ABSTRACT
Alginate is a linear polysaccharide produced mainly by brown algae in marine environments. Alginate consists of a linear block copolymer made up of two monomeric units, β-d-mannuronate (M) and its C-5 epimer α-l-guluronate (G). Alginate lyases are polysaccharide lyases (PL) that degrade alginate via a β-elimination reaction. These enzymes play an important role in marine carbon recycling and also have widespread industrial applications. So far, more than 1,774 alginate lyase sequences have been identified and are distributed into 7 PL families. In this review, the folds, conformational changes during catalysis, and catalytic mechanisms of alginate lyases are described. Thus far, structures for 15 alginate lyases have been solved and are divided into 3 fold classes: the β-jelly roll class (PL7, -14, and -18), the (α/α)n toroid class (PL5, -15, and -17), and the β-helix fold (PL6). These enzymes adopt two different mechanisms for catalysis, and three kinds of conformational changes occur during this process. Moreover, common features in the structures, conformational changes, and catalytic mechanisms are summarized, providing a comprehensive understanding on alginate lyases.
KEYWORDS: alginate, alginate lyase, structure, conformational change, catalytic mechanism
INTRODUCTION
Alginate is a linear polysaccharide present in the cell walls and intracellular material of brown algae and accounts for ∼40% of their dry weight (1). It is also produced by some bacteria belonging to the Pseudomonas and Azotobacter genera (2). Alginate consists of two monomeric units, β-d-mannuronate (M) and its C-5 epimer α-l-guluronate (G). These units are arranged in three kinds of blocks: homopolymeric G blocks (PG), homopolymeric M blocks (PM), and heteropolymeric blocks of alternating M and G units (PMG) (3, 4). Alginate is an important source of fixed carbon in marine ecosystems and has been used as a viscosifier, a stabilizer, and a gelling agent in the food, printing, and biomaterials industries. Furthermore, alginate oligosaccharides have widespread applications in agriculture and pharmacology.
Alginate lyases are polysaccharide lyases (PL) that degrade alginate by β elimination of the glycosidic 1-4 O-linkages between sugar monomers, producing unsaturated oligosaccharides containing a 4-deoxy-l-erythro-hex-4-enopyranosyluronic acid at the new nonreducing terminus (4). Alginate lyases have been isolated from various sources, including a variety of bacteria and fungi, as well as viruses, marine algae, and marine mollusks (4). Based on amino acid sequence analysis, alginate lyases fall into seven PL families (PL5, -6, -7, -14, -15, -17, and -18) in the Carbohydrate-Active enZYmes (CAZY) database (5). Further, they have been also classified into three distinct groups based on their substrate specificities: PM-specific lyases (EC 4.2.2.3), PG-specific lyases (EC 4.2.2.11), and bifunctional lyases that can degrade both PM and PG (EC 4.2.2.-). According to their modes of action, alginate lyases have been further divided into two classes. Endolytic alginate lyases degrade long alginate chains, releasing oligosaccharides as the main products (6, 7), whereas exolytic alginate lyases degrade small oligosaccharides into monomers and/or dimers from the ends of the chains (8–10). Endolytic alginate lyases have been successfully utilized in industry for the preparation of protoplasts and the production of biologically relevant oligosaccharides (11–13).
In recent years, research on alginate lyases has made rapid and remarkable progress, especially in structures and catalytic mechanisms (14–17). A number of three-dimensional structures of alginate lyases from various PL families have been determined, and the catalytic mechanisms of alginate lyases from all seven PL families have now been revealed. Two recent reviews on alginate lyases were published in 2011 and 2015 (18, 19) but did not introduce the structures and catalytic mechanisms of alginate lyases in detail and, more importantly, did not include the structures and catalytic mechanisms of alginate lyases reported after 2015 (16, 17). Thus, in this review, we discuss the different structures and catalytic mechanisms of alginate lyases in detail.
STRUCTURAL DIVERSITY OF ALGINATE LYASES
Great progress has been made in the structural determination of alginate lyases in the past several years. Currently, 15 alginate lyase structures have been solved, with at least one representative structure from each family, including two members in PL5, one in PL6, six in PL7, two in PL14, one in PL15, one in PL17, and two in PL18. These structures have shown that alginate lyases from different families may adopt similar folds. Based on the classification of polysaccharide lyases proposed by Garron and Cygler (20), alginate lyases can be classified into 3 fold classes: β-jelly roll (PL7, -14, and -18), (α/α)n toroid (PL5, -15, and -17), and right-handed β-helix (PL6).
β-Jelly roll class.
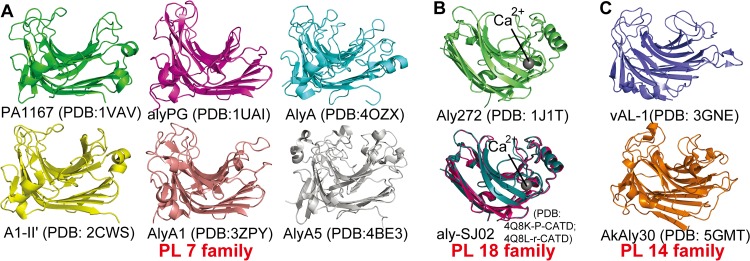
The β-jelly roll is the most common fold among alginate lyases. The β-jelly roll class contains 10 alginate lyase structures at the time of this publication, including 6 from PL7, 2 from PL14, and 2 from PL18 (Fig. 1). In general, this fold consists mainly of two curved antiparallel β-sheets (SA and SB) that are bent in the middle by nearly 90°, coming together to form a globular shape. The inner concave sheet (SA) shapes a cleft that harbors the catalytic site, and the alginate substrates enter the cleft and interact with the active-site residues. Loops linking the inner (SA) and outer (SB) sheets are variable in length and sequence (20). Three adjacent β-strands in the center of the SA sheet of PL7 lyases are well ordered, and the conserved residues on these strands play important roles in the catalytic reaction (21).
FIG 1.
Alginate lyase structures adopting the β-jelly roll fold. (A and B) Structural representatives of the PL7 (A) and PL18 (B) families. Representation of the aly-SJ02 structure includes both the P-CATD (pink) and r-CATD (deep teal). (C) Structures of alginate lyases in PL14.
