Abstract
Carcinoembryonic antigen (CEA), also referred as CEACAM5, is integral to the adhesion process during cancer invasion and metastasis and is one of the most widely used tumor markers for assisting the diagnosis of cancer recurrence and cancer metastasis. Antibodies against CEA molecules have been developed for detection and diagnostic applications following tumor removal. Single domain antibodies (sdAbs) against CEA isolated from dromedary and llama exhibited high specificity in binding to tumor cells. However, because these CEA sdAbs were not designed to be orientated when conjugated to surface sensors, there is potential for significant improvements in their activity and limit of detection. Herein we modified the CEA sdAbs with two different C-terminal fusions designed to aid with orientation by way of the tail’s charge and biotin binding. A fusion which incorporated the C-terminus addition of a positively charged tail (B5-GS3K) improved biosensor sensitivity to CEA while also retaining the sub-nanomolar binding affinity and thermal stability of the unmodified sdAb. Using our fabricated surfaces on bare gold chips and a multiplexed surface plasmon resonance imager (SPRi), we quantified the specific binding activities, defined as the percentage of bound epitopes to the total immobilized, of the sdAb fusions and anti-CEA mAb. Our results demonstrate that monovalent B5-GS3K exhibited significantly improved binding activity, approximately 3-fold higher than bivalent mAb.
Keywords: Biomedical engineering, Biochemistry, Pharmaceutical science, Organic chemistry
1. Introduction
Carcinoembryonic antigen (CEA), also referred to as CEACAM5, is a single polypeptide chain consisting of 641 amino acids. It is heavily glycosylated and contains 45–50% carbohydrates, which brings the molecular weight to 150–200 kDa. It is expressed on the cell surface and the protein links to the membrane through a glycosylphosphatidyl inositol (GPI) anchor (Stanners et al., 1995; Ordonez et al., 2007). In the presence of GPI-phospholipase D (PLD), GPI is cleaved and CEA released to the extracellular matrix (Pakdel et al., 2012). However, the regulation of secretion is not completely understood.
CEA belongs to the human cellular adhesion molecules (CEACAM) subfamily, which is one of two subfamilies within the human CEA family (Thompson et al., 1991; Hammarstrom, 1999; Duffy, 2001). It is likely that CEA is integral to the adhesion process during cancer invasion and metastasis (Kabel, 2017). CEA was first discovered in the extract of human malignant colorectal tissues (Gold and Freedman, 1965; Hammarstrom, 1999) and later was also found in other cancers, such as metastasized breast tumors (Geng et al., 2015), ovarian cancer, lung tumors (Grunnet and Sorensen, 2012), gastric and pancreatic tumors (Hatakeyama et al., 2013). CEA cannot be used as a primary and independent diagnostic tumor marker due to its lack of disease sensitivity and specificity. Nonetheless, CEA levels in the blood consistently rise in patients with cancer recurrence after treatment. Therefore, the CEA level is often monitored for cancer patients following initial treatment. In fact, it is one of the most widely used tumor markers for assisting the diagnosis of cancer recurrence and cancer metastasis. CEA is also believed to be integral to other processes involving cell migration such as wound healing (Obrink, 1997).
Improving CEA detection sensitivity will require high affinity ligands which are readily produced, resistant to denaturation, and can be orientated to optimize the probability of capture. Conventional antibodies (Abs) are the current detection ligands of choice, and while they exhibit the necessary specificity and affinity, it is not uncommon to find that only 10% or less of those conjugated to the sensor surface can actively bind analyte (Raphael et al., 2015). This loss of activity can be a result of incorrect orientation, denaturation due to multi-point attachment, as well as steric hindrances (or combinations of the above). Not only is this an inefficient use of antibody resources, but a surface comprised largely of inactive antibodies is more subject to non-specific binding from other solution constituents. Such non-specific binding events increase background levels, which reduce the probability of CEA capture due to steric hindrance and, in general, counteract surface blocking and deactivation steps commonly used to aid sensor surface specificity.
