ABSTRACT
Glutathione (l-γ-glutamyl-l-cysteinylglycine) (GSH), one of the key antioxidants in Sinorhizobium meliloti, is required for the development of alfalfa (Medicago sativa) nitrogen-fixing nodules. Glutathione exists as either reduced glutathione (GSH) or oxidized glutathione (GSSG), and its content is regulated by two pathways in S. meliloti. The first pathway is the de novo synthesis of glutathione from its constituent amino acids, namely, Glu, Cys, and Gly, catalyzed by γ-glutamylcysteine synthetase (GshA) and glutathione synthetase (GshB). The second pathway is the recycling of GSSG via glutathione reductase (GR). However, whether the S. meliloti GR functions similarly to GshA and GshB1 during symbiotic interactions with alfalfa remains unknown. In this study, a plasmid insertion mutation of the S. meliloti gor gene, which encodes GR, was constructed, and the mutant exhibited delayed alfalfa nodulation, with 75% reduction in nitrogen-fixing capacity. The gor mutant demonstrated increased accumulation of GSSG and a decreased GSH/GSSG ratio in cells. The mutant also showed defective growth in rich broth and minimal broth and was more sensitive to the oxidants H2O2 and sodium nitroprusside. Interestingly, the expression of gshA, gshB1, katA, and katB was induced in the mutant. These findings reveal that the recycling of glutathione is important for S. meliloti to maintain redox homeostasis and to interact symbiotically with alfalfa.
IMPORTANCE The antioxidant glutathione is regulated by its synthetase and reductase in cells. In the symbiotic bacterium S. meliloti, the de novo synthesis of glutathione is essential for alfalfa nodulation and nitrogen fixation. In this study, we observed that the recycling of glutathione from GSSG not only was required for redox homeostasis and oxidative stress protection in S. meliloti cells but also contributed to alfalfa nodule development and competition capacity. Our findings demonstrate that the recycling of glutathione plays a key role in nitrogen fixation symbiosis.
KEYWORDS: Sinorhizobium meliloti, glutathione reductase, redox homeostasis, symbiosis, reactive oxygen species
INTRODUCTION
Most organisms contain high concentrations of low-molecular-weight (LMW) thiols, which serve as redox buffers to protect cells against a variety of reactive chemical species, such as reactive oxygen species (ROS) (1, 2). ROS include hydrogen peroxide (H2O2), superoxide anions, and hydroxyl radicals. Glutathione is considered the most ubiquitous and potent LMW thiol antioxidant. Glutathione is present at millimolar concentrations in nearly all eukaryotic cells and many bacteria (including most Gram-negative bacteria) (3, 4). In cells, the content of glutathione is regulated by two pathways. The first pathway is the de novo synthesis pathway, in which two ATP-dependent enzymes, γ-glutamylcysteine synthetase (GshA) and glutathione synthetase (GshB), which are encoded by the gshA (GSH1) and gshB (GSH2) genes, respectively, sequentially catalyze the synthesis of glutathione from glutamate, cysteine, and glycine (5). Free glutathione exists in two different forms in cells, i.e., reduced glutathione (GSH), with a free thiol group, and oxidized glutathione (GSSG), with a disulfide bond between two identical molecules. Two GSH molecules are oxidized to form a GSSG molecule via a disulfide bond under oxidative stress, while glutathione reductase (GR) catalyzes the reduction of GSSG to GSH. This reduction represents the recycling synthesis pathway. In the yeast Saccharomyces cerevisiae, GSH is required as a reductant and may function to remove endogenously derived toxic metabolites. Yeast mutants lacking GSH showed a severely retarded growth phenotype and enhanced sensitivity to oxidative stress (6–8). Interestingly, GSH is not essential in Escherichia coli under normal growth conditions; moreover, transposon insertion mutants in which gshA was disrupted were unaffected in their resistance to compounds that cause oxidative stress (9).
Rhizobia, which are Gram-negative soil bacteria, can reduce N2 (nitrogen fixation) in specific legume root organs called nodules, via stringent symbiotic interactions. Notably, the establishment of Rhizobium-legume symbiosis depends on the capacity for rhizobial GSH synthesis. A GSH-deficient mutant of Rhizobium tropici induced Phaseolus vulgaris to form root nodules with early senescence (10). For Sinorhizobium meliloti, GSH was reported to be essential for free-living growth and symbiosis with alfalfa (11). Mutation of gshA led to a severe growth defect under normal conditions, and the mutant did not induce nodule formation in alfalfa plants. A gshB mutant induced the formation of alfalfa nodules with low nitrogen fixation capacity. However, whether the GR-catalyzed recycling synthesis of GSH from GSSG is required for redox homeostasis and symbiosis has not been determined to date. In this study, a gor null mutant was constructed in a S. meliloti 1021 background, and its free-living and symbiotic phenotypes were investigated. Our results indicate that the recycling of glutathione is important for S. meliloti to grow, to adapt to oxidative stress, and to interact symbiotically with host plants.
RESULTS
Construction of an S. meliloti gor mutant.