Nearly one-half of all known alginate lyase sequences have been classified into the PL7 family, and structural information is available for six of them, including PA1167 from Pseudomonas aeruginosa PAO1 (PDB code 1VAV) (22), AlyA from Klebsiella pneumoniae (PDB code 4OZX), alyPG from Corynebacterium sp. ALY-1 (PDB code 1UAI) (23), AlyA1 (PDB code 3ZPY), and AlyA5 (PDB code 4BE3) from Zobellia galactanivorans (24), and A1-II′ from Sphingomonas sp. A1 (PDB code 2CWS) (21) (Fig. 1A). Among these enzymes, PA1167, alyPG, AlyA1, and A1-II′ are endolytic, while AlyA5 is exolytic (24), and AlyA has not been characterized. PA1167 represents the first solved alginate lyase structure that adopts this fold (22). PA1167 consists of three short α-helices and two antiparallel β-sheets, SA and SB, that consist of eight and seven β-strands, respectively. A1-II′ is the best-studied enzyme in PL7 due to detailed analysis of the structures of mutant variants complexed with alginate tri- or tetrasaccharides (21, 25). Similar to the structure of PA1167, the overall structure of A1-II′ is composed of four helices and two β-sheets, including nine β-strands in SA and seven in SB. The overall structures of the other four enzymes (AlyA, AlyA5, alyPG, and AlyA1) resemble the two above. Except for the exolytic enzyme AlyA5, the architectures of the active-site centers of the PL7 alginate lyases are all wide open, corresponding to the endolytic character of PA1167, A1-II′, alyPG, and AlyA1, which also suggests that AlyA is most likely an endolytic enzyme. The structural determinant of the exolytic activity of AlyA5 was determined to be a long loop (residues 304 to 318) that blocks the catalytic groove at one end, leading to a pocket-like architecture.
Most PL18 alginate lyases contain an N-terminal extension that is excised during protein maturation. Thus far, all of the alginate lyases characterized from PL18 (AlyA, alyPEEC, and aly-SJ02) have been bifunctional and endolytic. The structures of two PL18 alginate lyases from Pseudoalteromonas, Aly272 and aly-SJ02, have been solved (Fig. 1B). Aly272 (PDB code 1J1T) was the first PL18 structure deposited in the PDB; unfortunately, no further research on this enzyme has been reported. Structural and functional analyses of aly-SJ02 were reported in 2014 (14). The mature aly-SJ02 enzyme exhibits the typical β-jelly roll fold. Akin to the active-site architectures of endolytic alginate lyases in PL7, three adjacent β-strands in aly-SJ02, i.e., SA2, SA3, and SA4, form a catalytic groove that is wide open at both ends. aly-SJ02 shows two main structural differences compared with alginate lyases from the PL7 family. First, according to the structures of the recombinant catalytic domain (r-CATD; PDB code 4Q8L) and the catalytic domain of the precursor (P-CATD, PDB code 4Q8K) of aly-SJ02, the unique N-terminal extension present in PL18 alginate lyase precursors functions as an intramolecular chaperone that mediates the correct folding of the catalytic domain. Second, a structural divalent metal ion, Ca2+, observed in both Aly272 and aly-SJ02, is far away from the active centers, which is important for maintaining the enzymatic activity of aly-SJ02, but does not directly participate in the catalytic reaction (14).
The PL14 family is the only family that contains alginate lyases from bacteria, eukaryotes, and viruses. In this family, four gastropod alginate lyases (AkAly30, LbAly28, HdAly, and HdAlex) and three PLs from the Chlorella virus (CL2, A215L, and vAL-1) have been characterized, but no data regarding the bacterial enzymes are available. At this time, the structures of two PL14 alginate lyases have been solved, including vAL-1 from Paramecium bursaria Chlorella virus CVK2 (PDB code 3GNE) (26) and AkAly30 from Aplysia kurodai (PDB code 5GMT) (16) (Fig. 1C). Among the eukaryotic enzymes, AkAly30, LbAly28, and HdAly are M specific and endolytic while HdAlex is exolytic. Most of the eukaryotic enzymes optimally degrade alginate under mesophilic conditions at neutral pH. Interestingly, two PLs of viral origin (CL2 and vAL-1) can function at alkaline pH. Wild-type (WT) vAL-1 prefers to exhibit alginate lyase activity at alkaline pH; however, at neutral pH, its activity on alginate decreases and the enzyme shows a preference for glucuronic acid containing polymers such as pectin. WT vAL-1 is comprised of two domains, an N-terminal domain (NTD) for cell wall attachment and a C-terminal catalytic domain. It is also reported that the N-terminal domain hinders the alginate lyase activity of WT vAL-1 at neutral pH (26). For vAL-1, only the structure of the C-terminal catalytic domain, vAL-1(S), has been solved, and it adopts a β-jelly roll fold. Although structurally similar to PL7 and -18 family lyases, vAL-1(S) has two additional small β-sheets, SC and SD, both of which consist of two β-strands. Interestingly, vAL-1(S) shows two pH-dependent modes of action toward alginate, functioning endolytically at pH 7.0 and exolytically at pH 10.0. These two modes of action of vAL-1(S) are greatly influenced by the electric charges in the active site at different pH levels. AkAly30 is an endotype alginate lyase and shares only 23% sequence identity with vAL-1. Despite the structural similarities with vAL-1(S), AkAly30 shows some differences in the conformations of two loops surrounding subsite −1, resulting in their different modes of action. A η4 loop in AkAly30 leads to the closed special arrangement of subsite −1, distinct from the corresponding loop in vAL-1(S). Moreover, the η2 loop of AkAly30 is cross-linked by a disulfide bond, which is not conserved in vAL-1(S). On this η2 loop, a Gly that is important for AkAly30 activity is replaced by Tyr in vAL-1(S) (16). The residues forming the catalytic clefts in PL14 enzymes are distinct from those in PL7 and PL18, including amino acids in both the loops and the β-strands (16). Particularly, conserved amino acids that are known to be crucial for catalysis in PL7 and PL18 lyases, including Arg, Gln, His, and Tyr, are replaced with Gly, Gly, Phe, and Phe, respectively, in vAL-1 and AkAly30 (26).
(α/α)n toroid class.
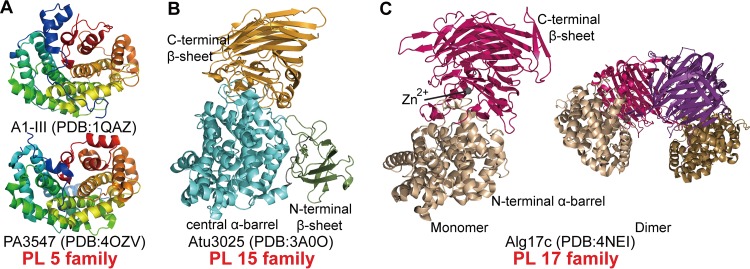
The alginate lyases in families PL5, -15, and -17 contain a common catalytic domain that adopts a barrel architecture formed by several antiparallel α-helices (helical hairpins). The hairpins are arranged counterclockwise (looking from the top of the hairpin), and their number, n, varies from three to seven. Hence, we classified these folds into the (α/α)n toroid class. There are four solved alginate lyase structures in this class, including two from PL5, one from PL15, and one from PL17. Among these enzymes, those in PL5 are single-domain proteins that have only a barrel catalytic domain (27) whereas those in PL15 and PL17 are multidomain proteins containing one or two other domains in addition to the catalytic domain (10, 15) (Fig. 2). The α-helices in the catalytic domain are divided into two layers, the inner and the outer helices, which form a deep tunnel-like cleft to accommodate the substrate. The inner helices run in approximately the same direction around the core of the barrel, whereas the outer ones are oriented in the opposite direction (27).
FIG 2.