Single domain antibodies (sdAbs) are the variable domains (VHHs) of heavy chain only antibodies, which exist only in camelids, such as dromedary and llama, as well as shark (termed IgNAR and shark-derived sdAb termed vNAR). Due to their small size, superior thermostability, great specificity and compatible affinity, they are a great match with the micro-sensor and nano-sensor platforms. Each VHH consists of three complementarity determining regions (CDRs) and four framework regions (FRs). SdAbs are small, stable, and soluble with a specificity and affinity comparable to conventional Abs. The ease of sdAb engineering and production is especially advantageous for tailoring the desirable function toward therapeutic and diagnostic reagents (Hamers-Casterman et al., 1993; Greenberg et al., 1995) as well as the corresponding control antibodies. Most sdAbs are very resistant to chemical and heat denaturation due to their intrinsic refolding ability. This inherent attribute makes sdAbs very attractive to be used as recognition elements on regenerative biosensor platforms. Biosensor platforms with immobilized sdAbs should be able to retain their antigen-binding capacity both in harsh environments and after multiple regeneration cycles. This is especially important for point-of-care diagnostics in more remote areas which require real-time results in the absence of refrigeration and rapid transport.
In addition, more sdAbs can be immobilized onto a biosensor chip surface than that of conventional Abs by virtue of their smaller size. This is of particular importance on nanosensor platforms with limited sensor surface area. The smaller size and ability to orient sdAbs has been shown to improve signal to noise ratios compared to that of conventional Abs when used to functionalized nanoplasmonic sensors (Raphael et al., 2015) as well as on larger commercial surface plasmon resonance (SPR) formats (Della Pia and Martinez, 2015). The later study demonstrated that with the same density of sdAbs and conventional Abs on a SPR chip, sdAbs had a lower limit of detection, suggesting that immobilized sdAbs can access binding pockets better than immobilized conventional Abs. However, sdAbs, like conventional Abs, randomly orient on sensor surfaces and modifications designed for directional immobilization have been shown to improve their activity (Even-Desrumeaux et al., 2010; Trilling et al., 2014) and reduce the chance that inactive antibodies are sites of non-specific binding. One convenient way to accomplish directional immobilization is to introduce fusions of sdAbs, which can be readily engineered onto the C-terminus to tailor the desired functionality without compromising their biochemical and physical properties. We previously used such a fusion approach to facilitate the proper orientation of immobilized sdAbs on a gold surface which had been chemically modified to be negatively charged. The fusion significantly improved both binding activity and sensor sensitivity for ricin detection (Raphael et al., 2015).
Antibodies against different CEACAM molecules have been developed for detection and diagnostic applications following tumor removal (Stocchi and Nelson, 1998). SdAbs against CEA were also isolated from dromedary and llama and showed binding to tumor cells (Cortez-Retamozo et al., 2004; Vaneycken et al., 2010; Behar et al., 2009). However, these CEA sdAbs were not designed to be orientated on sensor surfaces and need to be modified to orient properly for improved activity and limit of detection. Herein we modified the CEA sdAbs with two different C-terminal fusions designed to aid with orientation by way of the tail’s charge or through a biotin-rhizavidin mechanism (Liu et al., 2014). Using a multiplexed surface plasmon resonance imager (SPRi) we compared the binding kinetics of the sdAb fusion constructs, as well as that of anti-CEA mAb, and quantified their binding activities and sensitivities when crosslinked to the sensor surface. We demonstrate that the addition of a C-terminus, positively charged tail (B5-GS3K) improves biosensor sensitivity to CEA while also retaining the high binding affinity and thermal stability of the unmodified sdAb.
2. Materials and methods
2.1. Construction of recombinant anti-CEA sdAb clones
The DNA sequence for anti-CEA sdAb, referred to as B5, was obtained by reverse translating the protein sequence from the literature (Vaneycken et al., 2010) and synthesized by Eurofin MWG Operon (Louisville, KY). NcoI and XhoI sequences were added to 5′ and 3′ respectively. The NcoI and XhoI DNA fragments were cloned into pET 22b vectors with the corresponding sites. To construct B5-GS3K fusion, NcoI-NotI sdAb fragments cut from B5 sdAb from pET22b were inserted into GS3K-pet22b cut with the corresponding enzymes. A rhizavidin (Rhiz) fragment flanked with XhoI sites at both ends was fused to B5 sdAb pET22b cut with XhoI. The ligated clones were sent out for sequencing to screen the clone with right orientation of Rhiz fusion. The sequences for other recombinant clones were also confirmed by Sanger sequencing (Eurofin Genomics). All the restriction enzymes used in this study were purchased from New England Biolab unless specified, most of the reagents and media were obtained from Thermo Fisher scientific.