To investigate the roles of GR in free-living and symbiotic Rhizobium cells, a gor null mutant (gor19) was constructed in S. meliloti 1021 by using a plasmid insertion method (Fig. 1A). The mutant was identified by PCR, and the results showed that an ∼4.5-kb DNA fragment containing the suicide plasmid pK19mob2ΩHMB (3.5 kb) was amplified from the gor19 mutant, whereas an ∼1-kb DNA fragment was amplified from the parent S. meliloti 1021 strain (Fig. 1B). DNA sequencing indicated that the suicide plasmid pK19mob2ΩHMB was inserted after nucleotide 994 of the gor open reading frame (ORF) (1,392 bp). To determine whether the plasmid insertion in gor19 disrupted the entire transcript, total RNA was extracted and reverse transcription (RT)-PCR was performed. The results showed the absence of an entire gor transcript in the insertion mutant, whereas such a fragment was detected in the wild-type (WT) strain and in the complementation strain (cpgor19) (Fig. 1C). Finally, cell extracts were made from the wild-type strain and the gor19 strain and were assayed directly for GR activity. The extracts of the gor19 strain showed activity of <0.11 ± 0.01 nmol/s/mg protein, but the wild-type strain and complementation strain (cpgor19) extracts reduced glutathione at rates of 28.4 ± 0.88 nmol/s/mg protein and 75.8 ± 0.79 nmol/s/mg protein, respectively (Fig. 1D). These results show that the gor19 strain lacks GR activity.
FIG 1.
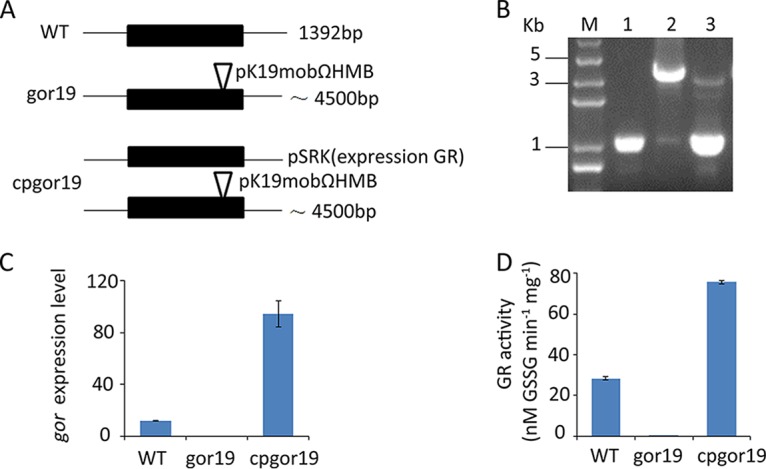
Strategies for plasmid insertion and characterization of the gor19 mutant. (A) Schematic representation of the location of the genomic locus before and after integration of the plasmid into the genome in the WT, gor19, and cpgor19 strains. WT, S. meliloti 1021; gor19, mutant with a plasmid insertion in gor; cpgor19, strain containing pSRK19 in the gor19 mutant. (B) Agarose gel with a DNA size marker and DNA fragments amplified with different genomic DNA. Lane M, DNA marker 2K Plus; lane 1, amplified gor PCR fragment from the S. meliloti 1021 (WT) strain; lane 2, amplified gor PCR fragment from the gor19 mutant; lane 3, amplified gor PCR fragment from the cpgor19 strain. (C) Real-time PCR analysis of gor expression levels in the WT, gor19, and cpgor19 strains. (D) GR activity of total protein in the WT, gor19, and cpgor19 strains. The data shown are the means of three experiments, with the error bars indicating the standard errors.
Growth defect of the S. meliloti gor mutant.
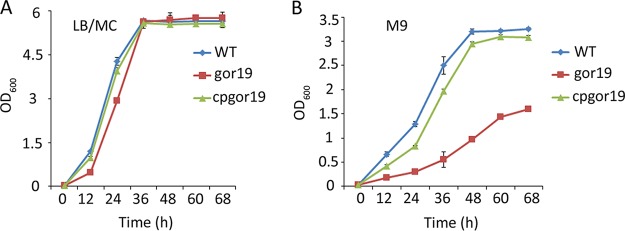
The gor mutant was first evaluated for growth defects. In rich broth (Luria-Bertani [LB] medium supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2 [LB/MC medium]), the mutant grew slightly more slowly (the logarithmic phase was extended) than the wild-type strain or the complemented strain (Fig. 2A). Striking growth arrest was observed in minimal broth (M9 medium) for gor19, as its growth rate was 50% less than that of the wild-type strain; growth was restored by constitutively expressing gor from a plasmid in the complemented strain (Fig. 2B). These results indicate that gor is required for the free-living growth of S. meliloti.
FIG 2.
Growth curves of the S. meliloti strains. At time zero, rhizobial cells were diluted with LB/MC medium or M9 medium to an OD600 of 0.01. The growth of the WT, gor19, and cpgor19 strains in LB/MC and M9 media was monitored by measuring the optical density at 600 nm. (A) Growth curves of free-living S. meliloti strains grown in LB/MC medium. (B) Growth curves of free-living S. meliloti strains grown in M9 medium. The data points represent the averages of triplicate samples, with the error bars indicating the standard errors.
Requirement for the S. meliloti gor gene for redox homeostasis.
