Abstract
The aim of the present study was to compare the expression of transcriptional coactivator with the PDZ-binding motif (TAZ) in pancreatic cancer (PC) patients, and to investigate the regulation mechanisms of TAZ in the proliferation of PC. PC tissues and matched peritumoral tissues, pancreatic juice and serum were collected from PC patients who underwent pancreatectomy between June 2012 and December 2015 at the Affiliated Hospital of Qingdao University (Qingdao, China). Pancreatic juice and serum were collected from patients with chronic pancreatitis as a control. The levels of taz mRNA expression in the samples were examined by reverse-transcription quantitative polymerase chain reaction, and the protein expression of TAZ was assessed by western blot analysis and ELISA. MicroRNAs (miRNAs) that regulate TAZ expression were also predicted by bioinformatics analysis and validated by dual luciferase reporter and rescue assays. In addition, the proliferation of PC cells was evaluated after transfection with TAZ small interfering RNA (siRNA) or its upstream miRNA agomir. Expression of TAZ was significantly increased in the PC tissues, pancreatic juice and serum of PC patients at the mRNA and protein levels compared with controls (P<0.05). Furthermore, TAZ was predicted and verified to be a target of miRNA (miR)-185, and miR-185 and TAZ were inversely expressed in samples from PC patients (P<0.05). In addition, TAZ siRNA or agomiR-185 transfection significantly inhibited human pancreatic adenocarcinoma cell proliferation (P<0.05). However, overexpression of TAZ in the agomiR-185 group rescued the inhibition (P<0.05). Finally, the expression of TAZ effector proteins, namely ankyrin repeat domain-containing protein and cysteine-rich 61, were upregulated in PC tissues (P<0.05), but repressed following transfection of PC cells with agomiR-185 (P<0.05). Thus, miR-185 may regulate the proliferation of PC by targeting TAZ, making it a promising diagnostic marker for PC.
Keywords: microRNA-185, pancreatic cancer, transcriptional coactivator with PDZ-binding motif, proliferation
Introduction
Pancreatic cancer (PC) is associated with high rates of morbidity and mortality. In the United States, the incidence and mortality rates of PC ranked 8 and 4th of all tumors, respectively (1,2). In China, the incidence of PC has been increasing yearly, and its incidence and mortality rates ranked 7 and 6th, respectively (3). Therefore, PC is a common tumor of the digestive system.
At present there is no widely accepted screening method for the early detection of PC. Surgical treatment is currently used to prolong the survival rate of patients with PC; however, the 5-year survival rate of patients typically remains low (15–25%) following radical resection of the tumor (4). Biomarkers are considered to be important in predicting the prognosis of PC (5–7), and numerous biological markers associated with PC have been identified by previous studies (8,9). In particular, transcriptional coactivator with PDZ-binding motif (TAZ), as a type of 14-3-3 binding protein, was identified in 2000 (10). TAZ is a costimulatory protein transcription factor that contains a WW domain, and is involved in the proliferation, migration and metastasis of tumors through modulation of its target genes, including ankyrin repeat domain-containing protein (ANKRD), cysteine-rich 61 (CYR61) and connective tissue growth factor (11–14). The oncogenic role of TAZ in PC has been verified (15,16). More recently, studies into microRNAs (miRNAs) have demonstrated that upregulation of numerous miRNAs may suppress the development, proliferation and metastasis of tumors by regulating the expression of TAZ (17,18). However, to the best of our knowledge, no experimental or clinical studies have documented the potential roles of miRNA (miR)-185 in PC.
The present study investigated the expression of TAZ at the mRNA and protein levels in the tumor tissues, blood and pancreatic fluid of PC patients and controls using reverse transcription-quantitative polymerase chain reaction (RT-qPCR), western blot analysis and ELISA. The miRNAs targeting TAZ were also predicted by bioinformatics methods, and TAZ was confirmed as a target of miR-185 by a dual-luciferase reporter assay. Furthermore, the expression of miR-185 was measured in the samples obtained from PC patients.
Materials and methods
PC tissue specimens
A total of 46 patients with PC who were diagnosed and received surgery between June 2012 and December 2015 at the Affiliated Hospital of Qingdao University (Qingdao, China) were enrolled. A total of 35 patients with chronic pancreatitis with non-neoplastic diseases were included as a control group. Among the 46 PC patients, there were 28 male and 18 female patients, and the median age was 56 years (ranging between 31 and 75 years). Among the 35 controls, there were 20 male and 15 female patients, and the median age of patients was 58 years (ranging between 25 and 77 years). All subjects were diagnosed and had no history of hormone medicine, radiotherapy and chemotherapy prior to surgery. The Tumor Node Metastasis (TNM) clinical stages of the 46 cases of PC were as follows: Stage I: 2 cases; stage II: 1 case; stage III: 12 cases and stage IV: 31 cases (19). Clinical information and medical records of patients were also collected. Prior written informed consent was obtained from all patients and the study was approved by the Ethics Review Board of the Affiliated Hospital of Qingdao University.
Sample collection
Paired human PC tissues and their matched peritumoral non-cancerous tissues were collected during surgery and stored in liquid nitrogen (−196°C) until further analysis. Serum was collected from the peripheral blood of PC and chronic pancreatitis patients. Briefly, 10–15 ml peripheral blood was kept at 4°C for 1–2 h, and serum in the upper layer was aspirated and centrifuged for 10 min at 400 × g and 4°C. The serum was then aliquoted and stored at −80°C. A total of 10 ml pancreatic juice was also collected from PC and chronic pancreatitis patients during endoscopic retrograde cholangiopancreatography examination and stored at −80°C.
Plasmid construction
The coding sequence (CDS) of the taz gene (10,20) was cloned into a PCDNA 3.1 plasmid (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) using a ClonExpress II One Step Cloning kit (C112-01; Vazyme Biotech Co., Ltd., Nanjing, China) according to the manufacturer's protocol. The primers used for the clone were as follows: Taz CDS forward, 5′-CTAGCGTTTAAACTTAAGCTTATGCCTCTGCACGTGAAGTGG-3′ and reverse, 5′-CCACACTGGACTAGTGGATCCCTATCTCCCAGGCTGGAGGTG-3′, and the Taz CDS sequence was confirmed by sequencing (Sangon Biotech Co., Ltd., Shanghai, China) (21).
