-salinity site; C, oyster composite; vcgC, virulence-correlating gene type C; Ami, amikacin; Aug2, amoxicillin-clavulanic acid; Amp, ampicillin; A/S2, ampicillin-sulbactam; Fep, cefepime; Fot, cefotaxime; Fox, cefoxitin; Taz, ceftazidime; Axo, ceftriaxone; Cep, cephalothin; Chl, chloramphenicol; Dox, doxycycline; Imi, imipenem; Mero, meropenem; Pip, piperacillin; Tet, tetracycline. Minus signs in the resistance profile column indicate susceptibility to all tested antibiotics, and those in the pathogenicity column indicate negativity for vcgC. Letters A to K on the left represent pulsed-field gel electrophoresis clusters. Ab, temperature-abused oyster; N-ab, non-abused oyster.
Characterization of V. parahaemolyticus and V. vulnificus isolated from oysters using pulsed-field gel electrophoresis. (i) PFGE analysis of V. parahaemolyticus isolates.
In this study, PFGE was performed using a restriction enzyme (SfiI) to study the genetic relatedness between V. parahaemolyticus isolates, selected based on their antimicrobial susceptibility profiles and location of isolation during the relaying trials from the harvesting and the two relaying sites (moderate- and high-salinity sites). A total of 58 PFGE banding patterns were generated from 64 V. parahaemolyticus strains isolated from high- and moderate-salinity relaying trials. The dendrogram revealed 12 clusters (A to L) with 60% pattern similarity (Fig. 3), indicating extensive genetic diversity among the isolates, even between those obtained from the same site, same relaying interval day, and with similar antibiotic susceptibility profiles. However, a few tested isolates showed a tendency to cluster based on their antimicrobial susceptibility profiles and the site of isolation. For example, clusters A (5 isolates), D (7 isolates), K (6 isolates), and F (5 isolates) showed mostly no resistance to any of the tested antibiotics and intermediate resistance to cephalothin. Clusters E (5 isolates) and G (4 isolates) showed resistance or intermediate resistance to one or more tested antibiotics. Overall, the PFGE profiles of V. parahaemolyticus were genetically diverse and had no relationship according to the sampling site (relaying process), antimicrobial susceptibility test results, or pathogenicity.
(ii) PFGE analysis of V. vulnificus isolates.
A dendrogram was constructed based on SfiI PFGE patterns (Fig. 4) to compare the banding profiles of V. vulnificus isolates with different antimicrobial susceptibility and pathogenicity profiles isolated from oysters during the moderate- and high-salinity relaying process. Out of the 58 isolates of V. vulnificus examined in this study, 47 isolates were successfully characterized by the PFGE, and approximately 19% of the isolates were not typeable and failed to yield discernible PFGE patterns. A total of 45 PFGE banding patterns were generated from 47 V. vulnificus isolates. These generated patterns were grouped into 11 clusters (A to K) with about 60% similarity (Fig. 4). V. vulnificus strains isolated from different sites with different antimicrobial susceptibility and pathogenicity profiles were distributed among all identified clusters. Similar to V. parahaemolyticus, the PFGE profiles of V. vulnificus were genetically diverse, and no genetic relationship was observed between the PFGE profile of this pathogen and the salinity relaying process.
DISCUSSION
In this study, we examined the presence of the vcgC gene in V. vulnificus as well as the presence of tdh and/or trh genes in V. parahaemolyticus isolated from oysters. Among all tested V. vulnificus isolates, 20.9% were vcgC+, and no significant differences in the prevalences of the virulence genes were observed at the relaying sites of differing salinity. Our results were comparable to the findings of Warner and Oliver (19), who found that 15.6% of the V. vulnificus isolates recovered from oyster samples from the eastern coast of North Carolina were vcgC+. We observed that only 0.72% of the V. parahaemolyticus isolates were tdh+, and only 2.2% were trh+. This is consistent with findings of DePaola et al. (30), who reported that tdh+ and/or trh+ isolates typically represent <1% of environmental V. parahaemolyticus strains, except in the Pacific Northwest. The prevalence of these strains may vary by sample type and detection method, as well as by location (31–33). Whitaker et al. (28) studied the effect of salinity on V. parahaemolyticus pathogenicity using the cytotoxicity assay, and they found that V. parahaemolyticus grown in 1% NaCl was significantly more cytotoxic than that grown in 3% NaCl. However, in this study, no clear differences in the prevalences of V. parahaemolyticus virulence genes at relaying sites of different salinity were observed; also, similar to our study, Johnson et al. (34) found that salinity was not a significant predictor of tdh density in oyster samples, but it was a significant predictor of the trh density.
