ABSTRACT
Nitrogen-fixing (N2) cyanobacteria provide bioavailable nitrogen to vast ocean regions but are in turn limited by iron (Fe) and/or phosphorus (P), which may force them to employ alternative nitrogen acquisition strategies. The adaptive responses of nitrogen fixers to global-change drivers under nutrient-limited conditions could profoundly alter the current ocean nitrogen and carbon cycles. Here, we show that the globally important N2 fixer Trichodesmium fundamentally shifts nitrogen metabolism toward organic-nitrogen scavenging following long-term high-CO2 adaptation under iron and/or phosphorus (co)limitation. Global shifts in transcripts and proteins under high-CO2/Fe-limited and/or P-limited conditions include decreases in the N2-fixing nitrogenase enzyme, coupled with major increases in enzymes that oxidize trimethylamine (TMA). TMA is an abundant, biogeochemically important organic nitrogen compound that supports rapid Trichodesmium growth while inhibiting N2 fixation. In a future high-CO2 ocean, this whole-cell energetic reallocation toward organic nitrogen scavenging and away from N2 fixation may reduce new-nitrogen inputs by Trichodesmium while simultaneously depleting the scarce fixed-nitrogen supplies of nitrogen-limited open-ocean ecosystems.
IMPORTANCE Trichodesmium is among the most biogeochemically significant microorganisms in the ocean, since it supplies up to 50% of the new nitrogen supporting open-ocean food webs. We used Trichodesmium cultures adapted to high-CO2 conditions for 7 years, followed by additional exposure to iron and/or phosphorus (co)limitation. We show that “future ocean” conditions of high CO2 and concurrent nutrient limitation(s) fundamentally shift nitrogen metabolism away from nitrogen fixation and instead toward upregulation of organic nitrogen-scavenging pathways. We show that the responses of Trichodesmium to projected future ocean conditions include decreases in the nitrogen-fixing nitrogenase enzymes coupled with major increases in enzymes that oxidize the abundant organic nitrogen source trimethylamine (TMA). Such a shift toward organic nitrogen uptake and away from nitrogen fixation may substantially reduce new-nitrogen inputs by Trichodesmium to the rest of the microbial community in the future high-CO2 ocean, with potential global implications for ocean carbon and nitrogen cycling.
KEYWORDS: nutrient colimitation, nitrogen fixation, Trichodesmium, global change, ocean acidification, microbial ecology, cyanobacteria, evolution, TMA, trimethylamine, high CO2, nutrient limitation, organic nitrogen
INTRODUCTION
Oceanic food webs and climate feedbacks are significantly influenced by carbon dioxide (CO2)- and nitrogen (N2)-fixing microbes, thereby contributing to both global productivity and biogeochemistry (1–3). However, only a few studies have investigated the physiological and evolutionary responses of the globally important, photoautotrophic, N2-fixing cyanobacterium Trichodesmium erythraeum strain IMS101 to high-CO2 conditions under multiple nutrient-limiting regimes (4–6). Simultaneous iron (Fe) and phosphorus (P) limitation of IMS101 under both low- and high-CO2 conditions has been found to yield higher growth rates, reduced cell sizes, and a unique Fe/P protein complement, compared to the limitation of either Fe or P alone (5, 7). This fitness advantage conferred by Fe/P “balanced limitation” contrasts with the long-standing Liebig limitation model (8) and has implications for global biogeochemical cycles in both the current and future oceans (3).
Oligotrophic populations are predicted to potentially experience longer periods of enhanced nutrient (co)limitation due to intensifying Fe stress under high-CO2 conditions (5, 9) and reduced vertical P supplies from increased density-driven stratification (3, 10). However, a major unknown is the potential change in future new-nitrogen inputs by globally distributed N2-fixing microbes (diazotrophs). Indeed, past research has demonstrated significant decreases in the growth and N2 fixation of marine diazotrophs under Fe limitation, with molecular evidence indicating iron reallocation via reduction in the photosystem I-to-photosystem II (PSI/PSII) ratio, decreases in metalloenzyme inventories, and increases in Fe stress PS antenna abundances (5, 11–13). Additionally, Trichodesmium has been demonstrated to take up both inorganic (e.g., nitrate and ammonia) and organic (e.g., amino acids) nitrogen species, thereby inhibiting N2 fixation (14, 15). However, because much of the low-latitude surface ocean is largely nitrogen limited (16), fixed N sources can be severely depleted, resulting in a marked dependence on Fe and P bioavailability to fuel N2 fixation (1, 3).
To date, nearly all diazotrophic nitrogen assimilation research has focused on the relationship between N2 fixation and nitrate, ammonia, and amino acid uptake, resulting in the view that ammonia is the preferred microbial nitrogen source for both diazotrophic and nondiazotrophic microbes (17). Identifying and measuring preferentially scavenged organic nitrogen species by marine microbes has been largely precluded by rapid biochemical turnover in situ. Furthermore, microbial consortia aggregating around microbial species of interest can also inhibit discernment of the microbial uptake of specific substrates (18).
In this study, functional genomics and molecular physiology provide strong evidence that following adaptation to high-CO2 conditions, Trichodesmium allocates the greatest biosynthetic investment to the acquisition of the organic nitrogen substrate trimethylamine (TMA) and potentially other organic nitrogen- and sulfur-containing compounds. This shift in nitrogen acquisition pathways under high-CO2 conditions is predicted to be mediated by large increases in bacterial flavin-containing monooxygenase (FMO) coupled with global shifts in transcription and translation. This indicates a fundamental change in both nitrogen and global cellular strategies whereby iron-rich biosynthetic pathways including N2 fixation and photosynthesis are significantly reduced under Fe-limited, high-CO2 conditions, in parallel with increased biosynthesis of the predicted TMA-oxidizing FMO enzyme. Additionally, N2 fixation is inhibited in the presence of exogenous TMA in a manner similar to that seen with nitrate and ammonia, with TMA simultaneously supporting growth rates equivalent to those seen during N2 fixation. Methylated amine compounds like TMA are products of protein putrefaction and degradation of quaternary amine osmoregulators (e.g., glycine betaine) (19) and are thus ubiquitous in the marine environment, representing a considerable pool of C and N with reported standing concentrations in the nano- to micromolar range (20, 21). Hence, future CO2 levels and limiting Fe may exacerbate cellular Fe stress, resulting in a fundamental metabolic shift whereby Trichodesmium reallocates resources away from N2 fixation and toward FMO-mediated organic nitrogen scavenging. The resulting metabolism change observed in Trichodesmium necessitates a reassessment of the relationship between new-nitrogen inputs and simultaneous removal by this globally important marine N2 fixer, as well as a broader characterization of the utilization of methylated amines like TMA by biogeochemically critical marine microbes.
RESULTS AND DISCUSSION
Nutrient-limited physiology under increased CO2.