Spatial arrangements of alginate lyase structures adopting the (α/α)n toroid fold. (A) The structures of A1-III and PA3547 from the PL5 family are depicted in rainbow colors from blue in the N-terminal region to red in the C-terminal region. (B) Structure of Atu3025 from the PL15 family. The C-terminal, central, and N-terminal domains are colored in orange, cyan, and green, respectively. (C) Representative structures of the Alg17c monomer (left) and dimer (right) from the PL17 family. The N- and C-terminal domains of the upper monomer are colored in wheat and hot pink, respectively. The N- and C-terminal domains of the second monomer are colored in sand and purple, respectively.
All PL5 alginate lyases characterized to date are endolytic and M-specific enzymes from bacteria. It is worth noting that Smlt1473 is the sole PL5 enzyme that is promiscuous and is able to use alginate, poly-d-glucuronic acid (GlcA), and hyaluronan as the substrates (28). The structures of two PL5 alginate lyases have been solved: A1-III from Sphingomonas sp. (PDB code 1QAZ) and PA3547 from Pseudomonas aeruginosa PAO1 (PDB code 4OZV) (Fig. 2A). A1-III is the most studied enzyme in the PL5 family and was the first alginate lyase shown to adopt an (α/α)n toroid fold (27, 29, 30). A1-III is comprised of 12 α-helices, six inners and five outers, which form an α6/α5 barrel consisting of four hairpin repetitions. Alginate molecules penetrate into the tunnel-like cleft, surrounded by the inner helices, to interact with residues of the catalytic site. PA3547 is structurally similar to A1-III with a root mean square deviation of 2.7 Å, and the main differences occur in the N- and C-terminal regions of these two enzymes. Although its structure has been determined and released, biochemical characterization of PA3547 has not been reported.
The PL15 family contains exolytic bacterial alginate lyases. Exotype alginate lyase Atu3025 (PDB code 3A0O) from Agrobacterium tumefaciens, the only solved structure in this family, is comprised of an N-terminal small β-sheet domain (residues 11 to 116), a central α/α-barrel domain (residues 128 to 491), and a C-terminal antiparallel β-sheet domain (residues 495 to 773) (Fig. 2B) (10). The N-terminal small β-sheet domain, whose function remains enigmatic, is made up of two antiparallel β-sheets consisting of seven β-strands. The central barrel domain is composed of 15 α-helices, 12 of which, including six inners and six outers, contribute to the formation of the (α/α)6-barrel catalytic domain. The C-terminal domain (CTD) is comprised of 18 β-strands, forming a three-layered antiparallel β-sheet sandwich structure. In contrast to the tunnel-like catalytic centers of PL5 alginate lyases, the active-site center of Atu3025 is located between the α/α-barrel and the C-terminal β-sheet domains, forming a deep pocket to accommodate alginate. The substrate is bound across the two domains, interacting predominantly with residues of the barrel domain but also with several residues of the C-terminal domain. Its exolytic mode action is related to the location of the bound substrates, which are introduced in more detail in “Conformational Changes during Catalysis” below.
Alginate lyases of the PL17 family are oligoalginate lyases that can degrade alginate and alginate oligosaccharides into monosaccharides by an exotypic mode of action. Alg17c (PDB code 4NEI) from Saccharophagus degradans 2-40 is the only PL17 alginate lyase for which a crystal structure has been determined (Fig. 2C) (15). Alg17c (PL17) and Atu3025 (PL15) are both exolytic enzymes; however, their structures are significantly different. Atu3025 is a three-domain monomeric protein, while Alg17c consists of only two domains and forms homodimers. Compared with Atu3025, Alg17c is also comprised of an α6/α6-barrel (residues 30 to 365) joined to a C-terminal antiparallel β-sheet (residues 370 to 735) but lacks the N-terminal β-sheet domain. The barrel domain of Alg17c is composed of 13 helices, 12 that form the α6/α6-barrel and 1 additional helix that is shored up against the side of the barrel, maintaining its rigidity. The C-terminal domain displays a three-layered antiparallel β-sheet structure and has four small helices near the top layer, interjecting between the two domains. Similar to what is seen in Atu3025, the substrate is also bound between the barrel and the β-sheet domains of the Alg17c monomer. Alg17c homodimerization results from the insertion of a β-turn (residues 660 to 672) from the β-sheet domain of one monomer into the barrel domain of the other monomer. In addition, a Zn2+ in Alg17c is located at the interface of the two domains, likely establishing the orientation of the residues interacting with the substrate (15).
β-Helix class.
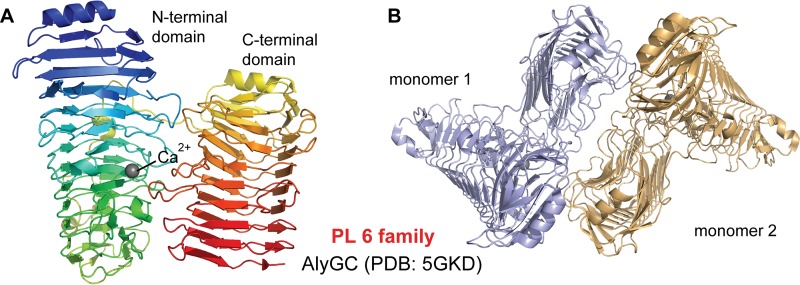
The PL6 family contains various PLs, including two dermatan sulfate lyases and alginate lyases with diverse substrate specificities and modes of action (17, 31–33). AlyGC from the PL6 family is an exotype G-specific alginate lyase and is now the first and only known alginate lyase that adopts a right-handed β-helix fold (Fig. 3) (21). The right-handed β-helix fold was first observed in a pectate lyase (pectate lyase C) from Erwinia chrysanthemi in 1993 (34) and has now been shown to be common among enzymes from PL1, -3, and -9 (35–37). AlyGC forms dimers in solution, and there is only a 2-fold rotation axis between the individual monomers. Each monomer of AlyGC is comprised of two domains, the N-terminal domain and the C-terminal domain, both of which adopt the β-helix fold. This is distinct from other reported proteins that share this fold but contain only a single β-helix domain, including proteins in PL1, -3, and -9 and chondroitinase B from PL6 (35–38). The architecture of the β-helix fold is composed of three β-sheets: PB1, PB2, and PB3. The turns (T) or loops between the two β-sheets are referred to as T1 (between PB1 and PB2), T2 (between PB2 and PB3), and T3 (between PB3 and PB1), and the arrangement of one coil of the β-helix is PB1-T1-PB2-T2-PB3-T3. PB1 and PB2 are nearly antiparallel, and PB3 is almost perpendicular to PB2 (38). The NTD of AlyGC consists of 12 PB1 strands, 14 PB2 strands, and 14 PB3 strands. The CTD consists of 9 PB1 strands, 11 PB2 strands, and 9 PB3 strands (17). Interestingly, the active site of AlyGC is located in the NTD but the CTD is also indispensable for the activity of AlyGC. The active-site center is a long cleft localized on the surface of the NTD, surrounded by loops and β-strands from the NTD and one loop (residues 627 to 638) from the CTD. One end of the cleft is blocked, leading to the exolytic mode of action of the enzyme (17). In addition, a Ca2+ ion is located at the catalytic center of each molecule, and its role in catalysis of PL6 enzymes is introduced in “Diversity in Catalytic Mechanisms” below.