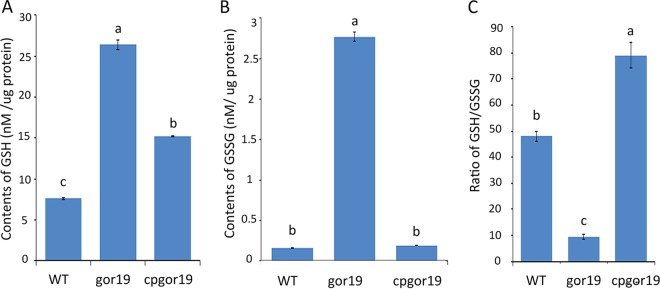
To evaluate the GSH and GSSG contents in the gor19 mutant, a liquid chromatography-mass spectrometry (LC-MS) method was used (see Materials and Methods). The data showed that the content of GSH was increased ∼3-fold in the mutant, compared to the parent strain, while the level of GSSG was ∼10-fold higher in gor19 than in S. meliloti 1021 (Fig. 3A and B). Thus, the GSH/GSSG ratio was 9:1 in gor19 and 49:1 in S. meliloti strain 1021, a decrease of ∼5-fold in the mutant compared with the parent strain (Fig. 3C). This defect could be rescued by constitutively expressing gor from a plasmid in the mutant (Fig. 3). These results indicate that the gor gene is associated with the production of GSH and GSSG and is required for redox homeostasis in S. meliloti cells.
FIG 3.
Accumulation of glutathione in S. meliloti. (A and B) High-performance liquid chromatography (HPLC) analysis of the GSH (A) and GSSG (B) contents in the WT, gor19, and cpgor19 strains. (C) GSH/GSSG ratios in the WT, gor19, and cpgor19 strains. The data represent the means ± standard errors for triplicate samples. Significant differences were identified by one-way analysis of variance (ANOVA), followed by Tukey's post hoc test, and are noted as different letters (P < 0.05).
Activation of gshA, gshB1, katA, and katB expression in the gor19 mutant.
The level of GSH was strikingly increased in the gor19 mutant, which prompted us to analyze the expression of gshA and gshB1 using quantitative RT-PCR (qRT-PCR). The data showed that the expression of both gshA and gshB1 was increased >3-fold in the mutant compared with the parent strain but that the wild-type levels could be restored by expressing gor from a plasmid in the complementation strain (Fig. 4). The expression of the catalase gene was also investigated, to further evaluate the redox status of the cells. qRT-PCR data showed that the expression levels of katA and katB were elevated 2-fold in gor19, compared with S. meliloti 1021, whereas the expression of katC was almost not detected in any of the strains. Expression of gor from a plasmid restored catalase expression in the mutant. These results indicate that the lack of GR causes oxidative stress in S. meliloti.
FIG 4.
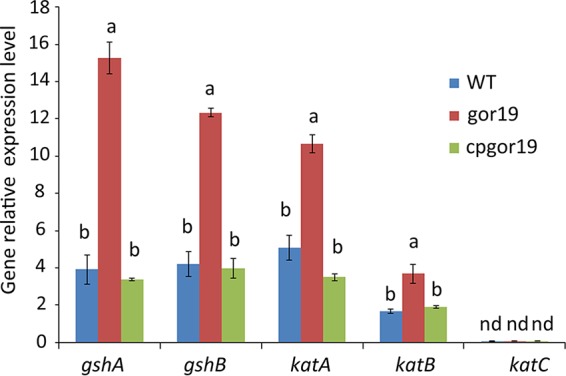
qRT-PCR analysis of the expression levels of various genes in the WT, gor19, and cpgor19 strains. The relative amount of mRNA for each gene was calculated using the threshold cycle (CT) method and was normalized to that of the S. meliloti rpsF gene. Error bars indicate standard deviations. This experiment was performed in triplicate. Significant differences were identified by one-way ANOVA, followed by Tukey's post hoc test, and are noted as different letters (P < 0.05). Vertical bars indicate the standard errors for three biological replicates. nd, not detected.
Requirement for the S. meliloti gor gene for adaptation to oxidative stress.
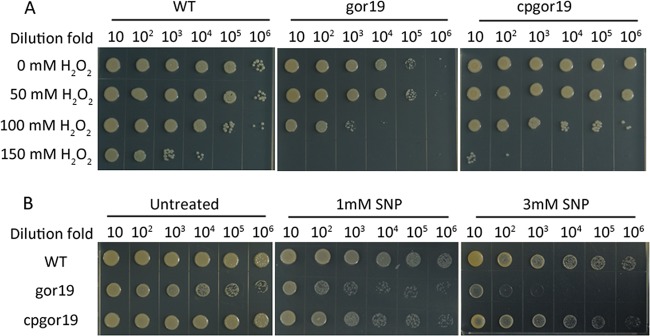
A Rhizobium etli strain with a Tn5 insertion mutation in GR was reported to be sensitive to oxidative stress (12). To examine the possibility that gor is required for the adaptation of S. meliloti to oxidative stress, sensitivity to H2O2 and sodium nitroprusside (SNP) was assayed in LB/MC medium. The zone of inhibition induced by H2O2 increased from 2.31 ± 0.04 cm for the WT S. meliloti 1021 strain to 2.69 ± 0.06 cm for the gor19 mutant, which could be rescued in the cpgor19 strain (2.32 ± 0.1 cm). Furthermore, the results of the cell sensitivity assay confirmed that the gor19 mutant was sensitive to 100 mM H2O2, whereas the parent and complemented strains were sensitive to 150 mM H2O2 (Fig. 5A). The sensitivity of the gor19 strain to SNP stress was also estimated, and we observed that the gor19 mutant was hypersensitive to 3 mM SNP compared with the S. meliloti 1021 and complemented cpgor19 strains (Fig. 5B). These data indicate that gor is essential for the adaptation of S. meliloti to oxidative stress.
FIG 5.