A comprehensive depiction of the experimental design used in this study can be found in Fig. S1 in the supplemental material, and proteomic results and their implications for nutrient limitation theory were presented previously 5. Here, we focus on steady-state, transcriptional/translational changes in nitrogen source-sink metabolism as they relate to other biogeochemically important pathways. Briefly summarizing Fig. S1, a single IMS101 population originally isolated from an individual Trichodesmium colony (22) was split into low-CO2 (380 μatm of CO2 [380-selected]) and high-CO2 (750 μatm of CO2 [750-selected]) treatments of six biological replicates each and grown for ∼7 years (∼1,000 and ∼1,500 generations for the 380- and 750-selected treatments, respectively). Semicontinuous culturing methods were used in modified trace metal-clean Aquil medium (devoid of ammonia or other fixed nitrogen) containing 25 μM EDTA, standard vitamins, and trace metals with 250 nM Fe and 10 μM phosphate (PO43−). Next, Fe/P-colimited treatments were generated by subculturing three biological replicates from each CO2 treatment and incubating them in the aforementioned medium except with colimiting levels of Fe (10 nM) and PO43− (0.25 μM) for >1 year. To then mimic episodic pulses of Fe or P typically experienced by Fe/P-colimited microbes in the oligotrophic ocean (3, 23, 24), either Fe or P was added at replete concentrations to subcultures of the Fe/P-colimited cell lines to generate P- and Fe-limited lines at both CO2 levels, respectively. Following a 2-month acclimation period to create steady-state conditions, the lines were sampled for molecular analyses.
Growth and N2 fixation rates were significantly increased at high-CO2 levels under both nutrient-replete (r750) and P-limited (750-P) conditions relative to low-CO2/nutrient replete (r380) and low-CO2/P-limited (380-P) treatments, respectively (Fig. 1). Conversely, no growth rate differences were observed in high-CO2/Fe-limited (750-Fe) and high-CO2/Fe/P-colimited (750-Fe/P) cell lines relative to the corresponding low-CO2/Fe-limited (380-Fe) and low-CO2/Fe/P-colimited (380-Fe/P) conditions, respectively. In a recent study, Hong et al. (9) observed a significant decrease in growth and N2 fixation rates under both replete (r750) and Fe-limited conditions at high CO2 concentrations (750-Fe) relative to the controls, unlike this study and a number of other previous replete (3–6, 10, 25–28) and high-CO2/Fe-limited studies (5). Hong et al. (9) attributed these discrepancies to ammonia contamination and metal toxicity in all other past studies, although ammonia was below the detection limit in our prepared Aquil medium and 25 μM EDTA was added to control for metal toxicity (29). One central difference between this study and nearly all others relative to that of Hong et al. (9) is that Trichodesmium cell lines were adapted (4–6) or acclimated (10, 26–28) to a low pH as a product of increasing CO2 partial pressure on seawater carbonate chemistry similar to pH reductions in situ. This contrasts with artificially controlling pH with short-term acid/base additions as in the study of Hong et al., which may also result in other unknown and uncontrolled effects on cell physiology. A second fundamental difference is that our Fe single-limitation treatments were generated by pulsing phosphorus into 7-year, high-CO2-adapted cell lines colimited for iron and phosphorus for ∼1 year, similar to persistent colimitation conditions observed in the oligotrophic ocean (23, 24).
FIG 1.
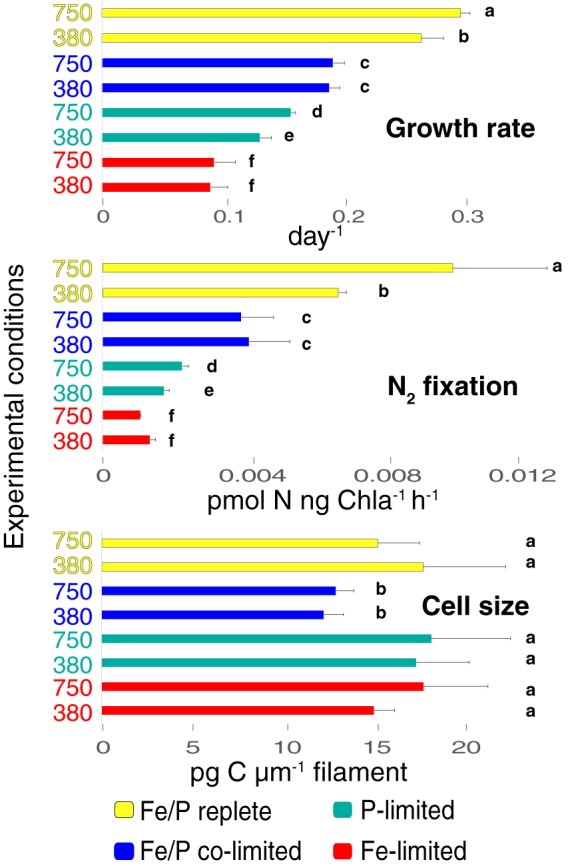
Cell physiology. Cell specific growth rates (day−1, top), N2 fixation rates (middle), and cell sizes (assessed by using the proxy carbon content per unit of filament length [pg of C/μm]) (bottom) are shown. Experimental conditions are shown to the left of all bars, which are color coded to indicate the nutrients used. Data are shown as means and standard errors (n = 6). Different letters denote statistical significance.
Looking ahead, it is predicted that increasing CO2 will impact ecosystems more rapidly than an increasing sea surface temperature (30), which in turn will affect the adaptive evolution of marine microbes and their contribution to elemental cycling. Hence, following long-term (∼7-year) adaptation of IMS101 to high-CO2 conditions (3, 5, 6), we investigated changes in IMS101 molecular physiology and nitrogen acquisition after ∼1 year of exposure to Fe/P colimitation, followed by additions of either Fe or P to create steady-state Fe and P single-limitation treatments.
Variable nitrogen assimilation strategies under nutrient colimitation in a future ocean.
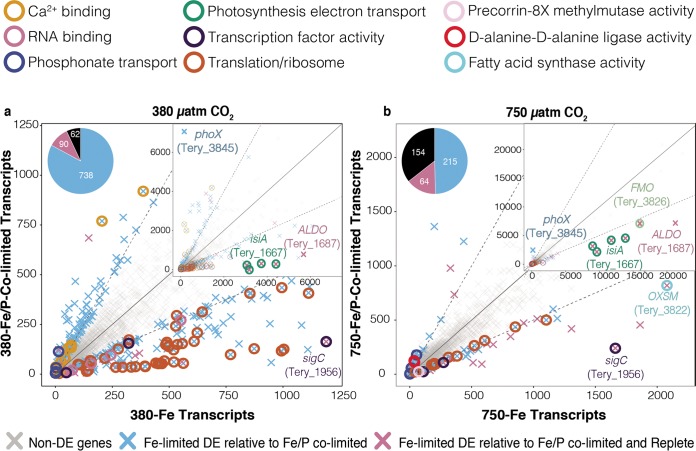
Below, we focus on the dynamics of global transcription and translation as they relate to changing nitrogen metabolic strategies under intensifying nutrient (co)limitation as a function of high-CO2 adaptation. A more in-depth discussion of other molecular changes in broad biochemical pathways can be found in the supplemental material. Genes in 380-Fe that were differentially expressed (DE) compared to those in r380, but not relative to those in 380-Fe/P, showed significantly different transcript levels in both 380-Fe and 380-Fe/P, respectively, relative to r380 (Fig. 2a; see also Fig. S2a and Data Set S1). This pool was enriched in Gene Ontology (GO) pathways involving nitrogen fixation (downregulated), cytochrome c oxidase activity (downregulated), copper ion binding (downregulated), nitrate/nitrite transport (upregulated), respiration (downregulated), glutamate synthase (upregulated), and amino acid transport (upregulated). These pathway enrichments indicate a downregulation of Fe-heavy, N2-fixing enzymes similar to other findings (5, 31) in conjunction with the upregulation of external nitrogen acquisition genes targeting inorganic (e.g., nitrate; Fig. 3) and/or possibly organic (e.g., amino acid transport) sources (Data Set S1).
FIG 2.