In this study, V. parahaemolyticus isolates belonging to different O serogroups were frequently found to be isolated from the same site (Fig. 3), indicating the high degree of divergence demonstrated by this pathogen (7, 35). Most of the V. parahaemolyticus isolates in this study (37%) were found to belong to the O3 serogroup. Consequently, given the increase of gastroenteritis outbreaks associated with serotype O3:K6 globally (36–39), the capsular (K) antigens of these isolates should be tested to clearly investigate the abundance of this serotype among the V. parahaemolyticus strains isolated from Chesapeake Bay region.
Most of the V. vulnificus isolates recovered during the relaying process were resistant to at least one antimicrobial, and almost 50% of them were resistant to two or more antimicrobials. Ten percent of the tested isolates were found to be completely susceptible to all antimicrobials tested. This result is comparable with that of a recent study conducted to investigate the antimicrobial susceptibility of V. vulnificus isolates recovered from recreational and commercial water of Chesapeake Bay. They reported that only 12.5% of the V. vulnificus isolates were susceptible to all tested antimicrobials (40). Similar to our findings, 45% of the environmental V. vulnificus isolates from a study in South Carolina and Georgia estuaries were resistant to three or more classes of antibiotic agents, including those that are usually prescribed for V. vulnificus infections (41).
The CDC-recommended antibiotics for treating V. vulnificus infections are doxycycline, cephalosporins (e.g., ceftazidime), fluoroquinolones (such as levofloxacin, ciprofloxacin, or gatifloxacin), and trimethoprim-sulfamethoxazole plus an aminoglycoside (29). Tetracycline or ciprofloxacin can be used only in cases of severe or prolonged Vibrio infections (42). In this study, V. vulnificus tested isolates exhibited varied degrees of resistance and intermediate resistance against most of these antimicrobial agents, while the quinolones (ciprofloxacin and levofloxacin) were the only recommended class of drug to which all V. vulnificus isolates were completely susceptible. Compatible with our findings, Wong et al. (43) recently studied the in vivo efficacy of antibiotics for lethal Vibrio species, and they found that treatment of V. vulnificus infection that includes a quinolone is associated with lower mortality. Shaw et al. (40) also reported that all tested V. vulnificus isolates were susceptible to quinolones. However, in contrast to our findings, all of their tested isolates were susceptible to tetracycline, and only 2% and 1% of their tested isolates exhibited intermediate resistance against ceftazidime and amikacin, respectively.
A large-scale study conducted to investigate the antimicrobial susceptibilities of both V. parahaemolyticus and V. vulnificus in Louisiana Gulf and retail oysters showed that V. vulnificus isolates were susceptible to the majority of tested antibiotics (20). However, the results of their study agreed with the present study and confirmed that fluoroquinolones were highly effective against V. vulnificus infection.
In our study, all V. parahaemolyticus isolates were susceptible or expressed low or intermediate resistance to the antibiotics (except for ceftazidime) recommended by the CDC for Vibrio infection treatment. In addition, approximately 8% of V. parahaemolyticus isolates showed resistances to two or more tested antibiotics, and approximately 54% were susceptible to all tested antibiotics. Shaw et al. (40) and Han et al. (20) reported for Chesapeake Bay and Gulf Coast oysters, respectively, a higher level of resistance among V. parahaemolyticus than among V. vulnificus isolates.
Our study showed no significant differences in the antimicrobial susceptibility profiles of V. vulnificus and V. parahaemolyticus isolated from the oysters before and during the relaying in both sites. This suggests that the salinity of the relaying sites did not impact the antimicrobial susceptibility profiles of the Vibrio isolates, although it reduced the overall concentration of these bacteria in oysters (26).