2.2. Preparation of recombinant anti-CEA sdAbs
The recombinant sdAb clones were transformed into E. coli Tuner (DE3) host and plated onto LB agar plates supplemented with Ampicillin and incubated at 37 °C overnight. The next day, a single colony was inoculated into 50 mL Terrific Broth (TB) and shook overnight at 25 °C. The culture was then transferred to a 500 mL TB culture to grow for 3 hours at RT before adding 0.5 mM IPTG to induce protein expression. After 2.5 hrs of IPTG induction, bacterial cells were pelleted down by centrifugation and subjected to osmotic shock and IMAC extraction according to a previously published protocol (Hayhurst et al., 2003; Turner et al., 2014). Following IMAC extraction, recombinant proteins were further purified from other protein contaminates through Superdex 75 10/300 GL columns (GE Healthcare) operated under a BioLogic DuoFlow chromatography system (Bio-Rad). Protein concentrations were estimated by measuring their absorbance at 280 nm.
2.3. Circular dichroism (CD) for measuring melting temperature (Tm)
Purified recombinant sdAbs were diluted to 22 μg/mL in deionized water and transferred into a quartz cuvette with a 1 cm path length. The cuvette was placed onto the holder of a Jasco J-815 Spectropolarimeter and CD measurement was performed at an ultraviolet wavelength between 200 and 210 nm. Samples were heated from 25 °C to 95 °C at a rate of 2.5 °C/min and an absorbance of denaturing graph was generated. A cooling stage at the same rate was used to generate the refolding graph after denaturation. The melting temperature is taken to be the inflection point of the S-shaped denaturation curve. The refolding ability is calculated as the change in CD magnitude upon cooling divided by the change in magnitude upon heating (expressed as a percentage).
2.4. Surface plasmon resonance measurements of CEA and anti-CEA sdAb binding kinetics
For these measurements, sensor surfaces were biofunctionalized with CEA and the antibodies were introduced in solution. In particular, multiplexed surface plasmon resonance imagery (SPRi) based binding kinetic measurements were performed using the ProteOn XPR36 (Bio-Rad) following a similar protocol to those described previously (Anderson et al., 2012). For testing the kinetics of the anti-CEA antibodies including recombinant sdAbs, mAb, and the control IgG, a commercial chip designed for amine coupling (BioRad GLC chip) was conjugated with purified CEA (Scripps, San Diego, CA). The CEA proteins were diluted to ∼20 μg/mL in 300 μL of 10 mM acetate buffer, pH 5. Standard EDC coupling chemistry provided by the manufacturer was utilized to attach the antigens to the chip with the exception that distilled water was maintained as the running buffer until after the coupling step. All experiments were performed at 25 °C. The binding of recombinant sdAbs and monoclonal Ab (mAb) was tested by flowing six concentrations (three-fold dilutions from 1000 to 0 nM) at 100 μL/min for 90 s over the antigen-coated chip and then monitoring dissociation for 420 s. The chip was regenerated using 0.085% phosphoric acid for 36 s, prior to any additional testing. The ordering of the antibody concentrations in the flow channels over the chip was alternated from left-to-right to right-to-left between tests to minimize artifacts due to incomplete regeneration. The data was corrected for inter spot signals and zero concentration, when appropriate. The results were analyzed using the accompanying ProteOn Manager 3.1 software using a global analysis Langmuir fit and bivalent analyte model for the mAb and sdAb-Rhiz.
2.5. Surface plasmon resonance measurements of sdAb coupling efficiencies and activity
For these measurements, recombinant sdAbs and mAbs were immobilized onto bare gold chips (Bio-Rad), which were pre-cleaned using hydrogen plasma ashing at 40 W, 300 m Torr in a mixture of 5% hydrogen and 95% argon. The cleaned surface was functionalized by immersing the chip in 3:1 ratio of SH-(CH2)8-EG3-OH (SPO): HS-(CH2)11-EG3-COOH (SPC) (Prochimia Surfaces Sp.) for 18 h to form a self-assembled monolayer (SAM) (Raghu et al., 2015). The chips were then rinsed with ethanol and dried under nitrogen gas. Next, the surface was activated with EDC/NHS in the SPRi instrument at a flow rate of 30 μl/min for 300 s. MAb for CEA (Scripps) and IgG mouse control antibodies were introduced at a concentration of 5 μg/mL whereas recombinant sdAbs, B5, B5-GS3K, C8-GS3K (negative control for GS3K), and B5-Rhiz were introduced with a concentration of 10 μg/mL The antibodies were diluted in phosphate buffer at a pH of 6.0 and their flow rate was 30 μl/min for 300 s. Unreacted SPC was deactivated with 0.1 M ethanolamine at a flow rate of 30 μl/min for 300 s. As a final analyte step, 100 nM of purified CEA (Scripps) was introduced to compare the activity and affinity of the various antibodies. All SPR measurements for the recombinant antibodies were performed using a Bio-Rad’s XPR36 protein analysis system at 25 °C.