Fe-limited versus Fe/P-colimited transcripts under 380 and 750 μatm of CO2. (a) Scatterplot of 380-Fe DE genes relative to 380-Fe/P (blue) and 380-Fe DE genes relative to both r380 and 380-Fe/P (magenta). The center diagonal is a 1:1 line, while the others are 2:1 and 1:2 lines, respectively. Insets are the same plot zoomed out to display highly expressed genes. (b) Similar scatterplot of 750-Fe DE genes relative to either 750-Fe/P (blue) or both r750 and 750-Fe/P (magenta) DE genes. Select genes are labeled. The differently colored circles represent genes within significantly enriched GO pathways under the two conditions.
FIG 3.
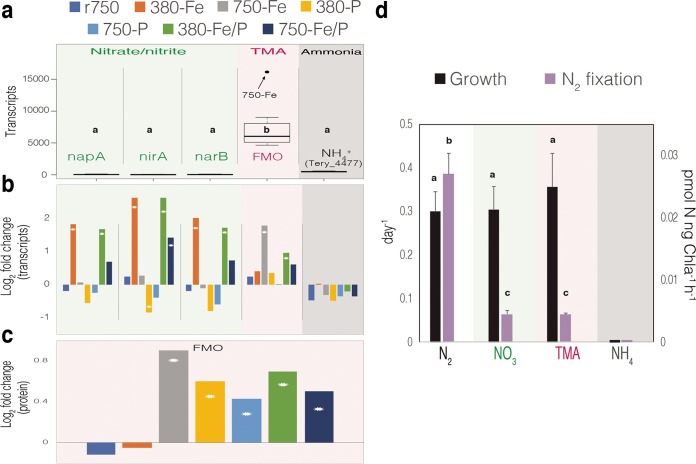
Distributions of nitrogen acquisition transcripts across treatments and log2 changes in transcript and protein abundance per treatment and physiological data of IMS101 grown on several nitrogen species. (a) The distribution of transcripts per gene across all treatments. Different letters signify significantly different mean values. (b) Log2 fold changes in gene transcript abundance relative to the r380 condition across experimental treatments. (c) Log2 fold changes in normalized FMO protein spectral counts relative to r380. For panels b and c, stars indicate statistically significant differences from r380. (d) Growth and N2 fixation rates of IMS101 grown without fixed nitrogen (N2) and in the presence of NO3, TMA, or NH4 at 20 μM. Different letters signify significantly different mean values.
While the 750-Fe pool of DE transcripts relative to r380 was enriched in GO pathways (Fig. S2) involved in nitrogen fixation (downregulated) and cytochrome c oxidase activity (downregulated), similar to 380-Fe relative to r380, genes involved in nitrate/nitrite transport had drastically lower expression levels in 750-Fe (gray bar) and 750-Fe/P (blue bar) than those in 380-Fe (orange bar) and 380-Fe/P (green bar), respectively (Fig. 3). This massive transcriptional reduction in inorganic nitrogen transport going from low- to high-CO2 conditions under Fe limitation regardless of the P concentration also co-occurred with ∼50% fewer genes being DE overall under high-CO2/low-Fe conditions, including a considerable reduction of protein synthesis transcripts (Fig. 2b; Fig. S2b and Data Set S1).
Interestingly, both transcript and protein abundances of a predicted TMA-oxidizing FMO (Tery_3826) harboring the FMO-specific motif (32) and a predicted transmembrane helix (according to the Integrated Microbial Genomes [IMG] annotation) sharply increased in 750-Fe (Fig. 2b and 3), along with the flavin biosynthesis gene ribD (Tery_1661; Data Set S1), relative to all other treatments. In fact, the FMO-encoding gene was one of the most highly expressed transcripts detected in the high-CO2, Fe-limited treatment. Furthermore, its general expression level was orders of magnitude greater than that of other nitrogen acquisition transcripts across all treatments (Fig. 2b and 3a). Transcripts of an annotated ammonia transporter gene (amT1; Tery_4477) remained unchanged across treatments, but it was consistently expressed in the top ∼6% of detected transcripts. Notably, although 380-Fe and 750-Fe growth rates were similar, aldO (Tery_1687; fructose-bisphosphate aldolase) transcript levels also drastically increased in 750-Fe along with the ferric uptake regulator (Tery_3404, fur) and Fe stress/photosynthesis antenna genes (Fig. 2b; Fig. S2b).
This global shift in transcript profiles, in conjunction with the downregulation of nitrate transport and the concurrent upregulation of FMO and ribD, suggests that high-CO2 conditions intensify iron limitation (5, 9), resulting in fundamental resource (e.g., riboflavin) reallocation toward increased biosynthetic investment in FMO-mediated, iron-lean organic nitrogen assimilation strategies. The large increase in FMO transcripts and proteins observed in 750-Fe suggests that Trichodesmium preferentially scavenges TMA and possibly other organic nitrogen and sulfur compounds under severe iron stress. Furthermore, FMO transcript and protein abundances are in the top ∼0.5% and ∼10% of all detected gene transcripts and protein products, respectively, irrespective of treatment, which indicates significant energetic investment in persistent FMO biosynthesis. Conversely, nitrate transport gene transcripts are only in the top ∼50 to 70% of the total transcripts, with no corresponding protein products detected in the proteome. Upon calculation of the total mean expression of nitrogen acquisition transcripts across all treatments, the gene for FMO was consistently expressed at significantly higher levels (P < 0.0001) than other nitrogen acquisition genes (Fig. 3a), providing further evidence of persistent and enriched biosynthesis of FMO. Taken together with the ammonia transporter expression in the top 6% of overall transcripts, these data support the notion that FMO-acquired compounds and ammonia are preferred relative to nitrate acquisition, as previously noted for ammonia (17). The TMA protein was significantly more abundant under high-CO2/low-Fe, low-P, and Fe/P colimitation conditions than under r380, r750, and 380-Fe conditions, suggesting enhanced energetic investment in TMA enzyme biosynthesis across various CO2/nutrient-limited global change scenarios (Fig. 3c). It is interesting that FMO transcript and protein levels considerably increased in 750-Fe but not 380-Fe, although Fe stress genes signaled iron limitation under both conditions. Strain IMS101 N2-fixing metabolism may be better adapted to low-CO2/low-Fe conditions because of prevalent Fe limitation in situ (24), thereby reducing the need to scavenge for nitrogen, while molecular metabolic restructuring induced by high-CO2/low-Fe conditions may require increased exogenous nitrogen for growth in a high-CO2 ocean. Since heterotrophs have been shown to grow on TMA as a sole C and N source (32), Trichodesmium may utilize TMA for both C and N (33, 34). It was also recently shown that an associated heterotrophic Alteromonas macleodii strain widely conserved in Trichodesmium metagenomic consortia has the genetic potential to cleave dimethylsulfoniopropionate to produce dimethylsulfide (DMS) (35), which may be further oxidized by the Trichodesmium FMO, as DMS has been shown to have nearly as high an affinity for FMO as that of TMA (32). Hence, TMA- and other FMO-mediated assimilation may be a more general compensatory strategy utilized by Trichodesmium for physiological maintenance in the face of fluctuating nutrients, which is evidenced by the significant increase in FMO protein abundance across nutrient-limited treatments (Fig. 3c, bottom).