Cell culture and transfection
The human PC cell lines HPAC and PANC-1, as well as 293T cells, were purchased from cell bank of Chinese Academy of Sciences (Shanghai, China). The cells were cultured in F12/Dulbecco's modified Eagle medium (DMEM) or RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.), respectively, supplemented with 10% heat inactivated fetal bovine serum (FBS; Sangon Biotech Co., Ltd.) and 1% penicillin-streptomycin. Cells were incubated in a humidified atmosphere with 5% CO2 at 37°C. One day before transfection, 3×105 cells were inoculated into 24-well plates with F12/DMEM or RPMI-1640 medium supplemented with 10% FBS. When cell confluence reached 70% at 37°C, cells were transfected with TAZ small interfering RNA (siRNA), agomiR-185 (5′-UGGAGAGAAAGGCAGUUCCUGA-3′) or, scramble RNA (5′-AAGACUAGGGAGUUUAGGACCG-3′; negative control, NC; RiboBio Co., Ltd., Guangzhou, China) using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Untransfected cells were used as a control. For a rescue assay, cells were cotransfected with agomiR-185 and Taz CDS-expressing plasmid or with agomiR-185 and the empty plasmid (control). Cells transfected with scramble RNA and empty plasmid were used as a NC. Cells were collected at 48 h post-transfection and stored at 37°C for further experiments. Sangon Biotech Co., Ltd. provided three TAZ siRNA sequences, and the first siRNA sequence with the highest knockdown efficiency was selected. The sequences of the three siRNA sequences were as below. No. 1, sense: 5′-GGGAAAGUGAACAUGAGUUdTdT-3′; antisense: 5′-AACUCAUGUUCACUUUCCCdTdT-3′; No. 2, sense: 5′-UGACGUCCUUCCUAACAGUdTdT-3′; antisense: 5′-ACUGUUAGGAAGGACGUCAdTdT-3′; No. 3, sense: 5′-GAGGAAUUCCAGCAUCUGAdTdT-3′; antisense: 5′-UCAGAUGCUGGAAUUCCUCdTdT-3′.
RNA extraction and RT-qPCR
Total RNA was isolated from HPAC/PANC-1 cells and patient samples using TRIzol® isolation reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. RNA was quantified using Nanodrop 2000 (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). mRNA (1 µg) was reverse transcribed using a TIANScriptII cDNA first strand cDNA synthesis kit (Tiangen Biotech Co., Ltd., Beijing, China) into cDNA. miRNA was reverse transcribed using a miRcute miRNA cDNA synthesis kit (Tiangen Biotech Co., Ltd.). qPCR was performed using a SuperReal PreMix (SYBR Green; Tiangen Biotech Co., Ltd.) for mRNA, or a miRcute miRNA qPCR detection kit (Tiangen Biotech Co., Ltd.) for miRNA. For mRNA amplification, the reaction mixture was incubated for 1 cycle at 95°C for 30 sec, followed by 40 cycles at 95°C for 10 sec and 62°C for 40 sec. For miRNA, the reaction mixture was incubated for 1 cycle at 95°C for 3 min, followed by 40 cycles at 95°C for 12 sec, 62°C for 35 sec and 72°C for 15 sec. For mRNA amplification, the primers used were as follows: For TAZ, forward, 5′-CTTGGATGTAGCCATGACCTT-3′ and reverse, 5′-TCAATCAAAACCAGGCAATG-3′; for β-actin, forward, 5′-CGGGAAATCGTGCGTGAC-3′ and reverse, 5′-CAGGAAGGAAGGCTGGAAG-3′; for ANKRD, forward, 5′-AGTAGAGGAACTGGTCACTGG-3′ and reverse, 5′-TGGGCTAGAAGTGTCTTCAGAT-3′; for CYR61, forward, 5′-CCTTGTGGACAGCCAGTGTA-3′ and reverse, 5′-ACTTGGGCCGGTATTTCTTC-3′; and for GAPDH, forward, 5′-CTCCTGCACCACCAACTGCT-3′ and reverse, 5′-GGGCCATCCACAGTCTTCTG-3′. For miRNA, the primers used were as follows: For miR-185, forward, 5′-TGGAGAGAAAGGCAGTTCCTGA-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; and for small nuclear U6, forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′. The relative expression levels were calculated using the 2−ΔΔCq method (22). The expressions of β-actin, GAPDH and U6 were used to normalize that of TAZ mRNA, ANKRD and CYR61 mRNA and miR-185, respectively. Each experiment was replicated three times.
MTT assay
PC cells were seeded into 96-well plates at a concentration of 2,000 cells/well in triplicate. After culturing by DMEM high glucose medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) and FBS (Sangon Biotech Co., Ltd.) for 24, 48 and 72 h, 20 µl MTT reagent (5 mg/ml: Beyotime Institute of Biotechnology, Beijing, China) was added, and cells were incubated for another 4 h. On the last day of incubation, the culture supernatant was removed and 150 µl dimethyl sulfoxide was added and incubated at 37°C for 4 h until a purple precipitate was visible. The absorbance was measured at 490 nm on a microplate reader. Cell growth curves were generated based on the absorbance values.
Western blot analysis
Proteins were extracted from HPAC/PANC-1 cells and patient samples by incubation with radioimmunoprecipitation assay buffer and the protease inhibitor phenylmethylsulfonyl fluoride (cat. no. KGP250; KeyGene Biotech, Jiangsu, China) according to the manufacturer's protocol. Protein concentration was determined using a bicinchoninic acid assay kit [RTP7102; Real-Times (Beijing) Biotechnology Co., Ltd., Beijing, China]. Next, 20 µg total protein per lane was separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Following blocking by 5% skimmed milk for 2 h at room temperature, the membranes were probed with the following primary antibodies: Rabbit anti-TAZ (1:1,000; ab84927) or rabbit anti-β-actin (1:5,000; ab129348; both from Abcam, Cambridge, MA, USA) at 4°C overnight. For detection, horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibodies (1:3,000; ab6721; Abcam) were used at room temperature for 1 h. Signal detection was conducted using an enhanced chemiluminescence reaction (ab65623; Abcam). The acquired images were analyzed using Image lab 3.0 software (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and relative protein expression was expressed as the densitometric value ratio of TAZ band density to β-actin band density. Each experiment was replicated 3 times.