Molecular typing of V. parahaemolyticus has been shown to be a useful tool for tracking the source of infection and detection of virulent strains, as well as for determining the geographical and host distribution of possible variants (44). Studies focusing on the interspecies variability and genetic relationships among the environmental isolates are rare and limited to specific geographic areas (45). In this study, PFGE was conducted to determine the genetic relatedness between V. parahaemolyticus isolates based on their antimicrobial susceptibility profiles and location (moderate- and high-salinity sites) during the relaying trials. The PFGE profiles of V. parahaemolyticus were genetically diverse, and no genetic relationship was noted among the sampling sites, antimicrobial susceptibility test results, pathogenicity, and O serogroups. A similar trend was also observed for the PFGE profiles of V. vulnificus isolates. Our results are similar to the results of a study conducted by Lewis et al. (25) in which V. vulnificus isolates recovered pre- and postdepuration were analyzed using the amplified fragment length polymorphism (AFLP) technique. All recovered isolates clustered at 40% or higher similarity, which indicates the high intraspecific diversity of this species. Previous investigators (4, 46) also found that the PFGE analysis demonstrated high heterogeneity among the V. parahaemolyticus isolated from seafood.
Conclusion.
This study provides baseline data on the phenotypic and genotypic characteristics of Vibrio spp. in oysters and the effect of relaying to high-salinity sites on these characteristics. Among the PCR-confirmed V. vulnificus and V. parahaemolyticus isolates recovered from oyster samples, 20.9% of V. vulnificus isolates were positive for the vcgC gene, while among V. parahaemolyticus isolates, only 0.7% and 2.2% were positive for the tdh and trh genes, respectively, and none of the tested isolates were positive for both genes. Analysis of the antibiotic resistance profiles of V. vulnificus and V. parahaemolyticus isolated from oysters indicated that V. vulnificus isolates possessed a higher rate of resistance or intermediate resistance, as well as a higher rate of multiple resistance to almost all tested antibiotics, including those recommended by the CDC for treating Vibrio infections. Also, V. parahaemolyticus showed high MICs for some of the Vibrio infection treatment antibiotics. No significant effect of the relaying process was observed on the antimicrobial resistance profiles or the presence of virulence genes of either V. vulnificus or V. parahaemolyticus isolates. Molecular characterization of the selected V. vulnificus and V. parahaemolyticus isolates showed a high genetic diversity, even among the isolates obtained from the same site and collection day.
MATERIALS AND METHODS
This study was conducted as part of an effort to examine the influence of high-salinity relay on Vibrio abundance in Chesapeake Bay and Maryland Coastal Bay oysters (Crassostrea virginica). Oysters were purchased from a commercial aquaculture facility (the salinity of the original harvest site was 10.3 to 16.3 ppt), and the harvested oysters were transported to the relaying sites (Pocomoke Sound, MD [moderate salinity, 12 to 18.7 ppt], and Chincoteague Bay, MD [high salinity, 28 to 32.5 ppt]). One hundred twenty oysters were placed in 2 ft by 3 ft oyster cages (3 cages at each site) and placed overboard. At each selected time interval (days 0, 7, 14, and 21), 3 composites were collected from each relaying site. Collected oysters were analyzed for presumptive V. parahaemolyticus and V. vulnificus, and abundances were determined using the alkaline peptone water (APW) 3-tube MPN series (26).
Isolation of V. vulnificus and V. parahaemolyticus from oyster samples.
To isolate V. parahaemolyticus colonies, from the top 1 cm of turbid (positive) APW tubes, a 3-mm loopful was streaked onto thiosulfate-citrate-bile-salt-sucrose (TCBS) agar, and for V. vulnificus isolation, turbid APW tubes were streaked on modified cellobiose polymyxin B-colistin (mCPC) agar. The inoculated plates were incubated at 35°C for TCBS and at 39°C for mCPC. Three to five positive colonies were picked from each MPN dilution (10−1 to 10−6) and frozen for further examination (26).
Approximately, a total of 400 colonies of each V. parahaemolyticus and V. vulnificus were isolated during the 3 relaying trials. In this study, we evaluated the pathogenicity, antimicrobial susceptibility, and genetic profiles of all retrieved viable V. parahaemolyticus and V. vulnificus colonies, as some of the isolates lost their viability during storage.
PCR assay for the virulence genes.