N2 fixation is strongly inhibited in the presence of 20 μM TMA, just as it is with 20 μM nitrate (Fig. 3d). However, Trichodesmium growth rates are virtually the same in the presence of 20 μM TMA as growth rates supported by N2 fixation, comparable to results obtained in the presence of 20 μM nitrate, indicating that both fixed nitrogen sources can support growth as efficiently as N2. In contrast, the presence of 20 μM ammonia was toxic, and the cells were unable to grow or fix N2 at all (Fig. 3d). One recent study observed enigmatically high growth and N2 fixation rates in the presence of 20 μM ammonia (9), contrasting with all other previous studies demonstrating drastic repression of N2 fixation at exogenous ammonia concentrations of >10 μM (15, 17, 36, 37).
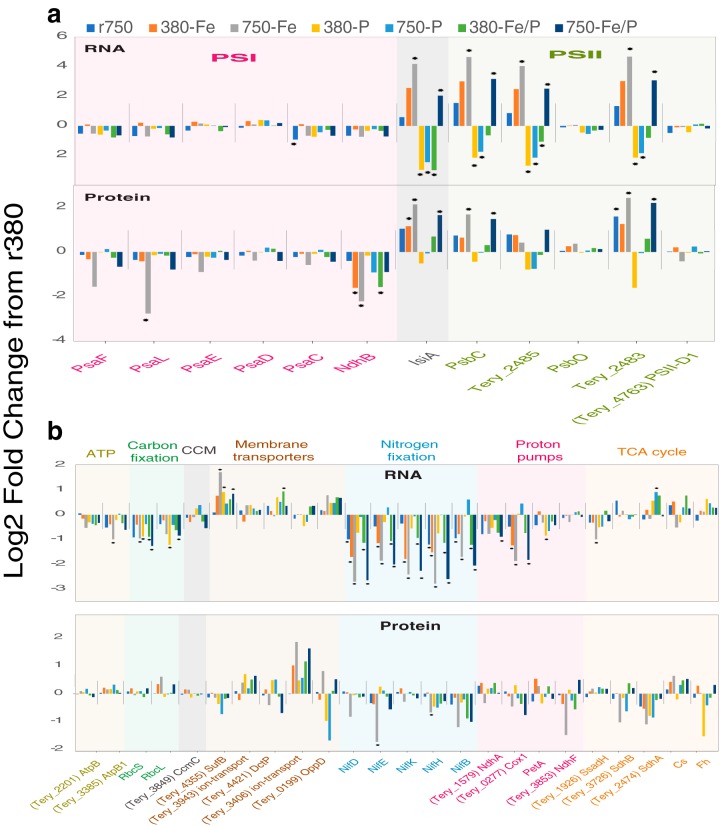
Although Hong et al. (9) observed decreases in the PSI/PSII ratio under 750-Fe conditions similar to those of this study (Fig. 4a) and others (5, 31, 38), they also observed increases in the iron-rich NifH protein under high-CO2/Fe-limiting conditions (750-Fe), contrasting with our observed decreases in multiple nitrogenase transcripts and protein subunits in this study (Fig. 4b) and another Trichodesmium iron-limited proteome (31). Although Hong et al. (9) suggested that more NifH is necessary in 750-Fe because of decreased enzyme efficiency, we observed a contrasting trend via a fundamental shift in nitrogen metabolic strategy, whereby iron-rich gene abundances (e.g., PSI and nif subunits) along with nitrate transport transcripts are significantly reduced under severe iron stress concomitantly with increases in TMA-scavenging FMO and riboflavin biosynthesis gene expression (750-Fe; Fig. 4; Fig. S2).
FIG 4.
Log2 fold changes in the normalized transcript and protein abundance of photosystem genes and genes from various other pathways. (a) Log2 fold changes in gene transcript abundance relative to the r380 condition across experimental treatments. (b) Log2 fold changes in normalized FMO protein spectral counts relative to r380.
Among the various forms of available exogenous nitrogen (e.g., amino acids, nitrate, TMA, ammonia), one possible reason why FMO may be preferred is that it can oxidize both nitrogen- and sulfur-containing compounds, including TMA, which may also serve as potential organic carbon sources. This may enable Trichodesmium to assimilate several macronutrients by minimizing energetic investment. Additionally, since Trichodesmium is physically colonized by a diverse microbial consortium (35, 39) and methylated amine compounds like TMA are ubiquitous as end products of, for example, protein putrefaction (32), Trichodesmium may have consistent access to organic nitrogen/carbon sources as a product of the biochemical decomposition of cellular components by members of its consortium. This contrasts with the lifestyles of other planktonic unicellular marine bacteria, which are typically associated with much less abundant and diverse consortia. When searching Trichodesmium consortium metatranscriptome sequences (35) for FMO via BLASTP (40), no significant hits containing the FMO-specific motif were observed, suggesting that it may not be widespread or expressed highly in consortia.
Although FMO homologs have been detected across global ocean data sets, including within the genomes of the ubiquitous SAR11 clade (32), only a few robust cyanobacterial and bacterial homologs to IMS101 were detected, with many of the remaining top high-scoring pairs deriving from metazoans (Fig. S3; high-scoring pair, ≥70% of the original Trichodesmium FMO sequence length; E value, ≤10−5), yielding an average identity of 27.9% (median, 27.8%) over all pairs. We did detect the FMO genes in another T. erythraeum genome (strain 2175) and a Trichodesmium metagenome from hand-picked natural colonies (41), demonstrating widespread retention and conservation of FMO. Interestingly, no homologs of other biogeochemically relevant marine cyanobacteria were detected. Taken together, the findings show that FMO-mediated scavenging may be ecologically advantageous for Trichodesmium in the oligotrophic ocean, given its potentially persistent availability via biochemical degradation and its relative scarcity in its physically attached consortia, as well as other sympatric cyanobacteria.
Fe-limited and Fe/P-colimited metabolism in the future ocean.
Our data suggest that interaction of limiting Fe with high-CO2 adaptation will intensify iron stress in Fe/P-colimited molecular metabolism. For example, known P stress genes (e.g., phoX, sphX, phnG), but not Fe stress genes (e.g., isiA), were significantly upregulated in 380-Fe/P compared to r380, suggesting that P stress may be more prominent than Fe stress under low-CO2/colimiting conditions (Fig. 4 and 5; Fig. S4 and S6 and Data Sets S2 and S3). In fact, isiA transcripts were significantly downregulated under both P-limited conditions, irrespective of CO2, as well as during 380-Fe/P colimitation (Fig. 4a). However, transcripts of Fe stress genes, including isiA, were significantly increased in 750-Fe/P relative to those in 380-Fe/P and this was accompanied by a significant decrease in the PSI/PSII ratio, indicating iron stress under high-CO2 conditions with colimitation (Fig. 4 and 5). Finally, this shift toward Fe stress is further evidenced by the high-confidence grouping of 750-Fe with the 380- and 750-Fe/P-colimited treatments (approximately unbiased [AU] bootstrap P value, >0.95) resulting from hierarchical clustering of genes exclusively DE (i.e., transcriptional complement; Data Set S3) in both 380- and 750-Fe/P (Fig. 6). Collectively, these data provide strong evidence of the intensification of Fe stress on cellular metabolism upon interaction with a high CO2 concentration, regardless of fluctuating P concentrations. Hence, iron-rich metabolic pathways that generate energy and fixed nitrogen will likely be impacted as cells utilize alternative iron- and energy-conserving strategies to acquire nutrients, with consequent reductions in new-nitrogen inputs.
FIG 5.
