Abstract
Autotrophic eukaryotes have evolved by the endosymbiotic uptake of photosynthetic organisms. Interestingly, many algae and plants have secondarily lost the photosynthetic activity despite its great advantages. Prototheca and Helicosporidium are non-photosynthetic green algae possessing colourless plastids. The plastid genomes of Prototheca wickerhamii and Helicosporidium sp. are highly reduced owing to the elimination of genes related to photosynthesis. To gain further insight into the reductive genome evolution during the shift from a photosynthetic to a heterotrophic lifestyle, we sequenced the plastid and nuclear genomes of two Prototheca species, P. cutis JCM 15793 and P. stagnora JCM 9641, and performed comparative genome analyses among trebouxiophytes. Our phylogenetic analyses using plastid- and nucleus-encoded proteins strongly suggest that independent losses of photosynthesis have occurred at least three times in the clade of Prototheca and Helicosporidium. Conserved gene content among these non-photosynthetic lineages suggests that the plastid and nuclear genomes have convergently eliminated a similar set of photosynthesis-related genes. Other than the photosynthetic genes, significant gene loss and gain were not observed in Prototheca compared to its closest photosynthetic relative Auxenochlorella. Although it remains unclear why loss of photosynthesis occurred in Prototheca, the mixotrophic capability of trebouxiophytes likely made it possible to eliminate photosynthesis.
Introduction
Acquisition of photosynthesis occurred in diverse eukaryotes by several endosymbiotic events wherein a photosynthetic organism was engulfed and integrated into a heterotrophic protist1,2. Phototrophic organisms can generate reduced carbon compounds in their plastids via the conversion of freely available light energy. Despite the great advantages, loss of photosynthesis has occurred in diverse lineages of organisms (e.g. apicomplexans, chlorophytes, cryptophytes, diatoms, dinoflagellates, euglenophytes, and Orobanchaceae species), along with heterotrophic free-living algae, holoparasitic plants, and pathogenic protists3. Such non-photosynthetic organisms survive by the uptake of organic carbon from the environment or host cells.
During the process of photosynthesis loss, plastids are generally reduced with regards to function, structure, and genome. Plastid genomes of non-photosynthetic organisms, except for Polytoma uvella4, are commonly smaller in size than that of the photosynthetic plastid genomes, because of the loss of genes related to photosynthesis, such as photochemical reaction complexes5. Particularly, the free-living green algae Polytomella6, the holoparasitic plant Rafflesia lagascae7, and the pathogenic alveolate Perkinsus marinus8 lack whole plastid genomes. Non-photosynthetic plastids lack the ability for light harvesting, photochemical reactions, and chlorophyll biosynthesis, whereas a part of the photosynthesis-related biosynthesis pathways is often retained. It has been reported that the nuclear genome of non-photosynthetic plastid-bearing organisms still encodes proteins for several plastid metabolic pathways, such as carbon fixation, fatty acid, terpenoid, tetrapyrrole, and isoprenoid biosynthesis9,10. Therefore, colourless plastids still possess some important functions other than those involved in photosynthesis.
Trebouxiophyte green algae include two non-photosynthetic genera, Prototheca and Helicosporidium, which are closely related to the photosynthetic genera, Chlorella and Auxenochlorella11–13. The genus Prototheca consists of free-living heterotrophic species, which exist in the soil and aqueous environments as ubiquitous organisms, and sometimes cause infections, termed protothecosis in animals, including humans14,15. The genus Helicosporidium is known to infect a variety of invertebrates; and in vitro axenic cultures are available for some strains16. Both Prototheca and Helicosporidium are believed to possess colourless plastids because of the presence of plastid genomes. Ultrastructural studies showed that Prototheca cells have a plastid-like structure surrounded by two membranes and filled by starch granules17,18. To date, complete plastid genomes of Prototheca wickerhamii and Helicosporidium sp. ATCC50920 have been reported19,20. The respective genomes encode 40 and 26 proteins, and lack most of the photosynthesis-related genes, though the plastid genome of P. wickerhamii contains six genes for ATP synthase. A comparative analysis revealed that the gene order excluding the absent genes is highly conserved in P. wickerhamii and its closest known photosynthetic relative Auxenochlorella protothecoides19. The plastid genome of Helicosporidium sp. is the smallest among the available plastid genomes of green algae20, and its gene order is diversified compared to Prototheca19. The nuclear genome of Helicosporidium sp. has been sequenced21, which revealed that many nuclear genes for the light-harvesting complexes, photosystems, and pigment biosynthesis have been lost; whereas part of photosynthesis-related functions, such as carbon fixation and terpenoid biosynthesis, have been retained.