3. Results and discussion
3.1. Preparation of recombinant anti-CEA sdAbs
SdAbs consisting of variable domains of heavy chain only antibodies are readily tailored toward the desired functions by site-directed mutations and fusions using standard molecular biological methods. They are soluble and easily produced from bacterial hosts. Recombinant anti-CEA sdAbs including B5 sdAb, B5-GS3K and B5-Rhiz were cloned into pet22b for protein expression and purification (Fig. 1). Rhiz peptide containing unpaired Cys allows for the B5-Rhiz to form dimers. Purified recombinant sdAbs had more than 95% purity after separation using gel extraction column based separation (data not shown). The yields for the B5 sdAb and B5-GS3K fusion was between 12–18 mg per liter of bacterial culture, while the yield for B5-Rhiz was approximately 3–5 mg per liter of culture. The yields were sufficient for all subsequent applications.
Fig. 1.
The fusion of C-terminal tails of CEA single domain antibodies. A. Unmodified CEA sdAb. B. CEA-GS3K consisting of C-terminal positively charged tail. C. CEA-Rhiz fusion has the C-terminal Rhizavidin (Rhiz), which forms dimers through the unpaired Cys (C in red font). All the recombinant proteins contain a 6 His tag at the end of the C-terminus.
3.2. Thermostability and refolding among the recombinant anti-CEA sdAbs
The stability of sdAbs is defined by their retention of binding activity after heating. Usually their high melting temperature (Tm), which is defined as an inflection point of the melting curve, and the intrinsic refolding capability of sdAbs contribute to stability against heat and other denaturants (Dumoulin et al., 2002). These intrinsic attributes allow sdAbs to work effectively in harsh environments and survive typically unsuitable pH cross-linking conditions if needed. Moreover, this ability allows biosensor platforms to regenerate multiple times while retaining analyte binding activity (Saerens et al., 2005). To test the sdAb refolding capability and intrinsic thermostability, circular dichroism (CD) was employed with Tm as the indicator. Our CD results show that all three recombinant sdAbs exhibited similar Tms, suggesting that C-terminal fusions do not alter the intrinsic melting property of the parental sdAb (Table 1). Likewise, C-terminal GS3K fusion did not alter the refolding capability of the parental sdAb. However, the C-terminal fusion to Rhiz decreased the refolding ability to 76%, which has the potential to negatively impact sdAb regeneration capabilities upon exposure to extreme temperatures and denaturants. Our data are consistent with previous observations that the fusion of longer peptides to sdAbs could affect the refolding more significantly (Liu et al., 2014).
Table 1.
Measurements for Tm and refolding of engineered sdAbs.
| Clone name | (°C) | Refolding |
|---|---|---|
| B5 sdAb | 67 ± 1 | 94% |
| B5-GS3K | 68 ± 1 | 92% |
| B5-Rhiz | 67 ± 1 | 76% |
3.3. Binding kinetics for the recombinant sdAbs
Binding affinity and specificity determine the performance of the recombinant sdAbs. The ProteOn XPR36 system, a multiplexed SPR-based imaging platform, was used to measure antibody association and dissociation rates to the immobilized CEA in real time. Purified CEA molecules (Scripps) were immobilized onto commercial SPR GLC chips (BioRad) at 20 μg/mL for high density surface conjugation using the EDC coupling chemistry described above, followed by flowing serial dilutions of each recombinant sdAb over the surface of the chip. The binding signals generated from commercial sensor surface is generally low and sdAbs exhibit only one-tenths of signals of conventional Ab due to their small size (Fig. S1). The association rate constant (ka) and dissociation rate constant (kd) for each sdAb and fusion were obtained using a one-to-one Langmuir model from which the equilibrium dissociation rate constant (KD = kd/ka) was calculated, with the exception of the mAb and B5-Rhiz fusion for which a bivalent model was used (BioRad XPR36 Software) as summarized in Table 2. Among the three recombinant sdAbs, B5-Rhiz exhibited a KD at least two orders of magnitude lower than the B5-sdAb and the conventional mAb, while the GS3K fusion had a similar KD to that of the sdAb alone indicating that C-terminus modification did not significantly alter its avidity. B5-Rhiz facilitates the formation of sdAb dimers capable of binding two CEA molecules simultaneously, thereby lowering the effective kd (Table 2, Fig. S1).