Sensitivity of the WT, gor19, and cpgor19 strains to oxidants. (A) H2O2 sensitivity of the WT, gor19, and cpgor19 strains grown in LB/MC medium. Cell cultures were adjusted to an OD600 of 0.5, treated for 30 min at 28°C with the indicated amount of H2O2, washed twice, resuspended in fresh LB/MC medium, and spotted onto LB/MC agar plates. The 10-fold serial dilutions are indicated above each column. The plates were incubated at 28°C for 4 days. (B) SNP sensitivity of the WT, gor19, and cpgor19 strains grown in LB/MC medium. Cell cultures were adjusted to an OD600 of 0.5 and spotted onto LB/MC agar in the absence or presence of 1 or 3 mM SNP. The 10-fold serial dilutions are indicated above each column. The plates were incubated at 28°C for 4 days.
Symbiotic defects in the S. meliloti gor mutant.
To determine the function of the S. meliloti gor gene during symbiotic interactions with an alfalfa host, the nodulation capacity of the gor19 mutant was first examined. The number of alfalfa nodules induced by gor19 was significantly decreased, compared with that induced by the parent strain, at 5 and 9 days postinoculation (dpi), but there was no significant difference in alfalfa nodulation with the mutant strain versus the parent strain at 14 dpi (Fig. 6D). To test the functionality of nodules, their nitrogen-fixing capacities and the dry weights of the alfalfa shoots were determined 4 weeks after inoculation with the WT, gor19, and cpgor19 strains. Using the acetylene reduction assay, it was found that the nitrogen-fixing capacity was severely affected in the gor19 mutant, with a reduction from 80.3 ± 4.8 nmol of C2H4/mg of nodules/min in WT S. meliloti 1021 or 74 ± 2.8 nmol of C2H4/mg of nodules/min in cpgor19 to 17.2 ± 2.8 nmol of C2H4/mg of nodules/min in the gor19 strain (Fig. 6C), which was consistent with the results of the dry weight of alfalfa shoots and the plant phenotype with gor19 (Fig. 6A and B). Finally, the competitive capacity of gor19 versus S. meliloti 1021 was evaluated. Coinoculation data from samples inoculated with WT/gor19 strain ratios of 1:1, 1:9, and 9:1 showed that nodule occupation proportions for the mutant were 6.5%, 13%, and 0%, respectively. These results indicate that infectivity and nitrogen fixation are affected by mutation of the gor gene.
FIG 6.
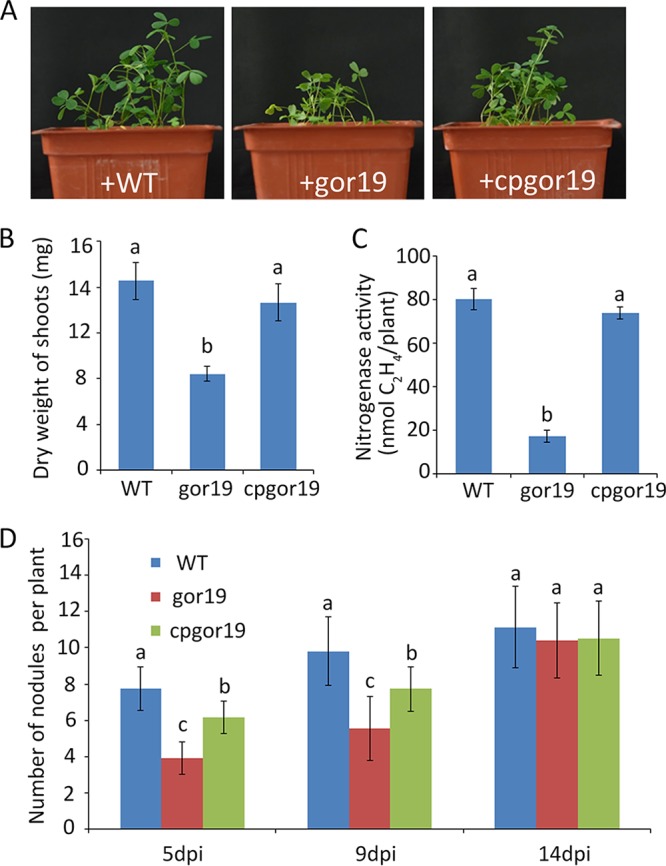
Deficiency of the gor19 strain in symbiosis with alfalfa. (A) Symbiotic phenotypes of the WT, gor19, and cpgor19 strains. (B) Dry weights of shoots inoculated with the WT, gor19, and cpgor19 strains. (C) Nitrogenase activity of alfalfa nodules per plant, as detected by C2H2 reduction, in plants inoculated with the WT, gor19, and cpgor19 strains. (D) Numbers of nodules per plant, assessed 5, 9, and 14 dpi. In panels B to D, the data represent means ± standard deviations (n = 32). The experiments were repeated four times, and a representative experiment is shown. Significant differences (P < 0.05) were identified by one-way ANOVA, followed by Tukey's post hoc test, and are noted as different letters (P < 0.05).