ELISA
Levels of TAZ protein in the serum and pancreatic fluid of patients were measured using a TAZ ELISA kit (FS-Ea-05614; Feng Shou Shi Ye Biotechnology Co., Ltd., Shanghai, China) according to the kit protocol. Briefly, 1:4 dilutions of serum samples and eight serial dilutions of standard substrate at a final volume of 50 µl were incubated overnight at 4°C. Following incubation with HRP-conjugated secondary antibody for 1 h, samples were washed 5 times and the chromogenic substrate solution was added. The reaction was stopped with H2SO4 and read at 450 nm by a Multiskan FC microplate reader (Thermo Fisher Scientific, Inc.).
Bioinformatics analysis
To verify the miRNAs that may regulate the expression of TAZ, bioinformatics software (23–27), namely miRanda (http://www.microma.org/rnicroma/home.do), TargetScan (www.targetscan.org), PiTa (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html), RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) and PicTar (http://pictar.mdc-berlin.de/) were used for a reliable prediction of the miRNAs that may target taz mRNA.
Dual-luciferase reporter gene assay
According to results of the bioinformatics prediction, a conservative miR-185 binding sequence of the 3′untranslated region (UTR) of taz mRNA (wild-type; 5′-CAGAUGUUCUCUCC-3′) or a mutant sequence (mutant; 5′-CAGAUGAAGAGAGG-3′) was cloned as previously described (28). Luciferase reporter plasmids were generated as previously described (28) by insertion of the wild-type or mutant taz sequences into the multiple cloning site (SpeI, 5′-AAGCTT-3′ and HindIII, 5′-ACTAGT-3′) of a pMIR-REPORT™ Luciferase plasmid (Thermo Fisher Scientific, Inc.) downstream of the luciferase reporter gene. 293T cells (1×105 per well) were transfected with 0.8 µg of the luciferase constructs and 100 nM agomiR-185 or NC RNA using Lipofectamine. A total of 10 ng pMIR-REPORT™ β-gal control plasmid (AM5795; Ambion; Thermo Fisher Scientific, Inc.) was transfected as an internal control to evaluate transfection efficiency. Luminescence was measured 24 h after transfection at 37°C using a Dual-Luciferase® Reporter Assay System (Promega Corporation, Madison, WI, USA) according to the manufacturer's instructions. Measurements of luminescence were conducted on a Glomax 20/20 Luminometer (Promega Corporation).
Statistical analysis
Data analysis was performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA) and expressed as the mean ± standard deviation. Normality tests were performed for all data. Differences between groups were evaluated for significance using one-way analysis of variance. Least significant difference or Student-Newman-Keuls tests were used when variances were equal, and Tamhane's T2 or Dunnett's T3 tests were used when variances were not equal. P<0.05 was considered to indicate a statistically significant difference.
Results
Taz mRNA is upregulated in the PC tissues, pancreatic juice and serum of PC patients
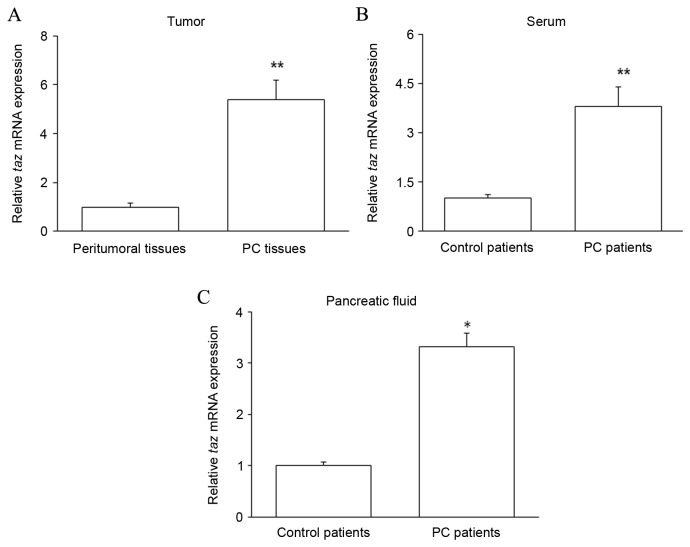
PC tissues and matched peritumoral tissues were collected from patients with PC. Pancreatic juice and serum were collected from PC patients and control patients with chronic pancreatitis. The expression of taz mRNA was assessed by RT-qPCR in the patient samples. The level of taz mRNA was significantly increased in PC tissues compared with that in peritumoral tissues (P<0.01; Fig. 1A). Furthermore, relative to that in control patients, taz mRNA expression was significantly increased in the serum (P<0.01; Fig. 1B) and pancreatic juice (P<0.05; Fig. 1C) of PC patients. These results suggest that high levels of TAZ expression are correlated with PC.
Figure 1.
RT-qPCR analysis of taz mRNA expression. The expression of taz mRNA was assessed by RT-qPCR in (A) PC tissues and matched peritumoral tissues from patients with PC, and (B) serum and (C) pancreatic juice samples from PC and control patients. *P<0.05 and **P<0.01 by one-way analysis of variance vs. controls. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; taz, transcriptional coactivator with PDZ-binding motif; PC, pancreatic cancer.