Table 2.
Measurements of binding kinetics.
| Antibodies | ka (M−1 s−1) | kd (s−1) | KD (M) |
|---|---|---|---|
| B5 sdAb | 5.7E + 5 | 5.5E-4 | 9.7E-10 |
| B5-GS3K | 4.9E + 5 | 3.9E-4 | 8.0E-10 |
| B5-Rhiz | 3.4E + 4 | 4E-9* | 1E-13* |
| CEA mAb | 9.3E + 5 | 2.8E-5 | 3.0E-11 |
Near lower limit of instrument sensitivity.
3.4. Coupling efficiency of sdAbs and mAb
The sdAbs and mAb were conjugated to the SPRi chip surface in parallel, each in a dedicated lane, using EDC/NHS crosslinking chemistry as described above. The relatively large response to mAb conjugation versus the B5 constructs is consistent with the mAb having a molecular weight approximately 10 times that of B5-GS3K and B5-sdAb, and approximately 5 times that of B5-Rhiz (Table 3, Fig. 2). Our results shows that the SPR response for mAb is approximately 6 fold higher than B5-sdAbs and 4-fold higher than B5-Rhiz and B5-GS3K. After normalizing the responses by molecular size, the coupling efficiency for the mAb is similar to that of the B5-sdAb and B5-Rhiz, and B5-GS3K has approximately 50% higher coupling efficiency than B5-sdAb (Fig. 2). B5-Rhiz was also immobilized through cross-linked biotins on the sensor surface, however, the SPR signal (RU) was low and resulted in a low CEA binding signal (data not shown). In contrast, when using a commercial GLC chip, our previous sdAb-Rhiz fusion studies exhibited higher SPR responses when conjugated through biotin than through direct covalent linking. We believe the SPC: SPO SAM prepared on the sensor surface in this study is different from the long alginated polymers on the commercial GLC chip surface and results in lower coupling through the biotin-Rhiz mechanism. Therefore, for a fair comparison of CEA binding activity measurements, cross-linking was used to couple the B5-Rhiz onto the prepared sensor surface. In general our prepared sensor surface generates much higher signals than commercial SPR GLC sensor surface (Fig. 3 vs Fig. S1).
Table 3.
Measurements of binding activity for each immobilized Ab.
| Antibody for CEA | XL (RU) | XA (RU) | pI/MWL* | Activity (%) | # of ligand on surface x (108/mm2) |
|---|---|---|---|---|---|
| B5 sdAb | 1200 | 525 | 6.35/14.7 | 3.6 | 480 |
| B5-GS3K | 805 | 910 | 7.84/16.0 | 14.3 | 322 |
| B5-Rhiz | 1363 | 839 | 5.27/29.4 | 5.0 | 272 |
| CEA mAb | 4389 | 621 | 7.0/155 | 5.4 | 351 |
MWL (kDa) represents the molecular weight of the ligand (antibody). The molecular weight for CEA used to calculate the activity was 180 kDa.
Fig. 2.
SPR sensograms showing the crosslinking of the various antibodies to the sensor surface. The concentrations used were 5 μg/mL for the mAb and 10 μg/mL for the sdAbs. The dashed line separates the association and dissociation phases.
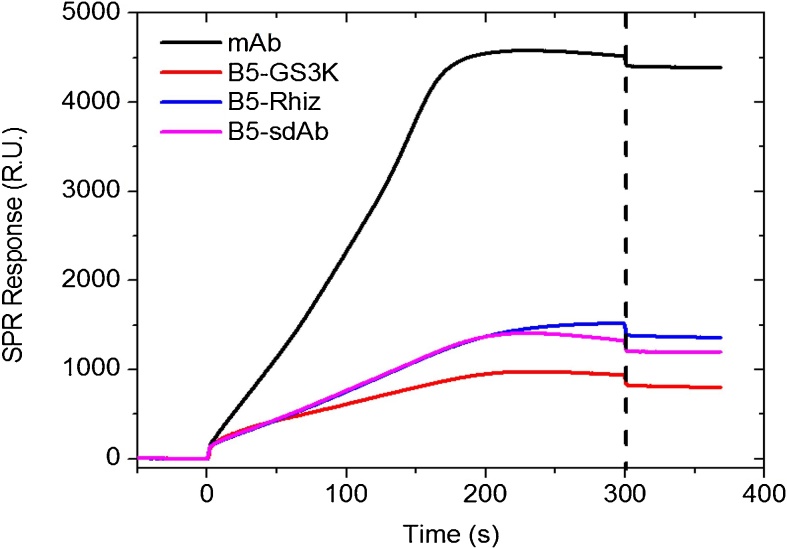
Fig. 3.