To gain further insight into the genome evolution during the shift from a photosynthetic to a heterotrophic lifestyle in trebouxiophytes, we sequenced the plastid and nuclear genomes of two Prototheca species, P. cutis (JCM 15793 strain) and P. stagnora (JCM 9641 strain). Our phylogenetic analyses using plastid- and nucleus-encoded proteins strongly suggest that independent losses of photosynthesis have occurred at least three times in Prototheca and Helicosporidium. Comparative analyses of the plastid and nuclear genomes revealed that the gene content for plastid functions was highly conserved among the non-photosynthetic lineages, and the photosynthesis-related genes have mostly disappeared. Our findings suggest that non-photosynthetic trebouxiophytes have convergently lost a similar set of genes related to photosynthesis.
Results and Discussion
Overview of plastid and nuclear genomes of P. cutis and P. stagnora
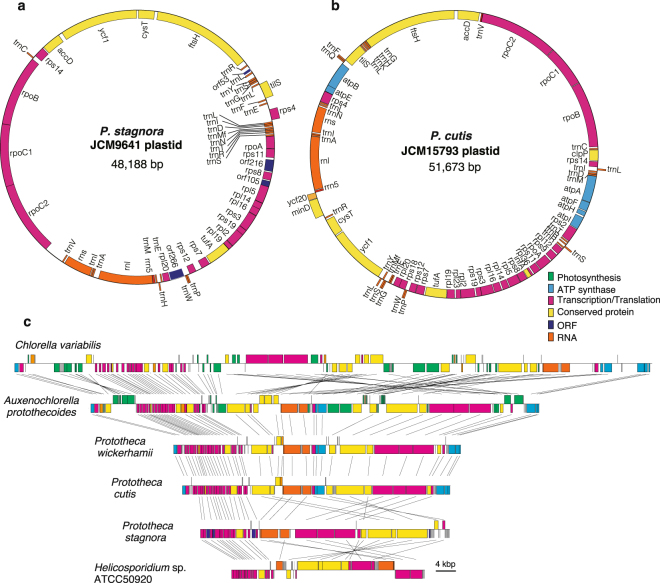
We sequenced the complete plastid and the draft nuclear genomes of two Prototheca species, P. cutis (JCM 15793 strain) and P. stagnora (JCM 9641 strain). The plastid genomes comprised 51.7 kb and 48.2 kb in P. cutis and P. stagnora, respectively (Fig. 1a,b); and these genomes were smaller than that of the plastid genome of P. wickerhamii (55.6 kb) and larger than that of Helicosporidium sp. (37.5 kb) (Table 1). Both plastid genomes were composed of relatively low GC (i.e. 29.7% in P. cutis and 25.7% in P. stagnora). The plastid genome of P. cutis was predicted to contain 72 genes, including 40 protein-coding genes, 29 tRNAs and 3 rRNAs; and its gene composition was almost identical to that of P. wickerhamii (Supplemental Table 1). In contrast, the P. stagnora plastid genome had 56 genes, including 28 protein-coding genes, 25 tRNAs, and 3 rRNAs. Both species lacked many plastid genes required for photosynthesis (e.g., genes for photosystem complexes, RubisCO large subunit, and chlorophyll biosynthesis). Although P. stagnora lacked all the photosynthesis-related genes, P. cutis retained six genes for the ATP synthase (atpA, atpB, atpE, atpF, atpH, and atpI) of the plastid similar to P. wickerhamii (Fig. 1b,c).
Figure 1.