TAZ protein is upregulated in the PC tissues, pancreatic juice and serum of PC patients
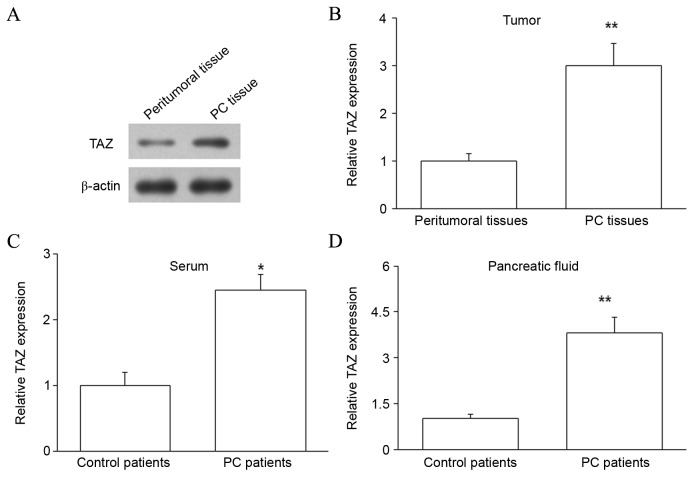
The expression of TAZ protein was subsequently assessed in PC tissues and matched peritumoral tissues by western blot analysis. The level of TAZ protein was upregulated in PC tissues compared with peritumoral tissues (P<0.01; Fig. 2A and B). Thus, TAZ mRNA and protein were upregulated in PC, indicating that increased TAZ expression may regulate the development of PC. The expression of TAZ protein in the pancreatic juice and serum of PC and control patients was also investigated by ELISA, and revealed that samples from PC patients contained higher levels of TAZ protein (P<0.05 for serum and P<0.01 for pancreatic juice; Fig. 2C and D). As blood is a common channel for tumor metastasis, upregulated expression of TAZ protein in the blood may influence the metastasis of PC. Furthermore, upregulated TAZ protein in the pancreatic juice may be due to the release of large quantities of DNA and proteins from the pancreas into the pancreatic juice.
Figure 2.
Analysis of TAZ protein expression in the PC tissues, serum and pancreatic juice of PC patients. (A) PC tissues and their matched peritumoral tissues were collected from patients with PC and the expression of TAZ protein was assessed by western blot analysis. β-actin expression was measured as an internal control. (B) The relative protein expression was calculated. (C) Serum and (D) pancreatic juice samples from PC and control patients were collected and the expression of TAZ protein in the samples was measured by ELISA. *P<0.05 and **P<0.01 by one-way analysis of variance vs. controls. TAZ, transcriptional coactivator with PDZ-binding motif; PC, pancreatic cancer.
TAZ is a target of miR-185
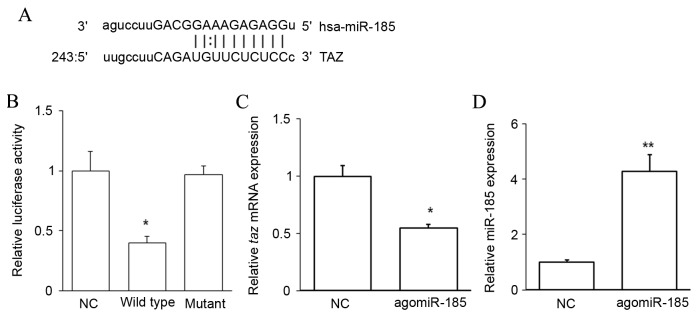
To evaluate the upstream regulator of TAZ, miRNAs complementary to the 3′UTR of TAZ were predicted using bioinformatics methods. An miR-185 binding site was identified in the 3′UTR of taz mRNA (Fig. 3A). A dual-luciferase reporter assay was performed to confirm this prediction. Cotransfection with agomiR-185 and a wild-type pMIR-REPORT-TAZ construct significantly decreased luciferase activity when compared with cells cotransfected with NC RNA and the wild-type pMIR-REPORT-TAZ construct (P<0.05). However, cotransfection with agomiR-185 and a mutant pMIR-REPORT-TAZ construct did not reduce luciferase activity (Fig. 3B). It was also observed that agomiR-185 transfection significantly downregulated the level of taz mRNA in HPACs (P<0.05 vs. NC; Fig. 3C) when miR-185 was successfully overexpressed (P<0.01 vs. NC; Fig. 3D). These results indicate that TAZ is a target of miR-185, and that miR-185 may regulate the expression of TAZ by binding to its 3′UTR and downregulating its expression at the mRNA level.
Figure 3.
TAZ is a target of miR-185. (A) Bioinformatics analysis identified a binding site for miR-185 in the 3′UTR of taz mRNA. (B) A wild type taz 3′UTR luciferase reporter construct (wild type) or mutant taz 3′UTR luciferase reporter construct (mutant) was cotransfected with agomiR-185, and wild type taz 3′UTR luciferase reporter construct was cotransfected with NC (NC). Luciferase activities were assayed 24 h post-transfection. (C) HPACs were transfected with agomiR-185 or NC, and the expression of taz mRNA was assessed by RT-qPCR. (D) Following transfection, miR-185 expression was measured by RT-qPCR. *P<0.05 and **P<0.01 by one-way analysis of variance vs. NC. TAZ, transcriptional coactivator with PDZ-binding motif; 3′UTR, 3′untranslated region; NC, negative control; HPAC, human pancreatic cancer cell line; ANOVA, analysis of variance; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Expression of miR-185 in PC patients
To further investigate the relationship between TAZ and miR-185 in PC, the expression of miR-185 was assessed by RT-qPCR in PC tissues and peritumoral tissues, and in the pancreatic juice and serum of PC and control patients. It was observed that miR-185 expression was significantly downregulated in PC tissues and samples from PC patients (P<0.05 for tissue and serum, P<0.01 for pancreatic juice; Fig. 4). Collectively, these data suggest that miR-185 may contribute to the regulation of PC by targeting taz mRNA and subsequently modulating the level of TAZ protein.
Figure 4.
RT-qPCR analysis of miR-185 expression. The expression of miR-185 was assessed by RT-qPCR in (A) PC tissues and matched peritumoral tissues from patients with PC, and (B) serum and (C) pancreatic juice samples from PC and control patients. *P<0.05 and **P<0.01 by one-way analysis of variance vs. controls. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR, microRNA; PC, pancreatic cancer.