SPR sensograms showing binding of 100 nM CEA antigen to the various antibodies. GS3K antibody (red) has 46% higher response than the commercial monoclonal antibody (black). The dashed line separates the association and dissociation phases.
3.5. Performance of surface immobilized recombinant sdAbs and fusions
After the ligand conjugation step, 100 nM of CEA was introduced to each lane as well as control lanes. The resulting SPR plots of CEA antigen against various antibodies indicate that immobilized B5-GS3K has the highest response to 100 nM CEA, 46% higher than the commercial anti-CEA mAb and approximately 2-fold higher binding signal than B5 sdAb alone (Fig. 3) as measured by the signal at the end of the dissociation phase. Care was taken to compare the response of surfaces with similar ligand binding site densities. The results are consistent with the notion that the positively charged tail preferentially orients downward to the negatively charged chip surface and orients the sdAb in a direction more accessible to CEA binding than the surfaces immobilized with other Abs (Raphael et al., 2015).
To further quantify the percentage of active analyte binding numbers of immobilized Abs, we calculated the specific activity defined as the moles of the antigen bound per mole of the immobilized Ab, expressed as a percentage (Wimalasena and Wilson, 1991), given by
where XA and XL are the SPR responses in RU for analyte and the ligand (Ab) respectively, MWA and MWL are the molecular weight of the analyte and the ligand respectively, and Sm is the number of binding sites for each ligand (Ab). This equation is independent of the sensor calibration constant due to the ratio of XA to XL.
The resulting activities calculated from Fig. 2, Fig. 3 show the B5-GS3K construct having considerably higher activity (Table 3). We then checked the reproducibility of these results, by repeating all functionalization steps three times with a specific focus on the antibodies with the highest activities from our initial tests (B5-GS3K and the mAb). From these experiments, the mean activities and standard deviations were 14.27 ± 0.6% and 5.4 ± 1.5% for B5-GS3K and the mAb, respectively, showing the improved activity of the B5-GS3K construct was statistically significant The limit of detection for the CEA antigen for both the mAb and B5-GS3K was approximately 300 pM, indicating that our B5-GS3K performs equally well or better when applied to SPR chips and control experiments showed no measurable non-specific binding (Fig. 4).
Fig. 4.
SPR sensograms showing binding of 100 nM CEA antigen to monoclonal CEA, B5-GS3K CEA and their respective control antibodies. The dashed line separates the association and dissociation phases.
4. Conclusion
Anti-CEA sdAbs were engineered by adding a C-terminal GS3K peptide or Rhiz biotin binding dimer forming protein to facilitate the orientation of immobilized Abs on carbohydrate functionalized gold surfaces. These recombinant sdAbs exhibited no significant changes in physical properties except for the Rhiz fusion which showed a 50% decrease of refolding capability compared to that of the parental sdAb. Both fusions exhibited increased binding affinity and activity compared to the parental sdAb. The GS3K fusion had higher binding activity when surface immobilized, while the Rhiz fusion had the lowest equilibrium dissociation rate constant. The GS3K fusion had similar or lower limits of detection to the CEA mAb and nearly 3-fold greater activity. We note that newer SPR instruments have a 50-fold lower noise floors than the one used here, making SPR a viable alternative to the sandwich assay techniques more typically used for CEA medical diagnostics. Furthermore, unlike the sandwich assays, the real-time nature of SPR provides kinetic rate constant information (ka, kd and KD) which give important additional evidence that the targeted analyte has been detected. These advantages, combined with their small size, make the sdAb fusions excellent candidates for incorporation into a wide range of surface-based sensors, including the growing selection of nanosensors.
Declarations
Author contribution statement
Jinny L. Liu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Deepa Raghu: Performed the experiments; Analyzed and interpreted the data.
George P. Anderson: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data.
Ellen R. Goldman: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Joseph A. Christodoulides: Contributed reagents, materials, analysis tools or data.
Marc Raphael: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Competing interest statement
The authors declare no conflict of interest.