Structure of the plastid genomes of P. stagnora and P. cutis. (a,b) Gene maps of the plastid genomes of P. stagnora and P. cutis, respectively. Genes are shown in different coloured boxes according to their putative functions. Genes on the outside of the maps are transcribed in the clockwise direction, and inner genes are transcribed in the counterclockwise direction. (c) Comparison of the gene order of the plastid genomes of C. variabilis, A. protothecoides, P. wickerhamii, P. cutis, P. stagnora, and Helicosporidium sp. Homologous genes are connected by straight lines as shown in the figure. Most of the photosynthesis-related genes (green) are absent in the non-photosynthetic lineages.
Table 1.
General features of the plastid and nuclear genomes of Prototheca spp., Helicosporidium sp., Auxenochlorella protothecoides, and Chlorella variabilis.
| Organisms | C. variabilis | A. protothecoides | P. wickerhamii | P. cutis | P. stagnora | Helicosporidium sp. |
|---|---|---|---|---|---|---|
| Plastid genomes | ||||||
| Reference | HQ914635.1 | Yan et al.19 | Yan et al.19 | This study | This study | de Koning and Keeling20 |
| Genome size (kb) | 124.6 | 84.6 | 55.6 | 51.7 | 48.2 | 37.5 |
| GC% | 33.9 | 30.8 | 31.2 | 29.7 | 25.7 | 26.9 |
| Genes | 115 | 111 | 72 | 72 | 56 | 54 |
| Proteins | 80 | 77 | 41 | 40 | 28 | 26 |
| Photosynthetic proteins* | 37 | 37 | 6 (atp) | 6 (atp) | 0 | 0 |
| tRNAs | 32 | 31 | 28 | 29 | 25 | 25 |
| rRNAs | 3 | 3 | 3 | 3 | 3 | 3 |
| Spacer (bp) | 460 | 119 | 122 | 54 | 98 | 36 |
| Nuclear genomes | ||||||
| Reference | Blanc et al. 2012 | Gao et al.46 | Not available | This study | This study | Pombert et al.21 |
| Assembly size (Mb) | 46.2 | 22.9 | 20.0 | 16.9 | 12.4 | |
| GC% | 67 | 63 | 60.3 | 71.4 | 61.7 | |
| Proteins | 9,791 | 7,039 | 6,884 | 7,041 | 6,035 | |
| Average exon size | 170 | 207 | 276.8 | 467.5 | 366 | |
| Average intron size | 209 | 246 | 204.4 | 290.3 | 168 | |
| Number of exons per gene | 7.3 | 5.7 | 5.4 | 4.0 | 2.3 | |
| Coding%** | 18.8 | 36.4 | 49.3 | 67.6 | 41.0 | |
*Excluding conserved genes ycf1, 3, 4, 12, 20. **Excluding intergenic regions, introns, and ncRNAs.
For the nuclear genomes, DNA short reads were assembled into 46 and 27 large scaffolds (>1 kb) and the total sizes were 20.0 and 16.9 Mb in P. cutis and P. stagnora, respectively. Completeness of the genome assembly was estimated using BUSCO22 by comparing with the whole proteins available in the eukaryote database. Both the genomes abundantly recovered core eukaryotic genes in P. cutis (92.4%) and P. stagnora (88.4%), similar to the genome sequence of A. protothecoides (85.2%). The putative nuclear genome sizes of Prototheca species were smaller than that of the photosynthetic relative Chlorella variabilis (46.2 Mb); however, it was slightly larger than the obligate parasite Helicosporidium sp. (12.4 Mb) (Table 1). In these organisms, the sizes of the plastid and nuclear genomes seem to be correlated with each other (Table 1). The nuclear genomes were predicted to encode 6,884 and 7,041 proteins in P. cutis and P. stagnora, respectively. These numbers were more than the nuclear genome of Helicosporidium sp. (6,035 proteins), less than that of C. variabilis (9,791 proteins), and comparable to that of A. protothecoides (7,039 proteins). Therefore, no obvious difference was observed in the number of protein-coding genes between photosynthetic and non-photosynthetic trebouxiophytes. However, gene-coding capacity displayed distinct levels among the five trebouxiophyte species; non-photosynthetic species (P. cutis, P. stagnora, and Helicosporidium sp.) showed higher rates (41 to 67.6%) than that of the photosynthetic relatives (36.4% for A. protothecoides and 18.8% for C. variabilis).