A total of 296 V. parahaemolyticus and 94 V. vulnificus PCR-confirmed samples (74% and 24%, respectively) of the total samples isolated during relaying trials (interval days, 0, 7, 14, and 21) (26) were subjected to further testing for the presence of pathogenic (tdh+ and/or trh+) genes using a real-time PCR V. parahaemolyticus multiplex assay (47). Real-time PCR was also used for the detection of virulence-correlated gene type C (vcgC) in V. vulnificus (41). Twenty-three microliters of a master mix consisting of 1× PCR amplification buffer (Invitrogen, Carlsbad, CA), 5.0 mM MgCl2 (Invitrogen), 400 nM each of the deoxynucleoside triphosphates (Roche, Indianapolis, IN), 200 nM (each) the trh and tdh forward and reverse primers (Integrated DNA Technologies, Coralville, IA), 75 nM (each) the tlh forward and reverse primers (Integrated DNA Technologies), 150 nM probe for tlh (Integrated DNA Technologies), 75 nM probe for tdh and trh (Applied Biosystems, Foster City, CA), and 2.25 U platinum Taq DNA polymerase (Invitrogen) was used. The remainder of the reaction mixture volume consisted of 2 μl of the template (boiled sample). Real-time PCR thermal cycling was conducted using a SmartCycler II system from Cepheid (Sunnyvale, CA). For the vcgC multiplex assay, the assay solution was composed of 20 μl of master mix and 5 μl of template. The following analysis settings were adjusted: “manual threshold fluorescence units” to 15 for the 6-carboxyfluorescein (FAM [tdh]), TET (trh), TxRed (tlh, total V. parahaemolyticus), and Cy5 (internal control) channels (26). Positive (for each targeted gene) and negative (PCR water) controls were included for each run.
Antimicrobial susceptibility test.
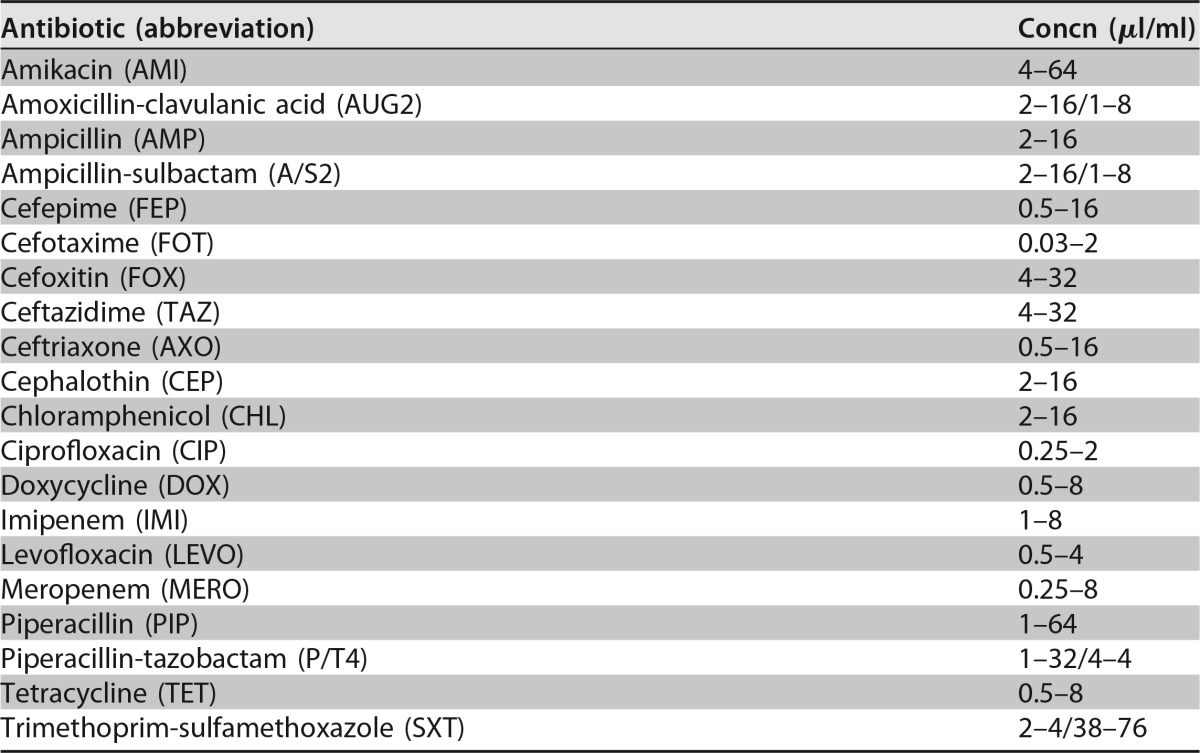
A total of 150 (38% of the collected colonies) V. parahaemolyticus (n = 92 [2 tdh+ and 2 trh+]) and V. vulnificus (n = 58 [9 vcgC+]) isolates were collected from oysters during the moderate- and high-salinity relaying trials (26). All isolates were tested for their susceptibility to 20 different antibiotics, chosen by clinical usage for these pathogens and aquaculture practices (Table 5), using the Sensititre microbroth dilution method, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (48). The MIC was determined as the lowest concentration of an antimicrobial that completely inhibited the growth of bacteria. Multidrug resistance was defined as an absence of susceptibility to two or more classes of antibiotics (40).