Suppression of TAZ or agomiR-185 transfection inhibits the proliferation of PC cells
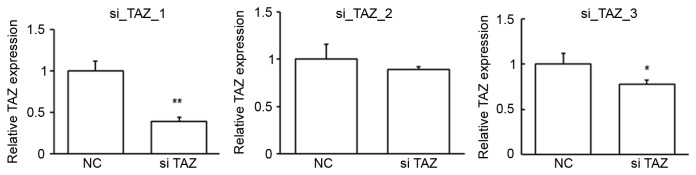
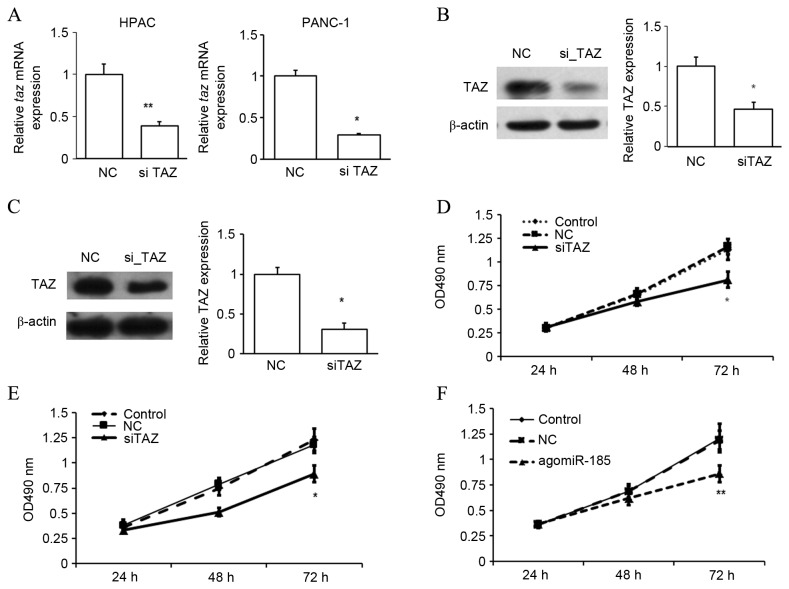
To determine whether TAZ regulated the proliferation of PC cells, TAZ expression was suppressed in HPACs and PANC-1 cells. To obtain the best suppression efficiency, the first siRNA was chosen for the following studies (Fig. 5). The repression of TAZ expression was confirmed by RT-qPCR (Fig. 6A) and western blot analysis (Fig. 6B and C). In an MTT assay, it was observed that repression of TAZ significantly inhibited the proliferation rate of HPACs (Fig. 6D) and PANC-1 cells (Fig. 6E) after 72 h (both P<0.05 vs. NC), suggesting a possible role of TAZ as an oncogene in the development of PC. As miR-185 was found to bind to the 3′UTR of taz mRNA, the potential role of miR-185 in regulating the development of PC was investigated. Similar to TAZ inhibition, it was observed that overexpression of miR-185 in HPACs significantly inhibited the rate of cell proliferation after 72 h, as determined by an MTT assay (P<0.01 vs. NC; Fig. 6F). These data indicate that miR-185 and TAZ may serve opposing roles in the proliferation of pancreatic cells.
Figure 5.
Inhibition efficiency of three TAZ siRNAs. HPAC cells were transfected with three TAZ siRNAs (siTAZ) or NC RNA (NC). The expression of Taz mRNA was assessed by reverse transcription-quantitative polymerase chain reaction. *P<0.05 and **P<0.01 vs. NC, by one-way analysis of variance. TAZ, transcriptional coactivator with PDZ-binding motif; siRNA, small interfering RNA; HPAC, human pancreatic cancer cell lines; NC, negative control; OD, optical density.
Figure 6.
Suppression of TAZ or overexpression of miR-185 inhibited pancreatic cell proliferation. (A-E) HPAC and PANC-1 cells were transfected with TAZ siRNA (siTAZ) or NC RNA (NC). (A) The expression of Taz mRNA was assessed by reverse transcription-quantitative polymerase chain reaction. The expression of TAZ protein in (B) HPAC and (C) PANC-1 cells was measured by western blot analysis. Cell proliferation was assessed by an MTT assay in (D) HPAC and (E) PANC-1 cells. (F) HPACs were transfected with agomiR-185 or NC, and cell proliferation was assessed by an MTT assay. *P<0.05 and **P<0.01 by one-way analysis of variance vs. NC. TAZ, transcriptional coactivator with PDZ-binding motif; miR, microRNA; HPAC and PANC-1, human pancreatic cancer cell lines; siRNA, small interfering RNA; NC, negative control.
TAZ works downstream of miR-185 to regulate cell proliferation
The aforementioned results indicate that miR-185 inhibits cell proliferation while TAZ promotes cell proliferation. In addition, miR-185 potentially targets TAZ and downregulates its expression. Therefore, whether miR-185 regulated cell growth through TAZ was subsequently investigated. HPACs were cotransfected with agomiR-185 and TAZ CDS-expressing plasmid, or with agomiR-185 and empty plasmid. Cells cotransfected with NC and empty plasmid were used as a NC. Transfection with agomiR-185 significantly decreased TAZ expression at the mRNA (P<0.05 vs. NC) and protein (P<0.01 vs. NC) levels, and this repression was rescued by cotransfection with TAZ expression plasmid (P<0.05; Fig. 7A and B). Results of an MTT assay also demonstrated that overexpression of miR-185 repressed cell proliferation at 48 and 72 h (P<0.05 and P<0.01, respectively, vs. NC), and upregulation of TAZ expression significantly rescued cell proliferation at 72 h (P<0.05; Fig. 7C). Thus, targeting of TAZ by miR-185 may inhibit the proliferation of PC cells.
Figure 7.
miR-185 targets TAZ to inhibit cell proliferation in HPACs. HPACs were cotransfected with agomiR-185 and a TAZ coding sequence-expressing plasmid, or with agomiR-185 and empty plasmid. Cells cotransfected with NC RNA and empty plasmid were used as a NC. (A) Expression of TAZ mRNA in cells was analyzed by reverse transcription-quantitative polymerase chain reaction. (B) Expression of TAZ protein was analyzed by western blot analysis, and β-actin expression was measured as an internal control. (C) Cell proliferation was analyzed by an MTT assay. *P<0.05 and **P<0.01 by one-way ANOVA vs. NC. #P<0.05 by one-way ANOVA vs. agomiR-185. miR, microRNA; TAZ, transcriptional coactivator with PDZ-binding motif; HPAC, human pancreatic cancer cell line; NC, negative control; ANOVA, analysis of variance.