FIG 3.
Structures of AlyGC depicting the β-helix fold. (A) The AlyGC monomer depicted with rainbow colors from blue in the N-terminal region to red in the C-terminal region. (B) Structure of the AlyGC dimer. Monomer 1 and monomer 2 are colored in light blue and wheat, respectively.
CONFORMATIONAL CHANGES DURING CATALYSIS
During the cleavage of alginate glycosidic bonds via β-elimination, conformational changes of the enzymes have been shown to always occur, such as lid loop movements, domain rotation movements, and subunit rotation movements. Some changes are specific, and others are common to the degradation of alginates and other polysaccharides.
Lid loop movements.
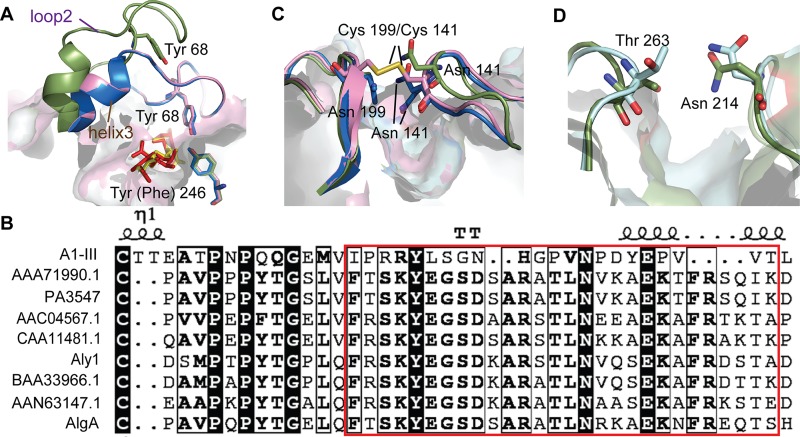
Lid loop movement during catalysis is a common feature in alginate lyases from PL5, -7, and -18. For PL5 alginate lyases, this movement occurs in just one loop, which is akin to a lid above the active-site center (29). This is in contrast to lyases in the PL7 and -18 families, which have two lid loops over their active-site centers (14, 25) (Fig. 4).
FIG 4.
Conformational changes of lid loops identified in alginate lyases. (A) Lid loop (residues 64 to 85) of PL5 A1-III. A structural representation of the apoenzyme (H192A; PDB 4E1Y) is shown in green. The two holoenzymes, Holo H192A (PDB 4F10) and Holo Y246F (PDB 4F13), are shown in blue and pink, respectively. The bound substrates are depicted by red/yellow sticks. (B) Residues of the loop region in PL5 alginate lyases. The GenBank accession numbers of the protein sequences are BAB03312.1 (A1-III), AAG06935.1 (PA3547), AEW23144.1 (Aly1), and AIY22644.1 (AlgA). (C) Flexible loops (loop1, residues 133 to 145, and loop2, residues 193 to 203) of PL7 A1-II′. Two ligand-free structures, 2CWS and 2Z42, are shown in green and blue, respectively. The mutant (N199C/N141C; PDB 2ZA9), shown in pink, has a disulfide bond linking the two flexible loops. (D) The flexible loops (loop1, Ala208-Gly217, and loop2, Ala260-Thr265) of PL18 aly-SJ02, with the P-CATD (PDB 4Q8K) and r-CATD (PDB 4Q8L), are depicted in green and blue, respectively.
The lid loop present in the PL5 alginate lyase A1-III (residues 64 to 85) consists of 16 residues (residues 64 to 79) from loop2 (residues 44 to 79), connecting helix2 and helix3, and 6 residues (residues 80 to 85) from helix3 (residues 80 to 105). The A1-III lid loop is able to migrate about 14 Å from an open form (apoenzyme, H192A) to a closed form (holoenzyme, H192A and Y246F). The loop of the apoenzyme protrudes into the solvent and bends at the middle of loop2 and in the front of helix3 when the enzyme binds a substrate (Fig. 4A). The conformational change of the lid loop from the open to the closed position results in coverage of the active-site cleft. This lid loop movement is a near-rigid-body motion with hinges around Ile64 and residues 82 to 85 (for details about the mechanism of this motion, see Gerstein and Krebs [39]). In the holoenzyme (H192A and Y246A), the closed lid loop interacts with the bound substrate, and the Tyr68 residue on this loop activates the catalytic residue Tyr246 through the formation of a hydrogen bond (Fig. 4A). Despite structural conservation, the amino acid sequence of the lid loop region is not strictly conserved among PL5 alginate lyases, and conservative replacements are found for Arg67 (Lys), Asn72 (Asp), Gly74 (Ala/Ser), and Val76 (Leu) (Fig. 4B).
Similar lid loop movements are observed in A1-II′ from PL7 (25) and aly-SJ02 from PL18 (14); however, each of these enzymes contains two mobile loops. The function of these two flexible loops was first identified in A1-II′ because the loops were found to be in different positions (open and closed forms) in two different ligand-free structures of the same protein (PDB codes 2CWS and 2Z42, respectively). Mobile loops L1 and L2 are located over the active-site cleft and interact through a hydrogen bond formed by two asparagine residues, Asn141 on L1 and Asn199 on L2. A double mutant, A1-II′ N141C/N199C, has a disulfide bond between two cysteine residues and retains little enzymatic activity, demonstrating that the flexibility of these loops is essential for access of the substrate to the active site and the concomitant release of products after the reaction (Fig. 4C). Almost all enzymes in PL7 have two loops corresponding to L1 and L2 of A1-II′, suggesting that lid loop movement during catalysis is a mechanism shared by PL7 enzymes (22–24). Similarly, two mobile loops over the substrate-binding pocket are present in the PL18 family lyase aly-SJ02 (14), and a hydrogen bond is formed between Asn214 on L1 and Thr263 on L2 (Fig. 4D). Molecular dynamic simulations indicated that the distance between the side chains of Asn214 and Thr263 increases from 3.2 Å in the closed state to 11.5 Å in the open state, making it possible for an alginate molecule to enter the substrate-binding pocket (14). The enlargement of space between the loops of A1-II′ and aly-SJ02 is predominantly caused by conformational changes in the side chains of the loop residues, which is distinct from the nearly-rigid-body motion of the A1-III lid loop. In addition, chondroitin and hyaluronate lyases in PL8 (40, 41) have also been shown to possess flexible loops involved in substrate binding and/or catalysis, indicating that flexible lid loops, as well as their movements, are a common mechanism more broadly shared by several PL families.
Domain rotation movements.