TABLE 5.
Concentration ranges of selected antibiotics tested for V. vulnificus and V. parahaemolyticus according to CLSI guidelines
Identification of the somatic (O) serotypes of V. parahaemolyticus isolates.
To determine the predominant O serotypes of the 92 viable and confirmed V. parahaemolyticus isolates recovered from oysters, the slide agglutination test was conducted using commercially available antisera that included 13 different O antigens, as described by the manufacturer (Denka Seiken Co., Ltd.).
PFGE.
To determine the effect of the relaying process on the genetic profile of the Vibrio isolates, V. parahaemolyticus (n = 64 [2 tdh+ and 2 trh+]) and V. vulnificus (n = 58 [9 vcgC+]) isolates with different antibiotic susceptibility profiles were selected. Isolates were inoculated onto tryptic soy agar (TSA) plus 1% NaCl and incubated overnight at 35°C. Plug preparation and PFGE were performed using the SfiI restriction enzyme, as described in the CDC PulseNet protocol for V. parahaemolyticus and V. vulnificus (49, 50). The gel was stained with ethidium bromide, and DNA bands were visualized with a UV light.
Statistical analysis.
Descriptive and inferential statistics were used to compare the frequencies of isolates testing positive for virulence genes and exhibiting intermediate resistance or resistance to antibiotics by site and/or day of collection during the relaying study. A Kruskal-Wallis one-way analysis of variance (ANOVA) was used to assess the significance of observed differences in the frequencies of antibiotic resistance of V. vulnificus and V. parahaemolyticus isolates collected from relaying sites with different salinities.
PFGE fingerprints were analyzed with the BioNumerics software 7.0 (Applied Maths) using standard band calling methods. Dendrograms were constructed using the Dice correlation, with 1.5% optimization and 1.5% tolerance, and the unweighted pair group method using average linkages (UPGMA) (50). Identification of clusters within a dendrogram was based on a 76% similarity. BioNumerics software version 7.0 (Applied Maths, Belgium) was used to analyze the DNA bands. Clustering was performed using UPGMA and the Dice correlation coefficient, with a position tolerance of 1.5%.
Supplementary Material
ACKNOWLEDGMENTS
We sincerely thank Kathy Brohawn and her group (Maryland Department of the Environment) for providing the oyster samples from the relaying sites. We thank the United States Department of Agriculture (USDA) for CBG award no. 2010-02370, USDA CBG award no. 2014-38821-22430, and the United States Department of Education Title III for funding.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01790-17.
REFERENCES
- 1.Centers for Disease Control and Prevention (CDC). 2016. National enteric disease surveillance: COVIS annual summary, 2014. Summary of human Vibrio cases reported to CDC, 2014. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/nationalsurveillance/pdfs/covis-annual-summary-2014-508c.pdf.