Phylogenetic analyses revealed multiple losses of photosynthesis in trebouxiophytes
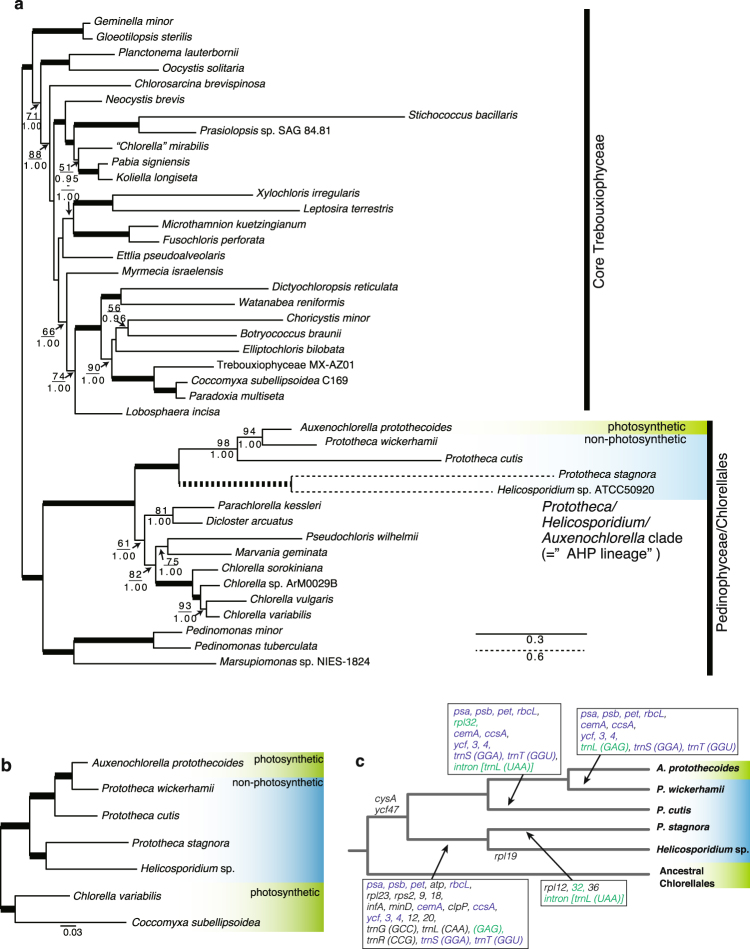
We performed phylogenetic analyses using plastid- and nucleus-encoded proteins to reveal the evolutionary scenario pertaining to the loss of photosynthesis in trebouxiophytes. We first collected 38 plastid-encoded proteins from 42 taxa of core Trebouxiophyceae, Chlorellales, and Pedinophyceae (Supplemental Tables 2 and 3), and constructed a maximum-likelihood (ML) tree. The tree showed that three Prototheca species, Helicosporidium sp., and A. protothecoides formed a monophyletic group with a robust statistical support (ML bootstrap support (BP) = 100 and Bayesian posterior probability (BPP) = 1.00) within the clade of Chlorellales (Fig. 2a). P. wickerhamii was closely related to A. protothecoides, and these two were found to be sister taxa to P. cutis. Monophyly of P. stagnora and Helicosporidium sp. was strongly supported (BP = 100, BPP = 1.00); and they were separated from the other three taxa at the basal position. Although these relationships were well resolved, the branch lengths of P. cutis, P. stagnora, and Helicosporidium sp. were much longer than the others. To assess the possibility of a long-branch attraction artefact, we also constructed a phylogenetic tree using 58 nucleus-encoded proteins of Prototheca, A. protothecoides, Helicosporidium sp., and two photosynthetic trebouxiophytes, C. variabilis and Coccomyxa subellipsoidea (Fig. 2b). The phylogenetic tree for the nucleus-encoded proteins was topologically identical to that for the plastid-encoded proteins, and each branch was strongly supported by 100% BP.
Figure 2.