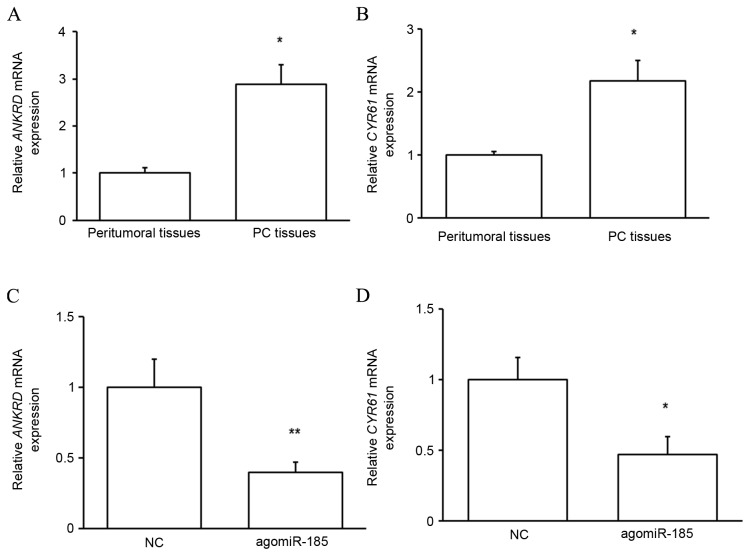
TAZ function in PC is dependent on its target proteins
To determine whether upregulated TAZ in PC tissues served functional roles, the expressions of two reported targets of TAZ, ANKRD and CYR61 (14), were evaluated. As predicted, the mRNA expression of the target proteins was significantly upregulated in PC tissues compared with that in peritumoral tissues (P<0.05; Fig. 8A and B), indicating that upregulated TAZ mediates the development of PC through its target proteins. It was subsequently determined whether miR-185 regulated the function of TAZ. As depicted in Fig. 8C and D, transfection of HPACs with agomiR-185 significantly reduced the expressions of ANKRD (P<0.01) and CYR61 (P<0.05) at the mRNA level. These results indicate that miR-185 indirectly regulates the growth of pancreatic cells by targeting TAZ expression and downstream protein function.
Figure 8.
Expression of TAZ downstream proteins. (A and B) PC tissues and their matched peritumoral tissues were collected from patients with PC and the levels of (A) ANKRD and (B) CYR61 mRNA in the samples were measured by RT-qPCR. (C and D) HPACs were transfected with agomiR-185 or NC, and the levels of (C) ANKRD and (D) CYR61 mRNA were measured by RT-qPCR. *P<0.05 and **P<0.01 by one-way analysis of variance vs. controls. TAZ, transcriptional coactivator with PDZ-binding motif; PC, pancreatic cancer; ANKRD, ankyrin repeat domain-containing protein; CYR61, cysteine-rich 61; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; HPAC, human pancreatic cancer cell line; NC, negative control.
Discussion
In the present study, the expression of TAZ at the mRNA and protein levels was analyzed in PC tissues, pancreatic juice and serum samples. Expression of miR-185, as the upstream regulator of TAZ, was also analyzed in these samples. In addition, the biological functions of TAZ and miR-185 in the development of PC were discussed.
The development of PC is a complex, multi-step process that involves multiple genes and epigenetic changes (29). With developments in biological therapy, it is useful to clarify the pathogenesis of PC and to search for novel genes as potential therapeutic targets. The hippo signaling pathway, first identified in the 1990s through genetic screens in overgrown Drosophila, is a conserved signaling pathway that serves essential roles in the regulation of organ growth, stem cell function, regeneration and tumor suppression (30,31). The transcriptional coactivator Yes associated protein (YAP) and TAZ interact with transcriptional enhancer associate domain 14 in the nucleus to promote biological functions downstream of the hippo signaling pathway (32,33). Previous studies of TAZ/YAP demonstrated that overexpression of TAZ/YAP induced the epithelial-mesenchymal transition, inhibited apoptosis and promoted proliferation of tumor cells (34,35). Furthermore, Xie et al (36) documented that TAZ was expressed in 66.8% of patients with non-small cell lung cancer, and its expression was significantly associated with poorer differentiation and prognosis. Similarly, Yue et al (37) identified the expression of TAZ protein in 77.4% of gastric cancer samples, and high-level expression of TAZ was observed in a higher percentage of gastric cancer samples with a histology of signet ring cell carcinoma rather than adenocarcinoma. A study by Wang and Tang (38) also demonstrated that YAP/TAZ expression was significantly higher in liver cancer tissues than that in adjacent normal tissues, and their expression was associated with Committee on Cancer Staging stage and alpha fetoprotein expression. In the present study, significantly higher levels of TAZ mRNA and protein were detected in PC tissues compared with matched peritumoral tissues, which was in accordance with former reports (15,16), indicating that TAZ is involved in the occurrence and development of PC. Thus, ectopic expression of TAZ may be a critical factor for the occurrence of PC. Indeed, the present in vitro assays in HPACs revealed that cell proliferation was inhibited after TAZ siRNA transfection.
To elucidate the regulatory mechanism of TAZ expression, a bioinformatics analysis was performed. MiR-185 is considered to be closely associated with TAZ, as miR-185 is a potential upstream regulator of TAZ. Previous results have demonstrated that miR-185 is a potential target in the genetic prevention, diagnosis and treatment of multiple diseases (39–41). For example, Kim et al (39) demonstrated that miR-185 effectively blocked cardiac hypertrophy signaling through multiple targets.. Furthermore, a study by Ma et al (40) revealed that miR-185 may inhibit cell proliferation and induce cell apoptosis by downregulating vascular endothelial growth factor A (VEGF-A) in Von Hippel-Lindau-inactivated clear cell renal cell carcinomas. Bao et al (41) also reported a role of miR-185 in the regulation of insulin secretion and β-cell growth in diabetes through its potential targeting of suppressor of cytokine signaling 3.
MiR-185 has also been implicated in the inhibition of breast cancer cell proliferation through its potential regulatory effects on the expression of c-Met and/or VEGF-A (42,43). In the present study, TAZ was identified as a target of miR-185 in a dual-luciferase reporter assay, and the interrelationship between miR-185 and TAZ expression was further evaluated. In the PC tissues, pancreatic juice and serum of PC patients, miR-185 and TAZ were inversely expressed. In addition, following agomiR-185 transfection in PC cells, the expression of taz mRNA was significantly decreased. Rescue assays also demonstrated that coexpression of TAZ with agomiR-185 alleviated the suppressive effects of agomiR-185 on cell proliferation, indicating that TAZ functions downstream of miR-185. Collectively, these results suggest that TAZ is a target of miR-185.