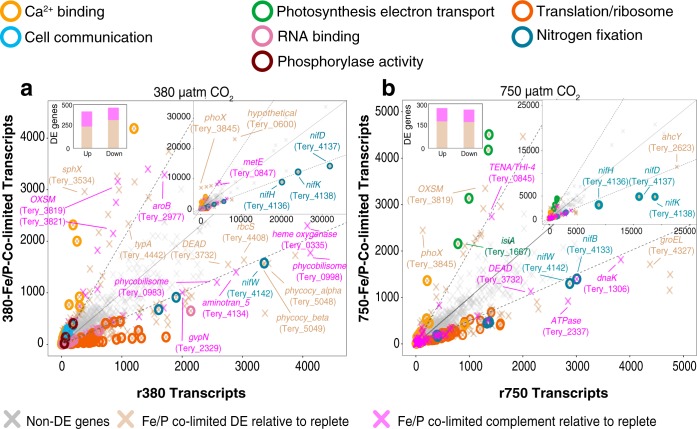
Replete versus Fe/P-colimited transcripts under 380 and 750 μatm of CO2. (a) Scatterplot of 380-Fe/P DE genes relative to r380 (tan) and 380-Fe/P DE genes relative to only r380 and no other 380 nutrient limitations (magenta). The center diagonal is a 1:1 line, while the others are 2:1 and 1:2 lines, respectively. The upper right insets are the same plots zoomed out to display highly expressed genes. (b) Similar scatterplot of 750-Fe/P DE genes relative to r750 (tan) and 750-Fe/P relative only to r750 and no other 750 nutrient limitations (magenta). Selected genes are labeled. The differently colored circles represent genes within significantly enriched GO pathways under the two conditions.
FIG 6.
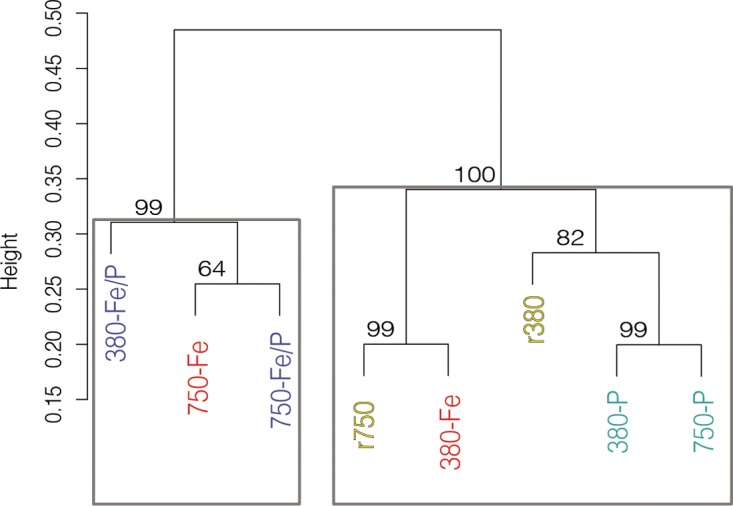
Hierarchical clustering of 380- and 750-Fe/P gene transcript complements. Hierarchical clustering of Bray-Curtis dissimilarities with multiscale bootstrap resampling calculated from normalized transcripts of all DE genes in both 380- and 750-Fe/P complements. Values at nodes are AU P values. Boxes highlight high-confidence clusters with AU P values of >0.95.
P-limited and Fe/P-colimited metabolism in present and future oceans.
Upon adaptation to high-CO2 conditions, both 750-P growth and N2 fixation increased relative to those in 380-P (Fig. 1), while most 750-P-limited transcript levels remained statistically unchanged between 380-P and 750-P. This is evidenced by the much smaller number (n = 4) of genes DE between 380-P and 750-P than those DE between 380-Fe and 750-Fe (n = 284; Fig. S5 and S6). Translation dominated the GO-enriched pathway pools for downregulated genes relative to both r750 only and 750-Fe/P (Fig. S4 and S6 and Data Set S2), as in 380-P. It is possible that increased N2 fixation rates of equivalently sized IMS101 cells under various CO2 conditions (e.g., r380 versus r750 and 380-P versus 750-P) are driven by factors contributing to altered enzymatic rates rather than by increases in enzyme abundance, as has been observed for temperature (42). This possibility is supported by either no change or decreases in N2 fixation transcripts and/or protein abundances (e.g., nifH, nifB, nifK) in replete, 7-year high-CO2-adapted IMS101 (r750) relative to replete, low-CO2 (r380) cells, despite higher N2 fixation rates in cells selected under high-CO2 conditions (Fig. 4) (5). For some N2 fixation genes, transcript increases were mirrored by increases in protein abundance (e.g., those encoded by nifB [Tery_4133] and nifK [Tery_4138]), though most remained unchanged in the proteomic data (e.g., those encoded by nifH [Tery_4136] and nifD [Tery_4137]). These trends suggest that different nitrogenase genes may be independently regulated at the transcriptional level or have different mRNA half-lives. Taken together, the results show that other regulatory mechanisms impacting enzymatic rates, but not abundances, may be more generally associated with changes in CO2-impacted physiology under P limitation.
Fe/P-colimited transcription in present and future oceans.
Photosynthesis-related (e.g., Tery_0983, Tery_0998, Tery_5048, Tery_5049, Tery_0728), ferredoxin (e.g., Tery_0916, Tery_4539, Tery_5051), heme-containing (e.g., Tery_1714, Tery_0335), and N2-fixing genes (Fig. 2, 4, and 5; Fig. S7) were all downregulated in 380-Fe/P. This indicates a potential reduction and/or reallocation of Fe away from these pathways in the colimited cells coinciding with reduced cell size (and therefore cellular elemental quotas), growth, and N2 fixation relative to those seen under replete conditions. Additionally, downregulation of GO-enriched pathways in 380-Fe/P involved in translational machinery and RNA binding (Fig. 5; Fig. S7) are consistent with observations in yeast and bacteria (i.e., stringent response), where proteins involved in cell growth and division (e.g., ribosomal proteins) have been observed to be downregulated in minimal (e.g., 380-Fe/P) versus rich (e.g., r380) medium (43, 44). Moreover, the upregulated 380-Fe/P (relative to r380) transcriptional complement also contained many genes involved in precursor metabolism, potentially allowing for greater cellular plasticity and metabolic flexibility under low-nutrient regimes (5). Accordingly, we identified a set of upregulated proteins specific to 380-Fe/P colimitation that included many involved in precursor metabolisms (i.e., isoprenoid biosynthesis), a finding consistent with prior observations in yeast maintained in minimal medium (44). For example, 3-dehydroquinate synthase (Tery_2977, aroB) is upregulated solely in 380-Fe/P and is part of the shikimate pathway that produces chorismate, a precursor of aromatic amino acids, including phenylalanine, tyrosine, and tryptophan (45) (Fig. S7).