Funding statement
This work was supported by the Naval Research Laboratory’s Institute for Nanoscience, Office of Naval Research, the Office of the Assistant Secretary of Defense for Research & Engineering (Vannevar Bush Award), and a National Research Council Research Postdoc Associateship Award.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
SPR sensorgrams showing CEA binding kinetics of various antibodies to a commercial SPR GLC chip surface. Following the crosslinking of 100 nM of CEA to the surface via amine groups, a serial of concentrations, 300, 100, 33, 11, 1.7 nM of B5 sdAb and B5-GS3K were then introduced and the corresponding binding kinetics graphs are indicated in A and B respectively. The binding kinetics graph for dimeric B5-Rhiz is shown in C, while monoclonal CEA Ab is shown in D.
References
- Anderson G.P., Legler P.M., Zabetakis D., Goldman E.R. Comparison of immunoreactivity of staphylococcal enterotoxin B mutants for use as toxin surrogates. Anal. Chem. 2012;84:5198–5203. doi: 10.1021/ac300864j. [DOI] [PubMed] [Google Scholar]
- Behar G., Chames P., Teulon I., Cornillon A., Alshoukr F., Roquet F., Pugniere M., Teillaud J.-L., Gruaz-Guyon A., Pelegrin A., Baty D. Llama single-domain antibodies directed against nonconventional epitopes of tumor-associated carcinoembryonic antigen absent from nonspecific cross-reacting antigen. FEBS J. 2009;276:3881–3893. doi: 10.1111/j.1742-4658.2009.07101.x. [DOI] [PubMed] [Google Scholar]
- Cortez-Retamozo V., Backmann N., Senter P.D., Wernery U., De Baetselier P., Muyldermans S., Revets H. Efficient cancer therapy with a nanobody-based conjugate. Cancer Res. 2004;64:2853–2857. doi: 10.1158/0008-5472.can-03-3935. [DOI] [PubMed] [Google Scholar]
- Della Pia E.A., Martinez K.L. Single domain antibodies as a powerful tool for high quality surface plasmon resonance studies. PLoS One. 2015;10:e0124303. doi: 10.1371/journal.pone.0124303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy M.J. Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clin. Chem. 2001;47:624–630. [PubMed] [Google Scholar]
- Dumoulin M., Conrath K., Van Meirhaeghe A., Meersman F., Heremans K., Frenken L.G.J., Muyldermans S., Wyns L., Matagne A. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Desrumeaux K., Baty D., Chames P. Strong and oriented immobilization of single domain antibodies from crude bacterial lysates for high-throughput compatible cost-effective antibody array generation. Mol. BioSyst. 2010;6:2241–2248. doi: 10.1039/c005279e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng B., Liang Man-Man, Ye Xiao-Bing, Zhao Wen-Ying. Association of CA 15-3 and CEA with clinicopathological parameters in patients with metastatic breast cancer. Mol. Clin. Oncol. 2015;3:232–236. doi: 10.3892/mco.2014.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold P., Freedman S.O. Demonstration of tumor-specific antigens in Human Carcinomata by immunological tolerance and absorption techniques. J. Exp. Med. 1965;121:439–462. doi: 10.1084/jem.121.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg A.S., Avila D., Hughes M., Hughes A., McKinney E.C., Flajnik M.F. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- Grunnet M., Sorensen J.B. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer. 2012;76:138–143. doi: 10.1016/j.lungcan.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- Hammarstrom S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 1999;9:67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- Hatakeyama K., Wakabayashi-Nakao K., Ohshima K., Sakura N., Yamaguchi K., Mochizuki T. Novel protein isoforms of carcinoembryonic antigen are secreted from pancreatic, gastric and colorectal cancer cells. BMC Res. Notes. 2013;6:381. doi: 10.1186/1756-0500-6-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst A., Happe S., Mabry R., Koch Z., Iverson B.L., Georgiou G. Isolation and expression of recombinant antibody fragments to the biological warfare pathogen Brucella melitensis. J. Immunol. Methods. 2003;276:185–196. doi: 10.1016/s0022-1759(03)00100-5. [DOI] [PubMed] [Google Scholar]
- Kabel A.M. Tumor markers of breast cancer: new perspectives. J. Oncol. Sci. 2017;3:5–11. [Google Scholar]