DISCUSSION
In S. meliloti, glutathione plays key roles in the maintenance of redox homeostasis and the symbiotic root nodulation of alfalfa, according to a study of gshA and gshB1 (11). In this study, we found that the pathway of GSH recycling from GSSG is important for the cellular redox status and alfalfa nodulation in S. meliloti. Our findings suggest a feedback regulatory mechanism for GSH biosynthesis in S. meliloti. The gor gene encodes an NADPH-dependent GR that belongs to a flavin adenine dinucleotide (FAD)-binding disulfide reductase superfamily. The crystal structure of the S. meliloti GR indicates that this enzyme consists of domain I (positions 1 to 141 and 295 to 335), domain II (positions 141 to 158 and 265 to 294), domain III (positions 159 to 264), and domain IV (positions 336 to 463) (see Fig. S1A in the supplemental material). According to the DNA sequencing data, the suicide plasmid disrupts the coding regions of domains I and IV of GR in the gor19 mutant (Fig. S1B), possibly preventing the dimerization of GR and NADPH binding.
Transposon insertion mutants of gor were reported to be involved in defense against diamide but not H2O2 in Rhizobium etli E3 (13). However, we found that GR in S. meliloti 1021 is required for adaptation to oxidative stress, such as treatment with H2O2 or SNP (Fig. 5), which could be due to changes in the cellular redox status. The results indicate that GR is required for cellular redox homeostasis.
Catalase activity is often used as a physiological marker to assess whether S. meliloti cells are under oxidative stress. Three distinct hydroperoxidases have been reported to be involved with H2O2 in S. meliloti, i.e., two monofunctional catalases (KatA and KatC) and one bifunctional catalase-peroxidase (KatB) (14, 15). The gor mutant cells maintained their oxidative status because the expression of katA and katB increased (Fig. 4). At the same time, the oxidation status induced the expression of gshA and gshB1 (Fig. 4). This finding is consistent with the observed GSSG/GSH ratio, although the production of GSH and GSSG was increased in the mutant (Fig. 3).
Our study demonstrated that the growth of an S. meliloti strain with a mutated gor gene was severely impaired in chemically defined minimal broth (M9 medium) supplemented with glucose as the sole source of carbon and NH4Cl as the sole source of nitrogen, whereas only a slightly reduced growth phenotype was observed in rich broth (LB/MC medium). These results indicate that inactive GR may affect the uptake and/or utilization of sources of carbon and nitrogen in S. meliloti 1021. In fact, it has been reported that glutathione affects the transport activity of Rhizobium leguminosarum 3841 and is essential for efficient nodulation. A gshB mutant of Rhizobium leguminosarum 3841 failed to utilize glucose, succinate, glutamine, and histidine efficiently and caused a 50% reduction in the dry weight of plants and nitrogen-fixing ability (16). Therefore, we hypothesized that the slow growth of gor19 would alter bacterial infection and plant nodulation. This possibility was confirmed by inoculation and nodulation assays (Fig. 6), although the difference was minimal. In fact, another insertion mutant of gor (with an insertion 491 bp from the ATG of gor) may carry a second mutation since the expression of GR could not restore the phenotype (G. Tang, L. Yu, J. Yan, and L. Luo, unpublished data). Taken together, our findings reveal that the GSH recycling pathway is essential for cellular redox homeostasis and root-nodule symbiosis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli DH5α and MT616 were grown in LB medium at 37°C. The S. meliloti 1021 (WT), gor19, and cpgor19 strains were grown at 28°C in LB medium supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2 (LB/MC medium). The following antibiotics were used at the indicated concentrations: streptomycin, 500 μg/ml; neomycin, 200 μg/ml; chloramphenicol, 10 μg/ml; kanamycin, 25 μg/ml; and gentamicin, 10 μg/ml. The plasmids and strains used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant propertiesa | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− supE44 ΔlacU169 (ϕ80lacZΔM15) hsdR17(rK− mK+) recA1 endA1 gyrA96 thi-1 relA1 | TaKaRa Corp. |
| MT616 | pro-82 thi-1 hsdR17 supE44 recA56 (pRK600); Cmr | 18 |
| S. meliloti strains | ||
| 1021 | Wild type from SU47; Strr | Laboratory store |
| gor19 | Same as 1021 but with gor::pK19mob2ΩHMB; Strr, Neor | This study |
| cpgor19 | gor19 strain with pSRK19; Strr, Neor, Gmr | This study |
| Plasmids | ||
| pK19 | pK19mob2ΩHMB; Kmr | 17 |
| pK19gor | pK19 with 300-bp PCR-amplified fragment of gor; Kmr | This study |
| pSRK | Broad-host-range expression vector; Gmr | 19 |
| pSRK19 | pSRK with gor ORF; Gmr | This study |
Cmr, chloramphenicol resistant; Strr, streptomycin resistant; Neor, neomycin resistant; Gmr, gentamicin resistant; Kmr, kanamycin resistant.
Construction of the gor19 and cpgor19 strains.
First, a gor (SMc00154) DNA fragment (0.3 kb) from S. meliloti 1021 was PCR amplified using the primers P1 and P2. Next, the DNA fragments were digested with HindIII and BsrGI, ligated with a similarly treated pK19 plasmid (17), and transformed into DH5α cells. The resulting plasmid construct (pK19gor) was conjugated into S. meliloti 1021 using the helper strain MT616 in a triparental mating (18). Cells containing a plasmid insertion in the gor gene were selected for on solid LB/MC medium containing streptomycin and neomycin. A bacterial culture started from a single colony on the selective medium was used to prepare the S. meliloti phage φM12 lysate. The lysate was used to transduce the plasmid insertion of gor into the S. meliloti 1021 background, to ensure that the mutant (gor19) carried a single plasmid insertion in the gor gene. To construct a plasmid for complementation of the gor19 mutant, a 1,436-bp DNA fragment of the gor ORF was PCR amplified from the genomic DNA of S. meliloti 1021 by using primers P3 and P4. The PCR product was cloned into pSRK (19), and the resultant plasmid was named pSRK19. The recombinant plasmid was transferred into the gor19 recipient strain by triparental mating using the MT616 helper strain. The gor19(pSRK19) exconjugants, named cpgor19, were selected for on LB/MC medium containing neomycin, spectinomycin, and gentamicin. The primers used in this study are shown in Table 2.