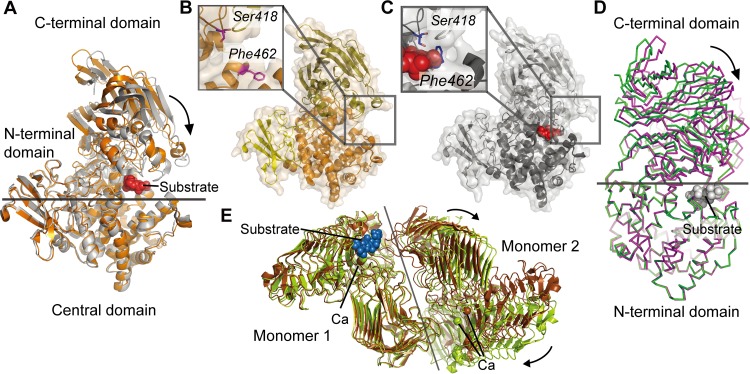
Domain rotation movements occur on the C-terminal β-sheet domains of alginate lyases from PL15 and PL17 during catalysis (Fig. 5A, B, C, and D). In the presence of substrate, the C-terminal β-sheet domain of Atu3025 (PL15) can rigidly rotate ∼10° compared with the apoenzyme (Fig. 5A). During the course of this hinge bending motion, the oligosaccharide substrate is embedded into the pocketlike active-site center. Phe462, located on one side of the pocket, then rotates to interact with Ser418 positioned at the opposite side of the active pocket (Fig. 5B and C) (10). Similarly, the β-sheet domain of Alg17c (PL17) undergoes a rigid body rotation of ∼12° upon ligand binding (Fig. 5D), leading to the rearrangement of residues around the active center to interact with the bound substrate (15).
FIG 5.
Domain and subunit rotation movements of alginate lyases from PL15, PL17, and PL6. (A) Structural alignment of the wild-type (WT) Atu3025 (PDB 3A0O; orange) from the PL15 family and its complex with an unsaturated trisaccharide, ΔGGG (PDB 3AFL; gray). Structures were aligned using the N-terminal domains. The ΔGGG substrate is shown as red spheres. (B and C) Conformational changes of the residues around the active centers in the WT structure (B) and the Atu3025-ΔGGG complex (C). (D) Structural alignment of WT Alg17c (PDB 4NEI; green) and its complex with ΔMMG (PDB 4OJZ; purple) from the PL17 family. Structures were aligned by the N-terminal domains. (E) Structural alignment of the dimers of the WT AlyGC (PDB 5GKD; brown) and its complex with the saturated tetrasaccharide MMMM (PDB 5GKQ; olive) from the PL6 family. Structures were aligned using one subunit. MMMM is shown as blue spheres.
Similar domain movement has also been reported in a PL8 hyaluronate lyase that adopts a similar modular architecture with an α/α-barrel linked to a β-sheet domain (40). Therefore, such domain movement during catalysis may be a common feature of enzymes that belong to the multidomain (α/α)n toroid class of PLs, despite the fact that enzymes from different PL families do not share a high degree of sequence similarity.
Subunit rotation movements.
Among all the alginate lyases, subunit rotation has been found only in the PL6 AlyGC (17). The two dimer subunits of AlyGC have a 2-fold rotation axis, and there are two identical active centers in each dimer. However, conformational alignment of WT AlyGC with a catalytically inactive mutant structure complexed with tetramannuronate showed that rotation occurs in one subunit of the dimer when a substrate is bound. This subunit rotation movement results in the enlargement of the entrance of one active-site center that binds the substrate, while the other remains small, indicating that only one monomer of dimeric AlyGC may bind substrate at any given time (Fig. 5E). The entrance state that shifts from “open” to “closed” may be related to the formation of a hydrogen bond between His192 from the NTD of one subunit and Asp526 from the CTD of the other. However, it is still unknown why AlyGC functions as a dimer but binds alginate substrates asynchronously.
DIVERSITY IN CATALYTIC MECHANISMS
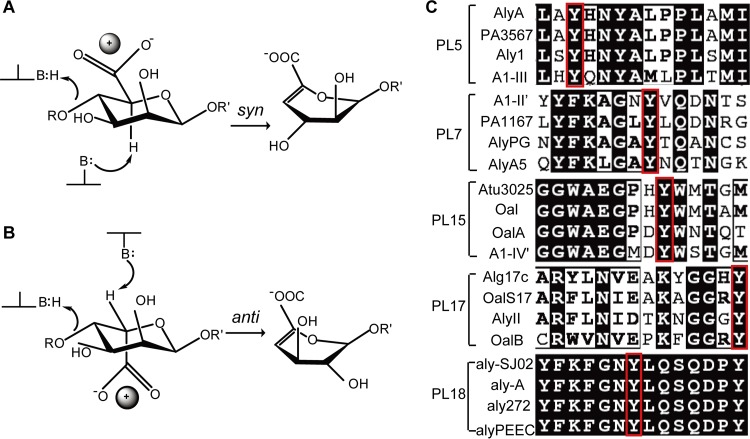
The β-elimination reaction that alginate lyases utilize to degrade alginate occurs in three steps: (i) the removal of the negative charge on the carboxylate anion to reduce the pKa of the H-5 proton; (ii) a general base-catalyzed abstraction of the proton on C-5 leading to an enolate intermediate; (iii) electron transfer from the carboxyl group resulting in the formation of a double bond between C-4 and C-5 (4, 42) and finally cleavage of the O-glycosidic bond (43, 44). This reaction requires the participation of a Brønsted base to accept the H-5 proton and a Brønsted acid as a proton donor (20). Based on the configurations at C-4 and C-5, the abstracted proton and the C-4 bridging oxygen can be either on the same side (syn configuration) or on opposite sides (anti configuration) of the uronic acid ring (20). For M residues, the C-5 proton and the departing oxygen on C-4 lie syn relative to each other. This is in contrast to G residues, where they lie anti relative to one another (43) (Fig. 6). The catalytic mechanisms of alginate lyases can be divided into two groups based on differences in the mechanism utilized for neutralization of the C-5 carboxyl and in the Brønsted bases and acids: (i) His (or Tyr)/Tyr β elimination and (ii) metal (Ca2+)-assisted β elimination. Available data indicate that the type of enzymatic mechanism used by PLs is conserved within each family.
FIG 6.
Catalytic mechanisms and amino acid conservation of catalytic residues of alginate lyases. (A and B) Elimination from d-mannuronate (A) and l-guluronate (B) via syn and anti pathways, respectively. The positively charged neutralizer (ball) can be either a metal ion or several amino acid residues. In the syn elimination reaction, the catalytic base and acid can be the same amino acid residue; however, in the anti reaction, the base and acid are two different residues. (C) The strictly conserved Tyr residue is used for catalysis in the PL5, -7, -15, -17, and -18 families. Tyr acts as the general catalytic acid in these enzymes. Four sequences of characterized alginate lyases in each family are aligned. The conserved Tyr is boxed in red.
His (or Tyr)/Tyr β elimination.
His (or Tyr)/Tyr β elimination has been widely adopted by alginate lyases, with the exception of members in the PL6 family. For these alginate lyases, the negative charge of the carboxylate anion is predominantly neutralized by Glu, His, Arg, or Asn, which forms a hydrogen bond with the acidic group to shift the equilibrium toward the enolate tautomeric form (43). In this case, a conserved Tyr serves as the general acid. Enzymes that utilize this mechanism can be subdivided based on their preferred catalytic base. Tyr/Tyr β elimination uses the same Tyr to serve as the general base, while enzymes that employ His (Tyr′)/Tyr β elimination utilize a His residue or another Tyr as the base.