Following 7 years of adaptation to high-CO2 conditions and 1 year of growth under high-CO2 Fe/P colimitation conditions (750-Fe/P), transcript levels had shifted across broad metabolic pathways, presumably orchestrating the maintenance of growth and N2 fixation similar to those seen under 380-Fe/P conditions (Fig. 5; Fig. S7). Of those genes that were exclusively DE during Fe/P colimitation, only ∼5% (n = 26; Data Set S3) from the 380-Fe/P complement (n = 313) remained in the 750-Fe/P complement (n = 174) (Data Set S3). In other words, these genes were either upregulated (n = 15) or downregulated (n = 11) only during 380-Fe/P and 750-Fe/P colimitation, relative to the respective replete reference conditions (r380 and r750). Shared upregulated genes include the universal stress global response regulator (Tery_2353, uspA), the aforementioned GTP-binding gene (Tery_1904), and a carbohydrate selective porin gene (Tery_0838, oprB). The consistent upregulation of these genes suggests that 380- and 750-Fe/P-colimited cellular physiology responds to environmental flux by consistently transcribing genes involved in general stress, biotimer regulation, and potential organic carbon uptake, as has been observed in other cyanobacteria (33, 34). Shared downregulated genes include that for a 5′,5″-P1,P4-tetraphosphate phosphorylase (Tery_0108, ap4A), which has been implicated in bacterial stress responses and gene regulation (46), and the mutator gene mutT (Tery_4056), which has been demonstrated to suppress transversion mutations (A·T to C·G) (47). Accordingly, Escherichia coli ΔmutT deletion mutants exhibited a 1,000-fold increase in unidirectional A·T-to-C·G transversion frequencies. Other studies have postulated that mutators can accelerate adaptive evolution under certain circumstances via increased mutation rates (48). Hence, the significant reduction of mutT transcription in long-term 380-Fe/P and 750-Fe/P treatments may help enable adaptive mutation by possibly derepressing transversion mutations in low-nutrient, stress-inducing environments.
Conclusions.
Here, we show a marked shift in nitrogen metabolism going from low-CO2/Fe-limited to high-CO2/Fe-limited regimes whereby iron-heavy pathways are significantly reduced, and cellular investment is reallocated toward a predicted FMO-mediated organic nitrogen scavenging, relative to acquisition pathways for inorganic nitrogen substrates. This fundamental iron-saving strategy may also enable simultaneous assimilation of several other required elements, including C and S from methylated amines like TMA. Our data also suggest that intensifying Fe stress under high-CO2 conditions may shift Fe/P-colimited metabolism into a more Fe-limited metabolic state. This trend further highlights the need for iron-saving metabolic strategies for nutrient limitation in a future high-CO2 ocean. Additionally, N2 fixation but not growth was inhibited in the presence of TMA. If interactive global change factors intensify nutrient limitation, leading to enhanced organic nitrogen scavenging and reduced N2 fixation by Trichodesmium, future work must consider the relationship between N2 fixation-mediated new-nitrogen inputs and simultaneous removal from the fixed nitrogen pool. The shifting balance between these two processes may have global implications for the role of Trichodesmium in future ocean biomes. Finally, transcript patterns in Fe/P-colimited metabolism also suggest reductions in iron-heavy pathways in exchange for increases in precursor-related genes that may aid cellular plasticity in response to various nutrient concentrations. Although limitations in this long-term experiment and others (49) include a steady-state environment lacking natural variability, as well as the use of a single strain of Trichodesmium, we believe that our results demonstrating molecular reallocations of nitrogen acquisition and iron-sparing expression provide robust indicators of potential in situ strategies some Trichodesmium populations may employ while under covarying nutrient-limited regimes. We highlight consistent molecular patterns in genes under Fe and/or P limitation while covarying CO2, which strengthens the broad trends outlined above. Future field work investigating colimitation should aim to compare in situ molecular patterns with those that we have identified in culture to try to help elucidate universal indicators of colimitation in the future ocean.
MATERIALS AND METHODS
Culturing methods.
Details of the nutrient-limited culturing methods used in this study can be found in reference 5 and in the supplemental material. For the growth experiment comparing TMA with other N sources, IMS101 was grown in four N treatments, (i) N2, (ii) 20 μM NaNO3, (iii) 20 μM TMA (C3H9N), and (iv) 20 μM NH4Cl. For all treatments, triplicate cultures were maintained semicontinuously at 26°C under a light irradiance intensity of 150 μmol of photons m−2 s−1 (12-h light/12-h dark cycle), cultures were diluted every other day, and 20 μM N sources were added after every dilution. Growth and nitrogen fixation rates were measured after 10 generations as described above and below.
Aquil medium was bubbled with 0.2-μm-filtered prepared air-CO2 mixtures (Praxair), and in-line high-efficiency particulate air filters were employed to avoid Fe contamination from particles in the gas tanks or lines. The pH was monitored daily, and dissolved inorganic carbon concentrations were measured at the final sampling. The acetylene reduction method was used to measure Trichodesmium N2 fixation rates (4, 14, 25). Samples from each experimental triplicate (20 ml) were incubated in gas-tight vials for 1 h after being injected with 2 ml of acetylene in 23 ml of headspace. The amount of ethylene produced was then analyzed by injecting a 200-μl aliquot of headspace gas into a gas chromatograph (model GC-8A; Shimadzu Scientific Instruments, Columbia, MD, USA). An ethylene accumulation-to-N2 fixation conversion ratio of 3 was used to calculate N2 fixation rates. Upon steady-state growth, Trichodesmium filament abundance and length were measured in a 1-ml phytoplankton counting chamber by epifluorescence microscopy and significant differences between nutrient conditions were calculated by two-way analysis of variance along with the Tukey test. For RNA sampling, cultures were swiftly and gently filtered onto 5-μm polycarbonate filters (Whatman) during the middle of the photoperiod, immediately flash frozen, and stored in liquid nitrogen until RNA extraction.
RNA isolation and extraction for Illumina sequencing.
For each replicate, samples for RNA extraction were taken concomitantly with proteome samples during the middle of the photoperiod, as described in reference 5. Briefly, cells were swiftly and gently filtered at 11 a.m. with 5-μm polycarbonate filters (Whatman), immediately flash frozen, and stored in liquid nitrogen until RNA extraction. RNA was extracted from two randomly chosen biological replicates per treatment with the Ambion MirVana microRNA isolation kit (Thermo Fisher Scientific) in an RNase-free environment in accordance with the manufacturer's instructions, followed by two incubations with the Ambion Turbo DNA-free kit to degrade trace amounts of DNA. RNA was then submitted to the University of California San Diego Institute for Genomic Medicine core for library preparation and sequencing (http://igm.ucsd.edu/genomics/services.shtml). Briefly, rRNA removal and library construction were done with the Ribo-Zero rRNA removal kit (Illumina) and the TruSeq Stranded RNA Library Prep kit (Illumina), respectively, and multiplexed libraries were sequenced by using Illumina Hi-Seq, yielding single-end 50-bp read libraries. All of the protein spectral data used in the above-described analyses can be found in Supplementary Data 4 of reference 5.
Expression analysis.
Differential expression was done as previously described (6), and detailed methods can be found in the supplemental material.
GO enrichment analysis.
As in reference 6, GO annotations for Trichodesmium were downloaded from the Genome2D web server (http://genome2d.molgenrug.nl). Next, the phyper function in R (R Core Team 2014) was used to test for significant enrichment of GO categories among the treatments and P values were corrected by the Benjamini-Hochberg method (50) with the p.adjust function (P ≤ 0.1). Finally, genes in enriched GO categories were manually checked.
Phylogenetic analysis.
FMO sequences for T. erythraeum strains IMS101 and 2175 were taken from the IMG website (https://img.jgi.doe.gov/), and Trichodesmium environmental metagenomic sequence data were downloaded from reference 41. The BLASTP algorithm was used to search sequences against the RefSeq protein database (51), in which all high-scoring pairs were retained if the aligned portion spanned >70% of the original query length with an E value of <10−5. Sequences were then aligned with MUSCLE v3.8.31 by using the default settings (52), and spurious sequences and poorly aligned regions were removed with trimAl 1.2rev59 (53). RAxML (54) was used for all maximum-likelihood phylogenetic analyses with the following settings: -f a -p 12345 -m PROTCATLG -N 100 -x 12345.