- 2.Mead PS, Slutsker L, Dietz V, McGai LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Emerg Infect Diseases 5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliver JD. 2006. Vibrio vulnificus, p 253–276. In Belkin S, Colwell RR (ed), Oceans and health: pathogens in the marine environment. Springer, New York, NY. [Google Scholar]
- 4.Liu F, Guan W, Alam MJ, Shen Z, Zhang S, Li L, Shinoda S, Shi L. 2009. Pulsed-field gel electrophoresis typing of multidrug-resistant Vibrio parahaemolyticus isolated from various sources of seafood. J Health Sci 55:783–789. doi: 10.1248/jhs.55.783. [DOI] [Google Scholar]
- 5.Newton A, Kendall M, Vugia DJ, Henao OL, Mahon BE. 2012. Increasing rates of vibriosis in the United States, 1996–2010: review of surveillance data from 2 systems. Clin Infect Dis 54(Suppl 5):S391–S395. doi: 10.1093/cid/cis243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States–major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alam M, Chowdhury WB, Bhuiyan NA, Islam A, Hasan NA, Nair GB, Watanabe H, Siddique AK, Hug A, Sack RB, Akhter MZ, Grim CJ, Kam KM, Luey CK, Endtz HP, Cravioto A, Colwell RR. 2009. Serogroup, virulence, and genetic traits of Vibrio parahaemolyticus in the estuarine ecosystem of Bangladesh. Appl Environ Microbiol 75:6268–6274. doi: 10.1128/AEM.00266-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishibashi M, Ohta K, Shimada T, Honda T, Sugiyama J, Miwatani Y. 2000. Current status of OK serotype combinations of Vibrio parahaemolyticus. Nippon Saikingaku Zasshi 55:539–541. (In Japanese.) doi: 10.3412/jsb.55.539. [DOI] [Google Scholar]
- 9.Wong HC, Lin CH. 2001. Evaluation of typing of Vibrio parahaemolyticus by three PCR methods using specific primers. J Clin Microbiol 39:4233–4240. doi: 10.1128/JCM.39.12.4233-4240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DePaola A, Kaysner CA, Bowers J, Cook DW. 2000. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl Environ Microbiol 66:4649–4654. doi: 10.1128/AEM.66.11.4649-4654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H. 2003. Analysis of the collective food poisoning events in Shanghai from 1990 to 2000. Chin J Nat Med 5:17–20. [Google Scholar]
- 12.Robert-Pillot A, Guenole A, Lesne J, Delesmont R, Fournier JM, Quilici ML. 2004. Occurrence of the tdh and trh genes in Vibrio parahaemolyticus isolates from waters and raw shellfish collected in two French coastal areas and from seafood imported into France. Int J Food Microbiol 91:319–325. doi: 10.1016/j.ijfoodmicro.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention (CDC). 2010. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. MMWR Morb Mortal Wkly Rep 59:418–422. [PubMed] [Google Scholar]
- 14.Klontz KC, Lieb S, Schreiber M, Janowski HT, Baldy LM, Gunn RA. 1988. Syndromes of Vibrio vulnificus infections. Clinical and epidemiologic features in Florida cases, 1981–1987. Ann Intern Med 109:318–323. [DOI] [PubMed] [Google Scholar]
- 15.Liu JW, Lee IK, Tang HJ, Ko WC, Lee HC, Liu YC, Hsueh PR, Chuang YC. 2006. Prognostic factors and antibiotics in Vibrio vulnificus septicemia. Arch Intern Med 166:2117–2123. doi: 10.1001/archinte.166.19.2117. [DOI] [PubMed] [Google Scholar]
- 16.Randa MA, Polz MF, Lim E. 2004. Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl Environ Microbiol 70:5469–5476. doi: 10.1128/AEM.70.9.5469-5476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reynaud Y, Pitchford S, de Decker S, Wikfors GH, Brown CL. 2013. Molecular typing of environmental and clinical strains of Vibrio vulnificus isolated in the northeastern USA. PLoS One 8:e83357. doi: 10.1371/journal.pone.0083357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokolova IM, Leamy L, Harrison M, Oliver JD. 2005. Intrapopulational variation in Vibrio vulnificus levels in Crassostrea virginica (Gmelin 1971) is associated with the host size but not with disease status or developmental stability. J Shellfish Res 24:502–508. [Google Scholar]
- 19.Warner EB, Oliver JD. 2008. Multiplex PCR assay for detection and simultaneous differentiation of genotypes of Vibrio vulnificus biotype 1. Foodborne Pathog Dis 5:691–693. doi: 10.1089/fpd.2008.0120. [DOI] [PubMed] [Google Scholar]