(i) Tyr/Tyr β elimination.
All PL5 family alginate lyases are M specific. The catalytic mechanism of A1-III was reported in 2001 (29, 30) and was the first reported catalytic mechanism for alginate lyases. Based on the structure of A1-III complexed with a trisaccharide cleavage product (ΔMMM, where ΔM is the unsaturated sugar residue) bound at subsites −3 to −1 from the nonreducing end, Tyr246 was predicted to act as both the catalytic base and the proton donor in a syn elimination mechanism. In this proposed mechanism, the negative charge on the C-5 carboxylate group is first neutralized by Arg239 and Asn191 so that the C-5 proton can be more easily removed. Next, Tyr246 extracts a proton from C-5 of mannuronic acid, forming a carboxylate dianion intermediate. Finally, Tyr246 donates a proton to the oxygen of the glycosidic bond, forming a double bond between C-4 and C-5. Similar to that of A1-III, aly-SJ02 is predicted to use the Tyr353 residue both as the catalytic acid and as the base, and the conserved residues in the active center, including Arg219, Lys223, Gln257, His259, Tyr347, and Lys349, recognize and stabilize the carboxyl group of the substrate (14).
(ii) His (Tyr′)/Tyr β elimination.
In the five structured PL7 alginate lyases, AlyA1 and alyPG are G specific, and PA1167 is MG specific. A1-II′ and AlyA5 are bifunctional, and both prefer to degrade PG. The first PL7 alginate lyase structure (PA1167) was reported in 2004 (22); however, the molecular mechanism of catalysis was not determined until a structure of an A1-II′ mutant complexed with an alginate tetrasaccharide was obtained in 2008 (25). Analysis of the structure of an A1-II′ mutant (H191N/Y284F) complexed with an alginate tetrasaccharide (GGMG) suggested that Gln189 functions as a neutralizer for the carboxyl group, His191 acts as the general base, and Tyr284 acts as the general acid. In addition, the His/Tyr β elimination mechanism has been widely adopted by polysaccharide lyases utilizing anti elimination, such as heparin lyase in the PL13 family (45). Atu3025, an exolytic and bifunctional alginate lyase, is the only PL15 alginate lyase whose catalytic mechanism has been clarified. Atu3025 uses His311 and Tyr365 as the catalytic base and acid, respectively, based on analysis of the structure of a mutant variant (H531A) in complex with an unsaturated alginate trisaccharide (ΔGGG). Positively charged residues, such as Arg199, Arg314, and His531, are proposed to play a role in stabilizing or neutralizing the negative charge on the carboxylate anion (10).
The molecular mechanism utilized by Alg17c from PL17 is slightly distinct from the typical His/Tyr β elimination. This exolytic bifunctional enzyme uses Tyr′/Tyr β elimination. Analysis of the structure of a catalytic mutant variant (Y258A) complexed with ΔMMG bound at subsites −1 to +2 indicated that Tyr450 is the general base that abstracts the proton at C-5, Tyr258 functions as the general acid, and Asn201 and His202 stabilize the negative charge of the carboxylate.
The catalytic mechanism(s) utilized by PL14 alginate lyases remains enigmatic. The viral enzyme, vAL-1, degrades alginate and poly-GlcA in a pH-dependent manner, and Lys197 and Ser219 are supposed to be crucial for vAL-1 activity at both pH 7.0 and 10.0, based on both structural and mutational analyses (16, 26, 46). For the M-specific eukaryotic enzyme, AkAly30, its substrate-binding models showed that Lys99 forms ionic bonds with the M residue at the +1 subsite, and Tyr140 and Tyr142 are coupled together and form a hydrogen bond network with the glycosidic bond (16, 26, 46). Although the molecular details are still unclear, these data implied that the catalytic mechanisms of vAL-1and AkAly30 are likely to be significantly different.
Despite significant variation in the protein folds of alginate lyases, the His (Tyr′)/Tyr mechanism is well adapted to both syn and anti types of elimination. The reaction can adopt a syn (Fig. 6A) or an anti (Fig. 6B) elimination depending on the identity of the sugar residue at the +1 subsite. Therefore, in the case of bifunctional alginate lyases, they can undertake both syn and anti elimination of the substrate (Fig. 6A and B). However, G-specific lyases can carry out only anti elimination (Fig. 6B), while M-specific lyases can perform only syn elimination (Fig. 6A). In this mechanism, Glx (Glu and Gln) and/or Asx (Asp and Asn) usually act as neutralizers that neutralize the carboxylic group of the uronate at the +1 subsite. Further, some positively charged residues, including His and Arg, assist the neutralizer(s) through a network of hydrogen bonds between the enzyme and the substrate. A Tyr residue, which is strictly conserved in alginate lyases utilizing this mechanism, functions as the Brønsted acid (Fig. 6C). In syn elimination, typified by A1-III from PL5, the enzymes prefer to use the same Tyr as the Brønsted base; however, enzymes that perform anti elimination prefer to utilize a His residue as the Brønsted base.
Metal-assisted β elimination.
Metal-assisted β elimination was first described for the PL6 AlyMG, which uses Ca2+-assisted β elimination for alginate degradation (33). Structural analysis of AlyGC indicated that a Ca2+ ion is coordinated by Asn181, Glu213, Glu215, and Glu184 in the active-site center, which are conserved in all characterized PL6 alginate lyases. Ca2+ is suggested to neutralize the carboxylic group of an alginate residue at the +1 subsite, similar to the role of Glx and Asx in enzymes using the His (Tyr)/Tyr mechanism (Fig. 6B). The interaction between Ca2+ and the carboxylic group likely enhances the reactivity of the C-5 proton (47). In this reaction, Lys220 and Arg241 function as the Brønsted base and acid, respectively (Fig. 6B). This metal-assisted β elimination mechanism is universal in enzymes adopting the β-helix fold (48).
Interestingly, by combining knowledge of the catalytic mechanisms, i.e., His (Tyr)/Tyr versus the metal-assisted mechanism, and the complexity of the three-dimensional (3-D) structures, we found that most of the reported PL enzymes utilizing a single residue as both the acid and base have a preference for M residues, such as A1-III from PL5 and aly-SJ02 from PL18. On the other hand, enzymes utilizing two different amino acid residues for catalysis, such as A1-II′ from PL7 and AlyGC from PL6, prefer G residues. M and G are epimeric; therefore, there may be some relevance between the catalytic residues and the sugar residue present at the +1 subsite for more-efficient electron transfer. However, due to the limited number of available structures and the paucity of experimental data, this hypothesis will require further data for confirmation.