Availability of data.
All of the transcriptome sequencing data used in this study have been deposited as raw fastq files in the NCBI Gene Expression Omnibus (55) and are accessible through GEO Series accession number GSE94951. All of the protein spectral data used in the above-described analyses can be found in Supplementary Data 4 of reference 5. Physiological and proteome data are archived through the U.S. National Science Foundation Biological and Chemical Oceanography Data Management Office (http://www.bco-dmo.org/dataset/649904).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by U.S. National Science Foundation grants OCE-1260233, OCE 1260490, and OCE 1657757 to D.A.H., E.A.W., and F.-X.F. and grant OCE OA 1220484 to D.A.H. as well as G. B. Moore Foundation grants 3782 and 3934 to M.A.S.
We have no competing interests to declare.
F.-X.F., D.A.H., and E.A.W. designed the research; N.G.W., M.D.L., X.C., and F.-X.F. performed the research; N.G.W., M.D.L., X.C., F.-X.F., D.A.H., M.A.S., and E.A.W. analyzed the data; and N.G.W., M.D.L., F.-X.F., D.A.H., M.A.S., and E.A.W. wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02137-17.
REFERENCES
- 1.Hutchins DA, Fu F. 2017. Microorganisms and ocean global change. Nat Microbiol 2:17058. doi: 10.1038/nmicrobiol.2017.58. [DOI] [PubMed] [Google Scholar]
- 2.Arrigo KR. 2005. Marine microorganisms and global nutrient cycles. Nature 437:349–355. doi: 10.1038/nature04159. [DOI] [PubMed] [Google Scholar]
- 3.Hutchins DA, Boyd PW. 2016. Marine phytoplankton and the changing ocean iron cycle. Nat Clim Chang 6:1072–1079. doi: 10.1038/nclimate3147. [DOI] [Google Scholar]
- 4.Hutchins DA, Walworth NG, Webb EA, Saito MA, Moran D, McIlvin MR, Gale J, Fu F-X. 2015. Irreversibly increased nitrogen fixation in Trichodesmium experimentally adapted to elevated carbon dioxide. Nat Commun 6:8155. doi: 10.1038/ncomms9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walworth NG, Fu F-X, Webb EA, Saito MA, Moran D, McIlvin MR, Lee MD, Hutchins DA. 2016. Mechanisms of increased Trichodesmium fitness under iron and phosphorus co limitation in the present and future ocean. Nat Commun 7:12081. doi: 10.1038/ncomms12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walworth NG, Lee MD, Fu F-X, Hutchins DA, Webb EA. 2016. Molecular and physiological evidence of genetic assimilation to high CO2 in the marine nitrogen fixer Trichodesmium. Proc Natl Acad Sci U S A 113:E7367–E7374. doi: 10.1073/pnas.1605202113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia NS, Fu F, Sedwick PN, Hutchins DA. 2015. Iron deficiency increases growth and nitrogen-fixation rates of phosphorus-deficient marine cyanobacteria. ISME J 9:238–245. doi: 10.1038/ismej.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Baar HJW. 1994. von Liebig's law of the minimum and plankton ecology (1899–1991). Prog Oceanogr 33:347–386. doi: 10.1016/0079-6611(94)90022-1. [DOI] [Google Scholar]
- 9.Hong H, Shen R, Zhang F, Wen Z, Chang S, Wenfang L, Kranz SA, Luo Y-W, Kao S-J, Morel FMM, Shi D. 2017. The complex effects of ocean acidification on the prominent N2-fixing cyanobacterium Trichodesmium. Science 356:527–531. doi: 10.1126/science.aal2981. [DOI] [PubMed] [Google Scholar]
- 10.Hutchins DA, Mulholland MR, Fu F. 2009. Nutrient cycles and marine microbes in a CO2-enriched ocean. Oceanography 22:128–145. doi: 10.5670/oceanog.2009.103. [DOI] [Google Scholar]
- 11.Saito MA, Bertrand EM, Dutkiewicz S, Bulygin VV, Moran DM, Monteiro FM, Follows MJ, Valois FW, Waterbury JB. 2011. Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc Natl Acad Sci U S A 108:2184–2189. doi: 10.1073/pnas.1006943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu F-X, Mulholland MR, Garcia NS, Beck A, Bernhardt PW, Warner ME, Sañudo-Wilhelmy SA, Hutchins DA. 2008. Interactions between changing pCO2, N2 fixation, and Fe limitation in the marine unicellular cyanobacterium Crocosphaera. Limnol Oceanogr 53:2472–2484. doi: 10.4319/lo.2008.53.6.2472. [DOI] [Google Scholar]
- 13.Chappell PD, Webb EA. 2010. A molecular assessment of the iron stress response in the two phylogenetic clades of Trichodesmium. Environ Microbiol 12:13–27. doi: 10.1111/j.1462-2920.2009.02026.x. [DOI] [PubMed] [Google Scholar]
- 14.Mulholland MR, Capone DG. 1999. Nitrogen fixation, uptake and metabolism in natural and cultured populations of Trichodesmium spp. Mar Ecol Prog Ser 188:33–49. doi: 10.3354/meps188033. [DOI] [Google Scholar]
- 15.Fu F-X, Bell PRF. 2003. Factors affecting N2 fixation by the cyanobacterium Trichodesmium sp. GBRTRLI101. FEMS Microbiol Ecol 45:203–209. doi: 10.1016/S0168-6496(03)00157-0. [DOI] [PubMed] [Google Scholar]
- 16.Moore CM, Mills MM, Arrigo KR, Berman-frank I, Bopp L, Boyd PW, Galbraith ED, Geider RJ, Guieu C, Jaccard SL, Jickells TD, La Roche J, Lenton TM, Mahowald NM, Marañón E, Marinov I, Moore JK, Nakatsuka T, Oschlies A, Saito MA, Thingstad TF, Tsuda A, Ulloa O. 2013. Processes and patterns of oceanic nutrient limitation. Nat Geosci 6:701–710. doi: 10.1038/ngeo1765. [DOI] [Google Scholar]
- 17.Post AF, Rihtman B, Wang Q. 2012. Decoupling of ammonium regulation and ntcA transcription in the diazotrophic marine cyanobacterium Trichodesmium sp. IMS101. ISME J 6:629–637. doi: 10.1038/ismej.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benavides M, Berthelot H, Duhamel S, Raimbault P, Bonnet S. 2017. Dissolved organic matter uptake by Trichodesmium in the Southwest Pacific. Sci Rep 7:41315. doi: 10.1038/srep41315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oren A. 1990. Formation and breakdown of glycine betaine and trimethylamine in hypersaline environments. Antonie Van Leeuwenhoek 58:291–298. doi: 10.1007/BF00399342. [DOI] [PubMed] [Google Scholar]
- 20.Van Neste A, Duce RA, Lee C. 1987. Methylamines in the marine atmosphere. Geophys Res Lett 14:711–714. doi: 10.1029/GL014i007p00711. [DOI] [Google Scholar]
- 21.Gibb SW, Hatton AD. 2004. The occurrence and distribution of trimethylamine-N-oxide in Antarctic coastal waters. Mar Chem 91:65–75. doi: 10.1016/j.marchem.2004.04.005. [DOI] [Google Scholar]
- 22.Prufert-Bebout L, Paerl HW, Lassen C. 1993. Growth, nitrogen fixation, and spectral attenuation in cultivated Trichodesmium species. Appl Environ Microbiol 59:1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mills MM, Ridame C, Davey M, LaRoche J, Geider RJ. 2004. Iron and phosphorus co-limit nitrogen fixation in the eastern tropical North Atlantic. Nature 429:292–294. doi: 10.1038/nature02550. [DOI] [PubMed] [Google Scholar]