- 20.Han F, Walker RD, Janes ME, Prinyawiwatkul W, Ge B. 2007. Antimicrobial susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus isolates from Louisiana Gulf and retail raw oysters. Appl Environ Microbiol 73:7096–7098. doi: 10.1128/AEM.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosche TM, Yano Y, Oliver JD. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus, which correlate with clinical or environmental isolation. Microbiol Immunol 49:381–389. doi: 10.1111/j.1348-0421.2005.tb03731.x. [DOI] [PubMed] [Google Scholar]
- 22.Crim SM, Iwamoto M, Huang JY, Griffin PM, Gilliss D, Cronquist AB, Cartter M, Tobin-D'Angelo M, Blythe D, Smith K, Lathrop S, Zansky S, Cieslak PR, Dunn J, Holt KG, Lance S, Tauxe R, Henao OL. 2014. Incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006–2013. MMWR Morb Mortal Wkly Rep 63:328–332. [PMC free article] [PubMed] [Google Scholar]
- 23.Baquero F, Martinez JL, Canton R. 2008. Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19:260–265. doi: 10.1016/j.copbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 24.WHO. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1. [Google Scholar]
- 25.Lewis M, Rikard S, Arias CR. 2010. Evaluation of a flow-through depuration system to eliminate the human pathogen Vibrio vulnificus from oysters. J Aquac Res Dev 1:103. [Google Scholar]
- 26.Parveen S, Jahncke M, Elmahdi S, Crocker H, Bowers J, White C, Gray S, Brohawn K. 2017. High salinity relaying to reduce Vibrio parahaemolyticus and Vibrio vulnificus in Chesapeake Bay oysters. J Food Sci 82:484–491. doi: 10.1111/1750-3841.13584. [DOI] [PubMed] [Google Scholar]
- 27.Chase E, Harwood VJ. 2011. Comparison of the effects of environmental parameters on growth rates of Vibrio vulnificus biotypes I, II, and III by culture and quantitative PCR analysis. Appl Environ Microbiol 77:4200–4207. doi: 10.1128/AEM.00135-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitaker WB, Parent MA, Naughton LM, Richards GP, Blumerman SL, Boyd EF. 2010. Modulation of responses of Vibrio parahaemolyticus 03:K6 to pH and temperature stresses by growth at different salt concentrations. Appl Environ Microbiol 76:4720–4729. doi: 10.1128/AEM.00474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Centers for Disease Control and Prevention (CDC). 2017. Management of Vibrio vulnificus wound infections after a disaster. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/disasters/disease/vibriofaq.html. [Google Scholar]
- 30.DePaola A, Ulaszek J, Kaysner CA, Tenge BJ, Nodstrom JL, Wells J, Puhr N, Gendel SM. 2003. Molecular serological and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food and clinical sources in North America and Asia. Appl Environ Microbiol 69:3999–4005. doi: 10.1128/AEM.69.7.3999-4005.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alam MJ, Tomochika KI, Miyoshi SI, Shinoda S. 2002. Environmental investigation of potentially pathogenic Vibrio parahaemolyticus in the Seto-Inland Sea, Japan. FEMS Microbiol Lett 208:83–87. doi: 10.1111/j.1574-6968.2002.tb11064.x. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Urtaza J, Varela-Pet J, Trinans J, Pazos Y, Garcia-Martin O. 2008. Environmental determinants of the occurrence and distribution of Vibrio parahaemolyticus in the raise of Galicia, Spain. Appl Environ Microbiol 74:265–274. doi: 10.1128/AEM.01307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parveen S, Hettiarachchi AK, Bowers CJ, Jones JL, Tamplin ML, McKay R, Beatty W, Brohawn K, DaSilva LV, DePaola A. 2008. Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters. Int J Food Microbiol 128:354–361. doi: 10.1016/j.ijfoodmicro.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 34.Johnson C, Flowers A, Noriea N, Zimmerman A, Bowers J, DePaola A, Grimes DJ. 2010. Relationships between environmental factors and pathogenic vibrios in the northern Gulf of Mexico. Appl Environ Microbiol 76:7076–7084. doi: 10.1128/AEM.00697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, Rammamurthy T, Wong HC, DePaola A, Kim YB, Albert MJ, Nishibuchi M. 2000. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol 38:578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ansaruzzaman M, Lucas M, Deen JL, Bhuiyan NA, Wang XY, Safa A, Sultana M, Chowdhury A, Nair GB, Sack DA, von Seidlein L, Puri MK, Ali M, Chaignat CL, Clemens JD, Barreto A. 2005. Pandemic serovars (O3:K6 and O4:K68) of Vibrio parahaemolyticus associated with diarrhea in Mozambique: spread of the pandemic into the African continent. J Clin Microbiol 43:2559–2562. doi: 10.1128/JCM.43.6.2559-2562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniels N, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond R, Thompson S, Wilson S, Bean N, Griffin P, Slutsker L. 2000. Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis 181:1661–1666. doi: 10.1086/315459. [DOI] [PubMed] [Google Scholar]