FUTURE PROSPECTS
Despite great progress toward elucidating the structures and catalytic mechanisms of alginate lyases, much work on these complex enzymes is still required. For example, Aly is an alginate lyase that remains unclassified in the CAZY database due to low sequence similarity with known PL families (49). Therefore, the structure and catalytic mechanism of this enzyme need to be defined to determine whether it represents another distinct family of PLs. As more novel alginate lyases are being reported, more information regarding the structures and catalytic mechanisms of these enzymes are anticipated. In addition, many alginate lyases are promiscuous and are able to degrade other polysaccharides beyond alginate. For example, Smlt1473 from PL5 can degrade poly-GlcA, hyaluronan, and PM. However, the molecular basis of this observed polyspecificity needs further exploration. Moreover, heparinase I from PL13, which has a β-jelly roll fold, is structurally similar to PL7 and PL18 alginate lyases. The PL6 family contains both alginate lyases and chondroitinase B lyases that share structural similarities. These similarities may indicate possible evolutionary relationships between polysaccharide lyases. Undisputedly, further research on these topics will broaden our understanding of alginate lyases and offer a better basis for their application.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation of China (grants 31630012, 31290230, 31290231, and 31670038), the National Key R&D Program of China (grant 2016YFA0601303), the Program of Shandong for Taishan Scholars (TS20090803), the AoShan Talents Cultivation Program (supported by Qingdao National Laboratory for Marine Science and Technology) (2017ASTCP-OS14), and the National Postdoctoral Program for Innovative Talents (BX201700145).
REFERENCES
- 1.Fischer FG, Dorfel H. 1955. Polyuronic acids in brown algae. Hoppe-Seylers Z Physiol Chem 302:186–203. doi: 10.1515/bchm2.1955.302.1-2.186. [DOI] [PubMed] [Google Scholar]
- 2.Rehm BH, Valla S. 1997. Bacterial alginates: biosynthesis and applications. Appl Microbiol Biotechnol 48:281–288. doi: 10.1007/s002530051051. [DOI] [PubMed] [Google Scholar]
- 3.Haug A, Larsen B, Smidsrød O. 1967. Studies on the sequence of uronic acid residues in alginic acid. Acta Chem Scand 21:691–704. doi: 10.3891/acta.chem.scand.21-0691. [DOI] [Google Scholar]
- 4.Wong TY, Preston LA, Schiller NL. 2000. Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu Rev Microbiol 54:289–340. doi: 10.1146/annurev.micro.54.1.289. [DOI] [PubMed] [Google Scholar]
- 5.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HT, Ko HJ, Kim N, Kim D, Lee D, Choi IG, Woo HC, Kim MD, Kim KH. 2012. Characterization of a recombinant endo-type alginate lyase (Alg7D) from Saccharophagus degradans. Biotechnol Lett 34:1087–1092. doi: 10.1007/s10529-012-0876-9. [DOI] [PubMed] [Google Scholar]
- 7.Chen XL, Dong S, Xu F, Dong F, Li PY, Zhang XY, Zhou BC, Zhang YZ, Xie BB. 2016. Characterization of a new cold-adapted and salt-activated polysaccharide lyase family 7 alginate lyase from Pseudoalteromonas sp. SM0524. Front Microbiol 7:1120. doi: 10.3389/fmicb.2016.01120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HT, Chung JH, Wang D, Lee J, Woo HC, Choi IG, Kim KH. 2012. Depolymerization of alginate into a monomeric sugar acid using Alg17C, an exo-oligoalginate lyase cloned from Saccharophagus degradans 2-40. Appl Microbiol Biotechnol 93:2233–2239. doi: 10.1007/s00253-012-3882-x. [DOI] [PubMed] [Google Scholar]
- 9.Park HH, Kam N, Lee EY, Kim HS. 2012. Cloning and characterization of a novel oligoalginate lyase from a newly isolated bacterium Sphingomonas sp. MJ-3. Mar Biotechnol (NY) 14:189–202. doi: 10.1007/s10126-011-9402-7. [DOI] [PubMed] [Google Scholar]
- 10.Ochiai A, Yamasaki M, Mikami B, Hashimoto W, Murata K. 2010. Crystal structure of exotype alginate lyase Atu3025 from Agrobacterium tumefaciens. J Biol Chem 285:24519–24528. doi: 10.1074/jbc.M110.125450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue A, Mashino C, Kodama T, Ojima T. 2011. Protoplast preparation from Laminaria japonica with recombinant alginate lyase and cellulase. Mar Biotechnol (NY) 13:256–263. doi: 10.1007/s10126-010-9290-2. [DOI] [PubMed] [Google Scholar]
- 12.Boyen C, Bertheau Y, Barbeyron T, Kloareg B. 1990. Preparation of guluronate lyase from Pseudomonas alginovora for protoplast isolation in Laminaria. Enzyme Microb Technol 12:885–890. doi: 10.1016/0141-0229(90)90027-N. [DOI] [Google Scholar]
- 13.Butler DM, Østgaard K, Boyen C, Evans LV, Jensen A, Kloareg B. 1989. Isolation conditions for high yields of protoplasts from Laminaria saccharina and L. digitata. J Exp Bot 40:1237–1246. doi: 10.1093/jxb/40.11.1237. [DOI] [Google Scholar]
- 14.Dong S, Wei TD, Chen XL, Li CY, Wang P, Xie BB, Qin QL, Zhang XY, Pang XH, Zhou BC, Zhang YZ. 2014. Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18. J Biol Chem 289:29558–29569. doi: 10.1074/jbc.M114.584573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park D, Jagtap S, Nair SK. 2014. Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases. J Biol Chem 289:8645–8655. doi: 10.1074/jbc.M113.531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin HM, Miyakawa T, Inoue A, Nishiyama R, Nakamura A, Asano A, Sawano Y, Ojima T, Tanokura M. 2017. Structure and polymannuronate specificity of a eukaryotic member of polysaccharide lyase family 14. J Biol Chem 292:2182–2190. doi: 10.1074/jbc.M116.749929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu F, Dong F, Wang P, Cao HY, Li CY, Li PY, Pang XH, Zhang YZ, Chen XL. 2017. Novel molecular insights into the catalytic mechanism of marine bacterial alginate lyase AlyGC from polysaccharide lyase family 6. J Biol Chem 292:4457–4468. doi: 10.1074/jbc.M116.766030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HS, Lee CG, Lee EY. 2011. Alginate lyase: structure, property, and application. Biotechnol Bioproc E 16:843–851. doi: 10.1007/s12257-011-0352-8. [DOI] [Google Scholar]
- 19.Zhu B, Yin H. 2015. Alginate lyase: review of major sources and classification, properties, structure-function analysis and applications. Bioengineered 6:125–131. doi: 10.1080/21655979.2015.1030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garron ML, Cygler M. 2010. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 20:1547–1573. doi: 10.1093/glycob/cwq122. [DOI] [PubMed] [Google Scholar]
- 21.Yamasaki M, Ogura K, Hashimoto W, Mikami B, Murata K. 2005. A structural basis for depolymerization of alginate by polysaccharide lyase family-7. J Mol Biol 352:11–21. doi: 10.1016/j.jmb.2005.06.075. [DOI] [PubMed] [Google Scholar]