TABLE 2.
Primers
| Primer | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| P1 | GGGTGTACAAGGACATCATCCAGTCGGT | Construction of pK19gor |
| P2 | CCCAAGCTTTGTTCTTGTATTCCGTCTCG | Construction of pK19gor |
| P3 | GGAATTCCATATGATGAGCGCTTTCGACTATGAC | Construction of pSRK19 |
| P4 | CTAGCTAGCTCCTACCAGAAAACGTGG | Construction of pSRK19 |
| P5 | TTATGCCTCGCAGTTTGC | Identification of cpgor19 |
| P6 | TCGGCTGATACATCGTGA | Identification of cpgor19 |
| P7 | ATGTGCTTCATCGAGACG | qRT-PCR of gor |
| P8 | GGCATTGACGACGAGCTT | qRT-PCR of gor |
| P9 | AACTGGCGGAAGGTGACAT | qRT-PCR of gshA |
| P10 | TTACCGAGGGCAAGCCTAA | qRT-PCR of gshA |
| P11 | GGAATCGTCAATGTCGGGGTC | qRT-PCR of gshB1 |
| P12 | GCTTGTCGGGCGTGTAGTGG | qRT-PCR of gshB1 |
| P13 | CATCGCCGAAGAAGACAA | qRT-PCR of katA |
| P14 | CGAAGACATCAGAGAAATGGTC | qRT-PCR of katA |
| P15 | CTGACCGCACCGGAAATGAC | qRT-PCR of katB |
| P16 | CATATCGAGCAGGTTGACGAAGAA | qRT-PCR of katB |
| P17 | GTCGGGCGCGTGAACTACC | qRT-PCR of katC |
| P18 | CCCCTGCTCCTCGGAAGG | qRT-PCR of katC |
Measurement of glutathione reductase activity.
Measurement of GR activity was performed as described by Davis et al. (20), with minor modifications. Strains were grown in LB/MC broth to early stationary phase (optical density at 600 nm [OD600] of 0.5); 1 ml was harvested and resuspended in 500 μl of 50 mM potassium phosphate buffer (pH 8). Cells were disrupted on ice with a Microson ultrasonic cell disruptor (Misonix, Farmingdale, NY) operating at 5 rpm for four periods of 15 s, and debris was removed by centrifugation for 5 min in an Eppendorf centrifuge. The protein concentration of the supernatant fraction was measured by the Lowry method. The reaction mixture contained 1.2 mM oxidized glutathione, 0.4 mM Na4NADPH, 0.6 mM 5,5′-dithiobis-2-nitrobenzoic acid, and 50 mM potassium phosphate buffer (pH 8), in a final volume of 1 ml, at 28°C. The reaction was started with the addition of supernatant and was monitored at 412 nm. Na4NADPH and GSSG were always freshly prepared.
Determination of growth curves.
The S. meliloti 1021 (WT), gor19, and cpgor19 strains were grown in LB/MC medium at 28°C, with shaking at 200 rpm, to an OD600 of 0.5. At time zero, 1 ml of rhizobial cells (OD600 of 0.5) was inoculated into LB/MC medium or M9 medium (50 ml). The growth of the S. meliloti 1021 (WT), gor19, and cpgor19 strains was monitored by measuring the optical density at 600 nm.
Assay of oxidative stress sensitivity.
The sensitivity of S. meliloti to SNP was assessed with a spotting assay. The S. meliloti 1021 (WT), gor19, and cpgor19 strains were grown in LB/MC medium at 28°C, with shaking at 200 rpm, to an OD600 of 0.5 and then were harvested by centrifugation. After the cultures were serially diluted 10-fold in LB/MC broth, aliquots (5 μl) were spotted onto LB/MC agar supplemented with 0, 1, or 3 mM SNP and the plates were incubated at 28°C for 4 days to determine the oxidant resistance of the strains. The sensitivity of S. meliloti to H2O2 was assessed as follows. Cells were adjusted to an OD600 of 0.5, treated with 0, 50, 100, or 150 mM H2O2 at 28°C for 30 min, washed twice with distilled water, resuspended in fresh medium, and spotted onto LB/MC agar. The plates were incubated at 28°C for 4 days. The growth inhibition of strain gor19 with H2O2 exposure was tested with the halo assay method (21), which is a long-term test of resistance to H2O2 exposure. Aliquots (300 μl each) of S. meliloti cultures with OD600 values of 0.5 were plated on LB/MC agar plates. Paper disks (10 mm in diameter) were impregnated with 3 μl of H2O2 (1 M) and placed in the center of the S. meliloti plates. After 2 to 3 days of incubation at 28°C, the diameters of the halos were analyzed.
Assay of glutathione levels in S. meliloti.