- 38.de Jesús Hernández-Díaz L, León-Sicairos N, Velazquez-Roman J, Flores-Villasenor H, Guadron-Llanos M, Martinez-Garcia JJ. 2015. A pandemic Vibrio parahaemolyticus O3:K6 clone causing most associated diarrhea cases in the Pacific Northwest coast of Mexico. Front Microbiol 6:221. doi: 10.3389/fmicb.2015.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Urtaza J, Simental L, Velasco D, DePaola A, Ishibashi M, Nakaguchi Y, Nishibuchi M, Carrera-Flores D, Rey-Alvarez C, Pousa A. 2005. Pandemic Vibrio parahaemolyticus O3:K6, Europe. Emerg Infect Dis 11:1319–1320. doi: 10.3201/eid1108.050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw KS, Rosenberg Goldstein RE, He X, Jacobs JM, Crump BC, Sapkota AR. 2014. Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Chesapeake Bay and Maryland coastal bays. PLoS One 9:e89616. doi: 10.1371/journal.pone.0089616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker-Austin C, McArthur JV, Lindell AH, Wright MS, Tuckfield RC, Gooch J, Warner L, Oliver J, Stepanauskas R. 2009. Multi-site analysis reveals widespread antibiotic resistance in the marine pathogen Vibrio vulnificus. Microb Ecol 57:151–159. doi: 10.1007/s00248-008-9413-8. [DOI] [PubMed] [Google Scholar]
- 42.Centers for Disease Control and Prevention (CDC). 2013. Vibrio species causing vibriosis. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/vibrio/index.html. [Google Scholar]
- 43.Wong KC, Brown AM, Luscombe GM, Wong SJ, Mendis K. 2015. Antibiotic use for Vibrio infections: important insights from surveillance data. BMC Infect Dis 15:226. doi: 10.1186/s12879-015-0959-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marshall S, Clark CG, Wang G, Mulvey M, Kelly MT, Johnson WM. 1999. Comparison of molecular methods for typing Vibrio parahaemolyticus. J Clin Microbiol 37:2473–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellingsen AB, Jorgensen H, Wagley S, Monshaugen M, Rorvik LM. 2008. Genetic diversity among Norwegian Vibrio parahaemolyticus. J Appl Microbiol 105:2195–2202. doi: 10.1111/j.1365-2672.2008.03964.x. [DOI] [PubMed] [Google Scholar]
- 46.Rojas MV, Matte MH, Droba M, Silva ML, Matte GR. 2011. Characterization of Vibrio parahaemolyticus isolated from oysters and mussels in São Paulo, Brazil. Rev Inst Med Trop São Paulo 53:201–205. [DOI] [PubMed] [Google Scholar]
- 47.Nordstrom JL, Vickery MCL, Blackstone GM, Murray SL, DePaola A. 2007. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Appl Environ Microbiol 73:5840–5847. doi: 10.1128/AEM.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clinical and Laboratory Standards Institute (CLSI). 2010. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline, 2nd ed CLSI document M45-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 49.Parsons MB, Cooper KL, Kubota KA, Puhr N, Simington S, Calimlim PS, Schoonmaker-Bopp D, Swaminathan B, Gerner-Smidt P, Ribot EM. 2007. PulseNet USA standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio parahaemolyticus. Foodborne Pathog Dis 4:285–292. doi: 10.1089/fpd.2007.0089. [DOI] [PubMed] [Google Scholar]
- 50.Kam KM, Luey CY, Parsons MB, Cooper KLF, Nair GB, Alam M, Islam MA, Cheung DTL, Chu YW, Ramamurthy T, Pazhani GP, Bhattacharya SK, Watanabe H, Terajima J, Arakawa E, Ratchtrachenchai OA, Huttayananont S, Ribot EM, GernerSmidt P, Swaminathan B, Vibrio parahaemolyticus PulseNet PFGE Protocol Working Group . 2008. Evaluation and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping Vibrio parahaemolyticus: an international multicenter collaborative study. J Clin Microbiol 46:2766–2773. doi: 10.1128/JCM.00424-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.