- 22.Yamasaki M, Moriwaki S, Miyake O, Hashimoto W, Murata K, Mikami B. 2004. Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7: a novel alginate lyase with a beta-sandwich fold. J Biol Chem 279:31863–31872. doi: 10.1074/jbc.M402466200. [DOI] [PubMed] [Google Scholar]
- 23.Osawa T, Matsubara Y, Muramatsu T, Kimura M, Kakuta Y. 2005. Crystal structure of the alginate (poly alpha-l-guluronate) lyase from Corynebacterium sp. at 1.2 A resolution. J Mol Biol 345:1111–1118. doi: 10.1016/j.jmb.2004.10.081. [DOI] [PubMed] [Google Scholar]
- 24.Thomas F, Lundqvist LC, Jam M, Jeudy A, Barbeyron T, Sandstrom C, Michel G, Czjzek M. 2013. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J Biol Chem 288:23021–23037. doi: 10.1074/jbc.M113.467217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogura K, Yamasaki M, Mikami B, Hashimoto W, Murata K. 2008. Substrate recognition by family 7 alginate lyase from Sphingomonas sp. A1. J Mol Biol 380:373–385. doi: 10.1016/j.jmb.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Ogura K, Yamasaki M, Yamada T, Mikami B, Hashimoto W, Murata K. 2009. Crystal structure of family 14 polysaccharide lyase with pH-dependent modes of action. J Biol Chem 284:35572–35579. doi: 10.1074/jbc.M109.068056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon HJ, Mikami B, Hashimoto W, Murata K. 1999. Crystal structure of alginate lyase A1-III from Sphingomonas species A1 at 1.78 A resolution. J Mol Biol 290:505–514. doi: 10.1006/jmbi.1999.2883. [DOI] [PubMed] [Google Scholar]
- 28.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream MA, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 9:R74. doi: 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikami B, Ban M, Suzuki S, Yoon HJ, Miyake O, Yamasaki M, Ogura K, Maruyama Y, Hashimoto W, Murata K. 2012. Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III. Acta Crystallogr D Biol Crystallogr 68:1207–1216. doi: 10.1107/S090744491202495X. [DOI] [PubMed] [Google Scholar]
- 30.Yoon HJ, Hashimoto W, Miyake O, Murata K, Mikami B. 2001. Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 A resolution. J Mol Biol 307:9–16. doi: 10.1006/jmbi.2000.4509. [DOI] [PubMed] [Google Scholar]
- 31.Li S, Wang L, Han F, Gong Q, Yu W. 2016. Cloning and characterization of the first polysaccharide lyase family 6 oligoalginate lyase from marine Shewanella sp. Kz7. J Biochem 159:77–86. doi: 10.1093/jb/mvv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maki H, Mori A, Fujiyama K, Kinoshita S, Yoshida T. 1993. Cloning, sequence analysis and expression in Escherichia coli of a gene encoding an alginate lyase from Pseudomonas sp. OS-ALG-9. J Gen Microbiol 139:987–993. doi: 10.1099/00221287-139-5-987. [DOI] [PubMed] [Google Scholar]
- 33.Lee SI, Choi SH, Lee EY, Kim HS. 2012. Molecular cloning, purification, and characterization of a novel polyMG-specific alginate lyase responsible for alginate MG block degradation in Stenotrophomas maltophilia KJ-2. Appl Microbiol Biotechnol 95:1643–1653. doi: 10.1007/s00253-012-4266-y. [DOI] [PubMed] [Google Scholar]
- 34.Yoder MD, Keen NT, Jurnak F. 1993. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science 260:1503–1507. doi: 10.1126/science.8502994. [DOI] [PubMed] [Google Scholar]
- 35.Alahuhta M, Chandrayan P, Kataeva I, Adams MW, Himmel ME, Lunin VV. 2011. A 1.5 A resolution X-ray structure of the catalytic module of Caldicellulosiruptor bescii family 3 pectate lyase. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:1498–1500. doi: 10.1107/S1744309111038449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenkins J, Shevchik VE, Hugouvieux-Cotte-Pattat N, Pickersgill RW. 2004. The crystal structure of pectate lyase Pel9A from Erwinia chrysanthemi. J Biol Chem 279:9139–9145. doi: 10.1074/jbc.M311390200. [DOI] [PubMed] [Google Scholar]
- 37.Pickersgill R, Jenkins J, Harris G, Nasser W, Robert-Baudouy J. 1994. The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat Struct Biol 1:717–723. doi: 10.1038/nsb1094-717. [DOI] [PubMed] [Google Scholar]
- 38.Huang W, Matte A, Li Y, Kim YS, Linhardt RJ, Su H, Cygler M. 1999. Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 A resolution. J Mol Biol 294:1257–1269. doi: 10.1006/jmbi.1999.3292. [DOI] [PubMed] [Google Scholar]
- 39.Gerstein M, Krebs W. 1998. A database of macromolecular motions. Nucleic Acids Res 26:4280–4290. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jedrzejas MJ, Mello LV, de Groot BL, Li S. 2002. Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate. J Biol Chem 277:28287–28297. doi: 10.1074/jbc.M112009200. [DOI] [PubMed] [Google Scholar]
- 41.Lunin VV, Li Y, Linhardt RJ, Miyazono H, Kyogashima M, Kaneko T, Bell AW, Cygler M. 2004. High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism. J Mol Biol 337:367–386. doi: 10.1016/j.jmb.2003.12.071. [DOI] [PubMed] [Google Scholar]
- 42.Preiss J, Ashwell G. 1962. Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosaccharides and 4-deoxy-L-erythro-5-hexoseulose uronic acid. J Biol Chem 237:309–316. [PubMed] [Google Scholar]
- 43.Ertesvag H. 2015. Alginate-modifying enzymes: biological roles and biotechnological uses. Front Microbiol 6:523. doi: 10.3389/fmicb.2015.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gacesa P. 1987. Alginate-modifying enzymes. A proposed unified mechanism of action for the lyases and epimerases. FEBS Lett 212:199–202. [Google Scholar]
- 45.Han YH, Garron ML, Kim HY, Kim WS, Zhang Z, Ryu KS, Shaya D, Xiao Z, Cheong C, Kim YS, Linhardt RJ, Jeon YH, Cygler M. 2009. Structural snapshots of heparin depolymerization by heparin lyase I. J Biol Chem 284:34019–34027. doi: 10.1074/jbc.M109.025338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacDonald LC, Berger BW. 2014. Insight into the role of substrate-binding residues in conferring substrate specificity for the multifunctional polysaccharide lyase Smlt1473. J Biol Chem 289:18022–18032. doi: 10.1074/jbc.M114.571299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garron ML, Cygler M. 2014. Uronic polysaccharide degrading enzymes. Curr Opin Struct Biol 28:87–95. doi: 10.1016/j.sbi.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 48.Charnock SJ, Brown IE, Turkenburg JP, Black GW, Davies GJ. 2002. Convergent evolution sheds light on the anti-beta-elimination mechanism common to family 1 and 10 polysaccharide lyases. Proc Natl Acad Sci U S A 99:12067–12072. doi: 10.1073/pnas.182431199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chavagnat F, Duez C, Guinand M, Potin P, Barbeyron T, Henrissat B, Wallach J, Ghuysen JM. 1996. Cloning, sequencing and overexpression in Escherichia coli of the alginatelyase-encoding aly gene of Pseudomonas alginovora: identification of three classes of alginate lyases. Biochem J 319(Part 2):575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]