- 24.Sohm JA, Webb EA, Capone DG. 2011. Emerging patterns of marine nitrogen fixation. Nat Rev Microbiol 9:499–508. doi: 10.1038/nrmicro2594. [DOI] [PubMed] [Google Scholar]
- 25.Hutchins DA, Fu F-X, Webb EA, Walworth N, Tagliabue A. 2013. Taxon-specific response of marine nitrogen fixers to elevated carbon dioxide concentrations. Nat Geosci 6:1–6. doi: 10.1038/ngeo1858. [DOI] [Google Scholar]
- 26.Kranz SA, Levitan O, Richter K-U, Prásil O, Berman-Frank I, Rost B. 2010. Combined effects of CO2 and light on the N2-fixing cyanobacterium Trichodesmium IMS101: physiological responses. Plant Physiol 154:334–345. doi: 10.1104/pp.110.159145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levitan O, Kranz SA, Spungin D, Prásil O, Rost B, Berman-Frank I. 2010. Combined effects of CO2 and light on the N2-fixing cyanobacterium Trichodesmium IMS101: a mechanistic view. Plant Physiol 154:346–356. doi: 10.1104/pp.110.159285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eichner MJ, Klawonn I, Wilson ST, Littmann S, Whitehouse MJ, Church MJ, Kuypers MM, Karl DM, Ploug H. 2017. Chemical microenvironments and single-cell carbon and nitrogen uptake in field-collected colonies of Trichodesmium under different pCO2. ISME J 11:1305–1317. doi: 10.1038/ismej.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchins DA, Fu F, Walworth NG, Lee MD, Saito MA, Webb EA. 2017. Comment on “The complex effects of ocean acidification on the prominent N2-fixing cyanobacterium Trichodesmium.” Science 357:eaao0067. doi: 10.1126/science.aao0067. [DOI] [PubMed] [Google Scholar]
- 30.Boyd PW, Cornwall CE, Davison A, Doney SC, Fourquez M, Hurd CL, Lima ID, McMinn A. 2016. Biological responses to environmental heterogeneity under future ocean conditions. Global Chang Biol 22:2633–2650. doi: 10.1111/gcb.13287. [DOI] [PubMed] [Google Scholar]
- 31.Snow JT, Polyviou D, Skipp P, Chrismas NAM, Hitchcock A, Geider R, Moore CM, Bibby TS. 2015. Quantifying integrated proteomic responses to iron stress in the globally important marine diazotroph Trichodesmium. PLoS One 10:e0142626. doi: 10.1371/journal.pone.0142626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Patel NA, Crombie A, Scrivens JH, Murrell JC. 2011. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc Natl Acad Sci U S A 108:17791–17796. doi: 10.1073/pnas.1112928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuart RK, Mayali X, Lee JZ, Everroad RC, Hwang M, Bebout BM, Weber PK, Pett-Ridge J, Thelen MP. 2016. Cyanobacterial reuse of extracellular organic carbon in microbial mats. ISME J 10:1240–1251. doi: 10.1038/ismej.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Francisco ÉC, Franco TT, Wagner R, Jacob-Lopes E. 2014. Assessment of different carbohydrates as exogenous carbon source in cultivation of cyanobacteria. Bioprocess Biosyst Eng 37:1497–1505. doi: 10.1007/s00449-013-1121-1. [DOI] [PubMed] [Google Scholar]
- 35.Lee MD, Walworth NG, McParland EL, Fu F-X, Mincer TJ, Levine NM, Hutchins DA, Webb EA. 2017. The Trichodesmium consortium: conserved heterotrophic co-occurrence and genomic signatures of potential interactions. ISME J 11:1813–1824. doi: 10.1038/ismej.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulholland MR, Ohki K, Capone DG. 2001. Nutrient controls on nitrogen uptake and metabolism by natural populations and cultures of Trichodesmium (Cyanobacteria). J Phycol 37:1001–1009. doi: 10.1046/j.1529-8817.2001.00080.x. [DOI] [Google Scholar]
- 37.Mulholland MR, Ohki K, Capone DG. 1999. Nitrogen utilization and metabolism relative to patterns of N2 fixation in cultures of Trichodesmium NIBB1067. J Phycol 35:977–988. doi: 10.1046/j.1529-8817.1999.3550977.x. [DOI] [Google Scholar]
- 38.Shi D, Kranz SA, Kim J-M, Morel FMM. 2012. Ocean acidification slows nitrogen fixation and growth in the dominant diazotroph Trichodesmium under low-iron conditions. Proc Natl Acad Sci U S A 109:E3094–E3100. doi: 10.1073/pnas.1216012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouco M, Haley ST, Dyhrman ST. 2016. Microbial diversity within the Trichodesmium holobiont. Environ Microbiol 18:5151–5160. doi: 10.1111/1462-2920.13513. [DOI] [PubMed] [Google Scholar]
- 40.Altschul S, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Walworth N, Pfreundt U, Nelson WC, Mincer T, Heidelberg JF, Fu F, Waterbury JB, Glavina del Rio T, Goodwin L, Kyrpides NC, Land ML, Woyke T, Hutchins DA, Hess WR, Webb EA. 2015. Trichodesmium genome maintains abundant, widespread noncoding DNA in situ, despite oligotrophic lifestyle. Proc Natl Acad Sci U S A 112:4251–4256. doi: 10.1073/pnas.1422332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toseland A, Daines SJ, Clark JR, Kirkham A, Strauss J, Uhlig C, Lenton TM, Valentin K, Pearson GA, Moulton V, Mock T. 2013. The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat Clim Chang 3:979–984. doi: 10.1038/nclimate1989. [DOI] [Google Scholar]
- 43.Carneiro S, Lourenço A, Ferreira EC, Rocha I. 2011. Stringent response of Escherichia coli: revisiting the bibliome using literature mining. Microb Inform Exp 1:14. doi: 10.1186/2042-5783-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 45.Herrmann KM. 1995. The shikimate pathway: early steps in the biosynthesis of aromatic compounds. Plant Cell 7:907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou W-T, Li W-Z, Chen Y, Jiang Y-L, Zhou C-Z. 2013. Structures of yeast Apa2 reveal catalytic insights into a canonical Ap4A phosphorylase of the histidine triad superfamily. J Mol Biol 425:2687–2698. doi: 10.1016/j.jmb.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 47.Akiyama M, Maki H, Sekiguchi M, Horiuchi T. 1989. A specific role of MutT protein: to prevent dG.dA mispairing in DNA replication. Proc Natl Acad Sci U S A 86:3949–3952. doi: 10.1073/pnas.86.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taddei F, Radman M, Maynard-Smith J, Toupance B. 1997. Role of mutator alleles in adaptive evolution. Nature 387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- 49.Collins S, Rost B, Rynearson TA. 2014. Evolutionary potential of marine phytoplankton under ocean acidification. Evol Appl 7:140–155. doi: 10.1111/eva.12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. [Google Scholar]
- 51.Tatusova T, Ciufo S, Federhen S, Fedorov B, McVeigh R, O'Neill K, Tolstoy I, Zaslavsky L. 2015. Update on RefSeq microbial genomes resources. Nucleic Acids Res 43:D599–D605. doi: 10.1093/nar/gku1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.