- Liu J.L., Zabetakis D., Walper S.A., Goldman E.R., Anderson G.P. Bioconjugates of rhizavidin with single domain antibodies as bifunctional immunoreagents. J. Immunol. Methods. 2014;411:37–42. doi: 10.1016/j.jim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Obrink B. CEA adhesion molecules: multifunctional proteins with signal-regulatory properties. Curr. Opin. Cell Biol. 1997;9:616–626. doi: 10.1016/S0955-0674(97)80114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez C., Zhai A.B., Camacho-Leal P., Demarte L., Fan M.M.Y., Stanners C.P. GPI-anchored CEA family glycoproteins CEA and CEACAM6 mediate their biological effects through enhanced integrin alpha5beta1-fibronectin interaction. J. Cell Physiol. 2007;210:757–765. doi: 10.1002/jcp.20887. [DOI] [PubMed] [Google Scholar]
- Pakdel A., Naghibalhossaini F., Mokarram P., Jaberipour M., Hosseini A. Regulation of carcinoembryonic antigen release from colorectal cancer cells. Mol. Biol. Rep. 2012;39:3695–3704. doi: 10.1007/s11033-011-1144-0. [DOI] [PubMed] [Google Scholar]
- Raghu D., Christodoulides J.A., Delehanty J.B., Byers J.M., Raphael M.P. A label-free technique for the spatio-temporal imaging of single cell secretions. J. Vis. Exp. 2015;105 doi: 10.3791/53120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raphael M.P., Christodoulides Joseph A., Byers Jeff M., Anderson George P., Liu Jinny L., Turner Kendrick B., Goldman Ellen R., Delehanty James B. Optimizing nanoplasmonic biosensor sensitivity with orientated single domain antibodies. Plasmonics. 2015;10:1649–1655. doi: 10.1007/s11468-015-9969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saerens D., Frederix F., Reekmans G., Conrath K., Jans K., Brys L., Huang L., Bosmans E., Maes G., Borghs G., Muyldermans S. Engineering camel single-domain antibodies and immobilization chemistry for human prostate-specific antigen sensing. Anal. Chem. 2005;77:7547–7555. doi: 10.1021/ac051092j. [DOI] [PubMed] [Google Scholar]
- Stanners C.P., DeMarte L., Rojas M., Gold P., Fuks A. Opposite functions for two classes of genes of the human carcinoembryonic antigen family. Tumour Biol. 1995;16:23–31. doi: 10.1159/000217925. [DOI] [PubMed] [Google Scholar]
- Stocchi L., Nelson H. Diagnostic and therapeutic applications of monoclonal antibodies in colorectal cancer. Dis. Colon Rectum. 1998;41:232–250. doi: 10.1007/BF02238254. [DOI] [PubMed] [Google Scholar]
- Thompson J.A., Grunert F., Zimmermann W. Carcinoembryonic antigen gene family: molecular biology and clinical perspectives. J. Clin. Lab. Anal. 1991;5:344–366. doi: 10.1002/jcla.1860050510. [DOI] [PubMed] [Google Scholar]
- Trilling A.K., Hesselink T., van Houwelingena A., Cordewener J.H.G., Jongsma M.A., Schoffelen S., van Hest J.C.M., Zuilhof H., Beekwilder J. Orientation of llama antibodies strongly increases sensitivity of biosensors. Biosens. Bioelectron. 2014;60:130–136. doi: 10.1016/j.bios.2014.04.017. [DOI] [PubMed] [Google Scholar]
- Turner K.B., Zabetakis D., Goldman E.R., Anderson G.P. Enhanced stabilization of a stable single domain antibody for SEB toxin by random mutagenesis and stringent selection. Protein Eng. Des. Sel. 2014;27:89–95. doi: 10.1093/protein/gzu001. [DOI] [PubMed] [Google Scholar]
- Vaneycken I., Govaert J., Vincke C., Caveliers V., Lahoutte T., De Baetselier P., Raes G., Bossuyt A., Muyldermans S., Devoogdt N. In vitro analysis and in vivo tumor targeting of a humanized, grafted nanobody in mice using pinhole SPECT/micro-CT. J. Nucl. Med. 2010;51:1099–1106. doi: 10.2967/jnumed.109.069823. [DOI] [PubMed] [Google Scholar]
- Wimalasena R.L., Wilson G.S. Factors affecting the specific activity of immobilized antibodies and their biologically active fragments. J. Chromatogr. 1991;572:85–102. doi: 10.1016/0378-4347(91)80475-r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SPR sensorgrams showing CEA binding kinetics of various antibodies to a commercial SPR GLC chip surface. Following the crosslinking of 100 nM of CEA to the surface via amine groups, a serial of concentrations, 300, 100, 33, 11, 1.7 nM of B5 sdAb and B5-GS3K were then introduced and the corresponding binding kinetics graphs are indicated in A and B respectively. The binding kinetics graph for dimeric B5-Rhiz is shown in C, while monoclonal CEA Ab is shown in D.