Phylogenetic tree and the evolutionary scenario of the plastid gene losses in Chlorellales. (a) Maximum Likelihood (ML) tree constructed using 38 plastid-encoded proteins. Bootstrap support (BP) is indicated above the lines, and Bayesian posterior probability (BPP) is indicated below the lines. BP <50 or BPP <0.95 are not shown. Bold lines represent 100% BP and 1.00 BPP. The dotted branches are shown in half-length. (b) ML tree of 58 nucleus-encoded proteins. All the nodes were supported with 100% BP. (c) Evolutionary scenario of gene losses in P. wickerhamii, P. cutis, P. stagnora, and Helicosporidium sp. Eliminated plastid genes are indicated on the tree. Genes shown in green and blue have independently disappeared two and three times in the AHP lineage. Loss of group-I intron is presented by intron [trnL(UAA)].
Previous studies have reported that the three trebouxiophyte genera, Prototheca, Helicosporidium, and Auxenochlorella, form a monophyletic group13,19,23,24, and are referred to as the AHP lineage24. Although phylogenetic relationships within the AHP lineage have remained controversial, our phylogenetic analyses depicted a more reliable relationship of the lineage; non-photosynthetic trebouxiophytes did not show monophyly, because the photosynthetic A. protothecoides branched within the AHP clade. This suggests that the loss of photosynthesis has occurred in Prototheca and Helicosporidium at least three times independently in P. wickerhamii, P. cutis, and the lineage of P. stagnora and Helicosporidium. Additionally, our phylogenetic analyses also proved that the three species of Prototheca are either poly- or paraphyletic, suggesting that the genus Prototheca will require emendation in the future.
Convergent reductive evolution of non-photosynthetic plastid genomes
The plastid genomes of P. wickerhamii, P. cutis, P. stagnora, and Helicosporidium sp. lacked 36, 37, 50, and 52 protein-coding genes compared to the photosynthetic relative A. protothecoides (Fig. 2c and Supplemental Table 1). The same set of 36 genes related to photosystem I and II complexes (psa and psb), cytochrome (pet), chlorophyll biosynthesis (chl), RubisCO large subunit (rbcL) and others (cemA, ccsA, ycf3, ycf4, and ycf12) was absent in all the four plastid genomes, whereas these genes were postulated to have been independently lost in each lineage based on the phylogenetic relationships. Additionally, 12 genes for ATP synthase (atp), translation (rps2, rps9, rps18, rpl23, and infA), and others (clpP and ycf20) were absent in P. stagnora and Helicosporidium sp. A few genes encoding ribosomal subunits were distinctly absent in the respective species; e.g. rpl12 and rpl36 were absent in P. stagnora and rpl19 was absent in Helicosporidium sp. As these plastid genes were not found in their nuclear genomes, they were probably lost in these organisms. Two to six tRNA genes were absent in the four plastid genomes, and trnS(GGA) and trnT(GGU) genes were absent in all the genomes. Additionally, P. cutis and P. stagnora were found to lack a group-I intron that is broadly conserved in the trnL genes of plastid genomes25,26. Although gene losses independently occurred in the respective lineages of Prototheca and Helicosporidium, they affected similar sets of genes. Hence, there might be convergent reductive evolution of non-photosynthetic plastid genomes in trebouxiophytes. In terms of gene order, plastid genomes of the AHP lineage showed many syntenic regions (Fig. 1c). Interestingly, the gene order of P. cutis and P. wickerhamii was almost identical, suggesting that these two Prototheca species have independently eliminated the same set of plastid genes, while retaining the genome structure (Fig. 1c). In contrast, the plastid genomes of P. stagnora and Helicosporidium sp. were highly rearranged. This is probably due to the differences in the evolutionary time during which respective lineages lost their photosynthetic ability.