To verify that upregulated TAZ in PC patients served functional roles in the development of PC, the expression of two TAZ target proteins associated with cancer, namely ANKRD and CYR61 (14), was investigated. The expression of ANKRD is typically induced in injured tissues, myometrial hypertrophy and denervated tissues, and is important in muscle metabolism and the maintenance of muscle tissue function (44). A previous study demonstrated that its functions mainly involve the regulation of transcription levels, cell cycling and apoptosis, cytoskeletal stability and endocytosis activities (45). CYR61 has been associated with cardiac cancer and is a biomarker of tumor metastasis (46). In the process of tumor formation, CYR61 promotes angiogenesis and enhances cell proliferation (47). Therefore, activation of ANKRD and CYR61 by upregulated TAZ in PC, as observed in the present study, may be a possible mechanism for the development of PC.
In conclusion, miR-185 regulated the development of PC by targeting taz mRNA, and the downregulation of TAZ protein expression resulted in the inhibition of cell proliferation. These functions of miR-185 and TAZ may have been achieved through the downstream effector proteins of TAZ. In future studies, it would be useful to detect the subcellular location and cell-specific expression of TAZ, and to specify the correlation of TAZ with clinicopathological indices of PC (age, gender, duke stages and TNM stages amongst others). In addition, further investigation into the underlying molecular mechanisms of the TAZ/miR-185 axis in PC may provide a theoretical basis for the prevention, diagnosis and treatment of PC.
Acknowledgements
The present study was supported by projects of the Medical and Health Technology Development Program in Shandong, China (grant no. 2013WS0270).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zeng H, Zhang S. The updated incidences and mortalities of major cancers in China, 2011. Chin J Can. 2015;34:502–507. doi: 10.1186/s40880-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tempero MA, Arnoletti JP, Behrman S, Ben-Josef E, Benson AB, III, Berlin JD, Cameron JL, Casper ES, Cohen SJ, Duff M, et al. Pancreatic adenocarcinoma. J Natl Compr Canc Netw. 2010;8:972–1017. doi: 10.6004/jnccn.2010.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le N, Sund M, Vinci A, GEMS collaborating group of Pancreas Prognostic and predictive markers in pancreatic adenocarcinoma. Dig Liver Dis. 2000;48:223–230. doi: 10.1016/j.dld.2015.11.001. 2016. [DOI] [PubMed] [Google Scholar]
- 6.Arnachellum RP, Cariou M, Nousbaum JB, Jezequel J, Le Reste JY, Robaszkiewicz M. Pancreatic Adenocarcinoma in the Finistère Area, france, between 2002 and 2011 (1002 Cases): Population characteristics, treatment and survival. Pancreas. 2016;45:953–960. doi: 10.1097/MPA.0000000000000594. [DOI] [PubMed] [Google Scholar]
- 7.Cho JH, Ryu JK, Song SY, Hwang JH, Lee DK, Woo SM, Joo YE, Jeong S, Lee SO, Park BK, et al. Prognostic validity of the American joint committee on cancer and the European neuroendocrine tumors staging classifications for pancreatic neuroendocrine tumors: A retrospective nationwide multicenter study in South Korea. Pancreas. 2016;45:941–946. doi: 10.1097/MPA.0000000000000586. [DOI] [PubMed] [Google Scholar]
- 8.Liu L, Xu HX, Wang WQ, Wu CT, Xiang JF, Liu C, Long J, Xu J, Fu de L, Ni QX, et al. Serum CA125 is a novel predictive marker for pancreatic cancer metastasis and correlates with the metastasis-associated burden. Oncotarget. 2016;7:5943–5956. doi: 10.18632/oncotarget.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alsubai J, Matters GL, McGovern CO, Liao J, Gilius EL, Smith JP. Germline mutation of the CCK receptor: A novel biomarker for pancreas cancer. Clin Transl Gastroenterol. 2016;7:e134. doi: 10.1038/ctg.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon YH, Park YH, Lee JH, Hong JH, Kim IY. Selenoprotein W enhances skeletal muscle differentiation by inhibiting TAZ binding to 14-3-3 protein. Biochim Biophys Acta. 2014;1843:1356–1364. doi: 10.1016/j.bbamcr.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, Zeng Q, Hong W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–2598. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 12.Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: Structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Ma K, Chen L, Zhu H, Liang S, Liu M, Xu N. TAZ promotes cell growth and inhibits Celastrol-induced cell apoptosis. Biosci Rep. 2016;36:pii: e00386. doi: 10.1042/BSR20160135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morvaridi S, Dhall D, Greene MI, Pandol SJ, Wang Q. Role of YAP and TAZ in pancreatic ductal adenocarcinoma and in stellate cells associated with cancer and chronic pancreatitis. Sci Rep. 2015;5:16759. doi: 10.1038/srep16759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie D, Cui J, Xia T, Jia Z, Wang L, Wei W, Zhu A, Gao Y, Xie K, Quan M. Hippo transducer TAZ promotes epithelial mesenchymal transition and supports pancreatic cancer progression. Oncotarget. 2015;6:35949–35963. doi: 10.18632/oncotarget.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M, Wang M, Xu S, Guo X, Jiang J. Upregulation of miR-181c contributes to chemoresistance in pancreatic cancer by inactivating the Hippo signaling pathway. Oncotarget. 2015;6:44466–44479. doi: 10.18632/oncotarget.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higashi T, Hayashi H, Ishimoto T, Takeyama H, Kaida T, Arima K, Taki K, Sakamoto K, Kuroki H, Okabe H, et al. miR-9-3p plays a tumour-suppressor role by targeting TAZ (WWTR1) in hepatocellular carcinoma cells. Br J Cancer. 2015;113:252–258. doi: 10.1038/bjc.