The extraction and quantification protocol for GSH was performed as described previously (22), with minor modifications. In brief, S. meliloti cells (OD600 of 2) from 50-ml cultures in LB/MC medium were prepared by centrifugation at 13,000 rpm for 1 min, washed twice with distilled water, and crushed in liquid nitrogen. The cell lysate was then suspended in 200 μl of distilled water. After centrifugation at 13,000 rpm for 10 min, one-half of each supernatant was used for analysis with a 6520 accurate-mass quadrupole time of flight LC-MS system (Agilent, San Jose, CA). The remaining supernatant was used to quantify total protein. The conditions for the purification and analysis of glutathione were as follows: column, Zorbax Extend-C18 (4.6 by 50 mm, 1.8 mm); injection volume, 5 μl; flow rate, 0.2 ml/min; solvent A, 0.1% trifluoroacetic acid (TFA) in water; solvent B, 0.1% TFA in methanol; detection wavelengths (diode array detector), 210, 254, 280, 320, 360, and 226 nm; mass range, m/z 50 to 400; nebulizer pressure, 40 lb/in2; drying gas, N2 (at 350°C and 9 liters/min); electrospray ionization voltages, 3,500 V (cap), 160 V (capillary and fragmentor), 65 V (skimmer), and 750 V (octupole radiofrequency voltage); and scanning mode, negative MS scan mode (2 GHz) with external dynamic range (to m/z 1,700). GSSG and GSH at 50 ng/μl were used as standards.
RNA extraction and real-time RT-PCR.
S. meliloti subcultures were grown to exponential phase (OD600 of 0.5) in 50 ml of fresh LB/MC medium with 1% of the overnight cultures. RNA was extracted using the TRIzol reagent (Invitrogen, Shanghai, China) (23). The RNA quality was assayed using 1% agarose gel electrophoresis. Next, 1 μg of total RNA was used to prepare cDNA by using random hexamer oligonucleotide primers and the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Dalian, China). Gene-specific primers were employed for real-time PCR using SYBR green supermix (TransGen, Shanghai, China). The expression of rpsF (a ribosomal protein gene) in each sample was used to normalize the expression of the gene(s) of interest. The real-time PCR primers used are listed in Table 2. The expression levels were calculated using the 2−ΔΔCT method.
Assay of nodulation occupancy and competitiveness.
Alfalfa (cv. aifeinite) was used as a host plant to test the nodulation of the S. meliloti strains. Seeds were surface sterilized and germinated on 1% agarose. Bacterial cells (1 ml, at an OD600 of 0.02) were plated on the surface of solid modified Fahraeus medium (24). Ten alfalfa seedlings were spread across each plate. The germinated alfalfa seeds were coinoculated with the wild-type S. meliloti 1021 and gor19 mutant strains at different ratios (1:1, 1:9, and 9:1), ultimately reaching 104 CFU per seed. The strains were isolated from nodules using both resistance to streptomycin (Str) and neomycin (Neo) and PCR screening at 3 weeks postinoculation (wpi).
Plant nodulation assay.
Sixteen germinated seedlings were transferred to a growth pot (10 by 10 cm) containing perlite and vermiculite at a ratio of 1:3 and supplied with modified Fahraeus medium. After 4 days of growth, plants were inoculated with 50 ml of Sinorhizobium meliloti culture (OD600 of 0.02) per pot. Different strains were used, including the WT, gor19, and cpgor19 strains. Each strain harbored a hemA-lacZ construct that was used to observe the infection events, which were examined 5, 9, and 14 dpi. To examine β-galactosidase activity, whole roots were fixed for 1 h with fixative solution (1% [vol/vol] glutaraldehyde in 1× phosphate-buffered saline [PBS] [pH 7.5]). The fixed samples were subsequently washed twice with 1× PBS and incubated overnight at room temperature in a solution composed of 0.2× PBS, 2.5 mM K3[Fe(CN)6], 2.5 mM K4[Fe(CN)6], and 0.8 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactoside in N,N-dimethylformamide. Stained samples were rinsed three times with 1× PBS and cleared using different concentrations of ethanol. The dry weights of the shoots of the WT, gor19, and cpgor19 strain-inoculated plants were examined 3 wpi.
Acetylene reduction assay.
Nitrogenase activity was assessed with the acetylene reduction assay as described previously (25). For this, 2 ml of acetylene was injected into sealed 20-ml tubes containing 4-week-old plants. Reduction of acetylene to ethylene was measured by flame ionization gas chromatography (Shanghai Analytic Instrument Factory, Shanghai, China) after 30 min of incubation at 28°C. For each sample, at least 30 plants, divided into three replicates, were analyzed.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Natural Science Foundation of China (grants 31370277 and 31570241 to L.L., grant 31500058 to G.T., and grant 31500197 to L.Y.), the Shanghai Key Program of Basic Research (grant 14JC1402300 to L.L.), and the Shanghai Key Program of Supporting (grant 15230500100 to L.L.). Research by G.T. was also supported by the China Postdoctoral Science Foundation (grant 2015M571539).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01937-17.
REFERENCES
- 1.Dalle-Donne I, Milzani A, Gagliano N, Colombo R, Giustarini D, Rossi R. 2008. Molecular mechanisms and potential clinical significance of S-glutathionylation. Antioxid Redox Signal 10:445–473. doi: 10.1089/ars.2007.1716. [DOI] [PubMed] [Google Scholar]
- 2.Roos G, Messens J. 2011. Protein sulfenic acid formation: from cellular damage to redox regulation. Free Radic Biol Med 51:314–326. doi: 10.1016/j.freeradbiomed.2011.04.031. [DOI] [PubMed] [Google Scholar]
- 3.Fahey RC, Brown WC, Adams WB, Worsham MB. 1978. Occurrence of glutathione in bacteria. J Bacteriol 133:1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meister A, Anderson ME. 1983. Glutathione. Annu Rev Biochem 52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 5.Meister A. 1974. Glutathione, metabolism and function via the γ-glutamyl cycle. Life Sci 15:177–190. doi: 10.1016/0024-3205(74)90206-9. [DOI] [PubMed] [Google Scholar]
- 6.Wu AL, Moye-Rowley WS. 1994. GSH1, which encodes gamma-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol Cell Biol 14:5832–5839. doi: 10.1128/MCB.14.9.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant CM, MacIver FH, Dawes IW. 1996. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr Genet 29:511–515. doi: 10.1007/BF02426954. [DOI] [PubMed] [Google Scholar]
- 8.Grant CM, MacIver FH, Dawes IW. 1997. Glutathione synthetase is dispensable for growth under both normal and oxidative stress conditions in the yeast Saccharomyces cerevisiae due to an accumulation of the dipeptide gamma-glutamylcysteine. Mol Biol Cell 8:1699–1707. doi: 10.1091/mbc.8.9.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg JT, Demple B. 1986. Glutathione in Escherichia coli is dispensable for resistance to H2O2 and gamma radiation. J Bacteriol 168:1026–1029. doi: 10.1128/jb.168.2.1026-1029.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muglia C, Comai G, Spegazzini E, Riccillo PM, Aguilar OM. 2008. Glutathione produced by Rhizobium tropici is important to prevent early senescence in common bean nodules. FEMS Microbiol Lett 286:191–198. doi: 10.1111/j.1574-6968.2008.01285.x. [DOI] [PubMed] [Google Scholar]
- 11.Harrison J, Jamet A, Muglia CI, Van de Sype G, Aguilar OM, Puppo A, Frendo P. 2005. Glutathione plays a fundamental role in growth and symbiotic capacity of Sinorhizobium meliloti. J Bacteriol 187:168–174. doi: 10.1128/JB.187.1.168-174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tate R, Cermola M, Riccio A, Diez-Roux G, Patriarca EJ. 2012. Glutathione is required by Rhizobium etli for glutamine utilization and symbiotic effectiveness. Mol Plant Microbe Interact 25:331–340. doi: 10.1094/MPMI-06-11-0163. [DOI] [PubMed] [Google Scholar]
- 13.Tate R, Ferraioli S, Filosa S, Cermola M, Riccio A, Iaccarino M, Patriarca EJ. 2004. Glutamine utilization by Rhizobium etli. Mol Plant Microbe Interact 17:720–728. doi: 10.1094/MPMI.2004.17.7.720. [DOI] [PubMed] [Google Scholar]
- 14.Jamet A, Kiss E, Batut J, Puppo A, Herouart D. 2005. The katA catalase gene is regulated by OxyR in both free-living and symbiotic Sinorhizobium meliloti. J Bacteriol 187:376–381. doi: 10.1128/JB.187.1.376-381.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamet A, Mandon K, Puppo A, Herouart D. 2007. H2O2 is required for optimal establishment of the Medicago sativa/Sinorhizobium meliloti symbiosis. J Bacteriol 189:8741–8745. doi: 10.1128/JB.01130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng G, Karunakaran R, East AK, Munoz-Azcarate O, Poole PS. 2017. Glutathione affects the transport activity of Rhizobium leguminosarum 3841 and is essential for efficient nodulation. FEMS Microbiol Lett 364:fnx045. doi: 10.1093/femsle/fnx045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 18.Finan TM, Hartweig E, LeMieux K, Bergman K, Walker GC, Signer ER. 1984. General transduction in Rhizobium meliloti. J Bacteriol 159:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan SR, Gaines J, Roop RM II, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis NK, Greer S, Jones-Mortimer MC, Perham RN. 1982. Isolation and mapping of glutathione reductase-negative mutants of Escherichia coli K12. J Gen Microbiol 128:1631–1634. [DOI] [PubMed] [Google Scholar]
- 21.Luo L, Qi MS, Yao SY, Cheng HP, Zhu JB, Yu GQ. 2005. Role of oxyR from Sinorhizobium meliloti in regulating the expression of catalases. Acta Biochim Biophys Sin (Shanghai) 37:421–428. doi: 10.1111/j.1745-7270.2005.00055.x. [DOI] [PubMed] [Google Scholar]
- 22.Lu D, Tang G, Wang D, Luo L. 2013. The Sinorhizobium meliloti LysR family transcriptional factor LsrB is involved in regulation of glutathione biosynthesis. Acta Biochim Biophys Sin (Shanghai) 45:882–888. doi: 10.1093/abbs/gmt083. [DOI] [PubMed] [Google Scholar]
- 23.Tang G, Wang Y, Luo L. 2014. Transcriptional regulator LsrB of Sinorhizobium meliloti positively regulates the expression of genes involved in lipopolysaccharide biosynthesis. Appl Environ Microbiol 80:5265–5273. doi: 10.1128/AEM.01393-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker DG. 2001. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14:695–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- 25.Luo L, Yao SY, Becker A, Ruberg S, Yu GQ, Zhu JB, Cheng HP. 2005. Two new Sinorhizobium meliloti LysR-type transcriptional regulators required for nodulation. J Bacteriol 187:4562–4572. doi: 10.1128/JB.187.13.4562-4572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.