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuttle RM, Haugen B, Perrier ND. Updated American Joint Committee on Cancer/Tumor-Node-Metastasis Staging System for Differentiated and Anaplastic Thyroid Cancer (Eighth Edition): What Changed and Why? Thyroid. 2017;27:751–756. doi: 10.1089/thy.2017.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Yue Z, Xiong W, Sun P, You K, Wang J. TXNIP overexpression suppresses proliferation and induces apoptosis in SMMC7221 cells through ROS generation and MAPK pathway activation. Oncol Rep. 2017;37:3369–3376. doi: 10.3892/or.2017.5577. [DOI] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors; Proc Natl Acad Sci USA; 1977; pp. 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Duan LJ, Ding M, Hou LJ, Cui YT, Li CJ, Yu DM. Long noncoding RNA TUG1 alleviates extracellular matrix accumulation via mediating microRNA-377 targeting of PPARγ in diabetic nephropathy. Biochem Biophys Res Commun. 2017;484:598–604. doi: 10.1016/j.bbrc.2017.01.145. [DOI] [PubMed] [Google Scholar]
- 24.Welten SM, Bastiaansen AJ, de Jong RC, de Vries MR, Peters EA, Boonstra MC, Sheikh SP, La Monica N, Kandimalla ER, Quax PH, Nossent AY. Inhibition of 14q32 MicroRNAs miR-329, miR-487b, miR-494, and miR-495 increases neovascularization and blood flow recovery after ischemia. Circ Res. 2014;115:696–708. doi: 10.1161/CIRCRESAHA.114.304747. [DOI] [PubMed] [Google Scholar]
- 25.Shah VV, Soibam B, Ritter RA, Benham A, Oomen J, Sater AK. Data on microRNAs and microRNA-targeted mRNAs in Xenopus ectoderm. Data Brief. 2016;9:699–703. doi: 10.1016/j.dib.2016.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F, Lu J, Peng X, Wang J, Liu X, Chen X, Jiang Y, Li X, Zhang B. Integrated analysis of microRNA regulatory network in nasopharyngeal carcinoma with deep sequencing. J Exp Clin Cancer Res. 2016;35:17. doi: 10.1186/s13046-016-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joladarashi D, Garikipati VNS, Thandavarayan RA, Verma SK, Mackie AR, Khan M, Gumpert AM, Bhimaraj A, Youker KA, Uribe C, et al. Enhanced cardiac regenerative ability of stem cells after ischemia-reperfusion injury: Role of human CD34+ cells deficient in MicroRNA-377. J Am Coll Cardiol. 2015;66:2214–2226. doi: 10.1016/j.jacc.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Zeng L, Chen Y, Lian G, Qian C, Chen S, Li J, Huang K. Pancreatic cancer epidemiology, detection, and management. Gastroenterol Res Pract. 2016;2016:8962321. doi: 10.1155/2016/8962321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halder G, Johnson RL. Hippo signaling: Growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway-an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 32.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: An updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 35.Varelas X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 2014;141:1614–1626. doi: 10.1242/dev.102376. [DOI] [PubMed] [Google Scholar]
- 36.Xie M, Zhang L, He CS, Hou JH, Lin SX, Hu ZH, Xu F, Zhao HY. Prognostic significance of TAZ expression in resected non-small cell lung cancer. J Thorac Oncol. 2012;7:799–807. doi: 10.1097/JTO.0b013e318248240b. [DOI] [PubMed] [Google Scholar]
- 37.Yue G, Sun X, Gimenez-Capitan A, Shen J, Yu L, Teixido C, Guan W, Rosell R, Liu B, Wei J. TAZ is highly expressed in gastric signet ring cell carcinoma. Biomed Res Int. 2014;2014:393064. doi: 10.1155/2014/393064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang YP, Tang DX. Expression of Yes-associated protein in liver cancer and its correlation with clinicopathological features and prognosis of liver cancer patients. Int J Clin Exp Med. 2015;8:1080–1086. [PMC free article] [PubMed] [Google Scholar]
- 39.Kim JO, Song DW, Kwon EJ, Hong SE, Song HK, Min CK, Kim DH. miR-185 plays an anti-hypertrophic role in the heart via multiple targets in the calcium-signaling pathways. PLoS One. 2015;10:e0122509. doi: 10.1371/journal.pone.0122509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X, Shen D, Li H, Zhang Y, Lv X, Huang Q, Gao Y, Li X, Gu L, Xiu S, Bao X, et al. MicroRNA-185 inhibits cell proliferation and induces cell apoptosis by targeting VEGFA directly in von Hippel-Lindau-inactivated clear cell renal cell carcinoma. Urol Oncol. 2015;33:169. e1–11. doi: 10.1016/j.urolonc.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 41.Bao L, Fu X, Si M, Wang Y, Ma R, Ren X, Lv H. MicroRNA-185 targets SOCS3 to inhibit beta-cell dysfunction in diabetes. PLoS One. 2015;10:e0116067. doi: 10.1371/journal.pone.0116067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu P, DU F, Yao M, Lv K, Liu Y. MicroRNA-185 inhibits proliferation by targeting c-Met in human breast cancer cells. Exp Ther Med. 2014;8:1879–1883. doi: 10.3892/etm.2014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang R, Tian S, Wang HB, Chu DP, Cao JL, Xia HF, Ma X. MiR-185 is involved in human breast carcinogenesis by targeting Vegfa. FEBS Lett. 2014;588:4438–4447. doi: 10.1016/j.febslet.2014.09.045. [DOI] [PubMed] [Google Scholar]
- 44.Miller MK, Bang ML, Witt CC, Labeit D, Trombitas C, Watanabe K, Granzier H, McElhinny AS, Gregorio CC, Labeit S. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J Mol Biol. 2003;333:951–964. doi: 10.1016/j.jmb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei J, Yu G, Shao G, Sun A, Chen M, Yang W, Lin Q. CYR61 (CCN1) is a metastatic biomarker of gastric cardia adenocarcinoma. Oncotarget. 2016;7:31067–31078. doi: 10.18632/oncotarget.8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuonen F, Secondini C, Ruegg C. Molecular pathways: Emerging pathways mediating growth, invasion, and metastasis of tumors progressing in an irradiated microenvironment. Clin Cancer Res. 2012;18:5196–5202. doi: 10.1158/1078-0432.CCR-11-1758. [DOI] [PubMed] [Google Scholar]