ATP synthase genes in non-photosynthetic plastids
Despite being non-photosynthetic, P. cutis and P. wickerhamii retained several photosynthesis-related genes in the plastid genomes, such as the ATP synthase genes (atpA, atpB, atpE, atpF, atpH, and atpI). Transcripts of these genes have been detected by reverse transcription PCR and Northern blot analysis in P. wickerhamii27. We further confirmed that the five ATP synthase genes (atpA, atpB, atpE, atpH, and atpI) were transcribed in P. cutis at a similar level to other plastid genes (rpL5, rpoB, and rpoC) by reverse transcription quantitative PCR (Supplemental Fig. 1). We found that the nuclear genome of P. cutis carried three genes for the other subunits of the plastid ATP synthase (atpC, atpD, and atpG). Therefore, P. cutis has a full set of ATP synthase genes, which are completely absent in P. stagnora and Helicosporidium sp. To evaluate the differences in the selective pressures on the ATP synthase genes between the photosynthetic and non-photosynthetic plastid genomes, we calculated their dN/dS ratios. The average dN/dS ratio between the photosynthetic C. variabilis and the non-photosynthetic P. cutis or P. wickerhamii was 0.021 or 0.040, which was not significantly different from the ratio between C. variabilis and A. protothecoides (0.010), and C. variabilis and C. subellipsoidea (0.007) (p > 0.05, paired t-test) (Supplemental Table 4). Hence, there is no indication that the ATP synthase genes have been exposed to peculiar selective pressures during the non-photosynthetic lifestyle. Therefore, we considered that the remaining genes for ATP synthase in Prototheca might have some function.
Plastid ATP synthase genes were also found in the non-photosynthetic plastids of the cryptophyte Cryptomonas paramecium28 and the diatom Nitzschia sp.29. It has been proposed that ATP hydrolysis in the non-photosynthetic plastids may produce a proton gradient between the thylakoids and stroma that is involved in the protein translocation to the thylakoids by the twin arginine translocator (Tat) system29. Although the photosynthetic relative A. protothecoides has a candidate gene for the plastid TatC protein (XP_011401675), no genes for the Tat system were found in the plastid and nuclear genome of Prototheca by our BLAST searches. These facts implied that the ATP synthase of the Prototheca plastid might have some unknown functions that is not related to the thylakoid Tat system; and this function is not indispensable in Prototheca, because P. stagnora completely lacks all genes required for the plastid ATP synthase.
Loss of nucleus-encoded plastid-targeted proteins
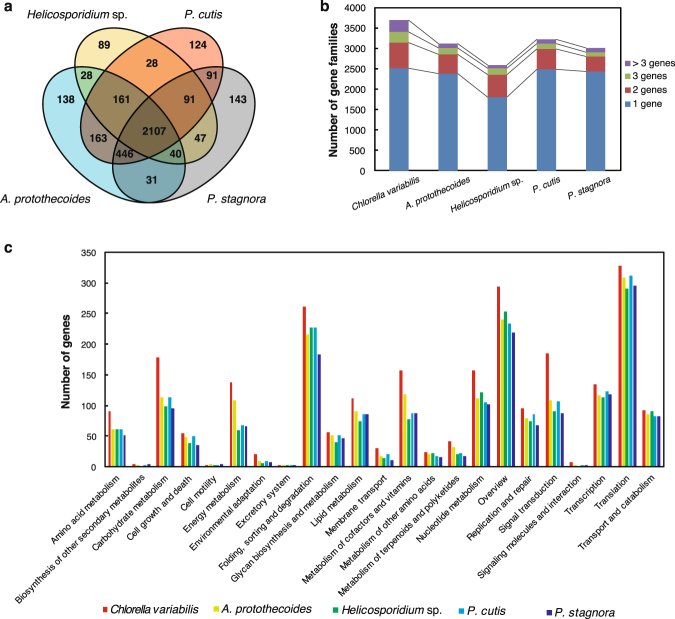
The nuclear genome sizes of P. cutis (20.0 Mb) and P. stagnora (16.9 Mb) were predicted to be smaller than that of their photosynthetic relatives, A. protothecoides (22.9 Mb), and C. variabilis (46.2 Mb) (Table 1). The nuclear genome of Helicosporidium sp. (12.4 Mbp) is the smallest among the available nuclear genomes of the AHP lineage, mainly because of a contraction of the gene family complexity rather than the loss of genes for certain functional categories21. To evaluate the complexity of gene families in Prototheca, we defined the orthologous genes in the respective nuclear genomes of P. cutis, P. stagnora, and Helicosporidium sp., as well as in A. protothecoides and C. variabilis using the TreeFam database30. We estimated 3,211 and 2,996 gene families in P. cutis and P. stagnora, respectively, which were similar to the number found in A. protothecoides (3,114 gene families) and smaller than that found in C. variabilis (3,688 gene families) (Fig. 3a,b; Supplemental Table 5). We also compared the number of genes according to the KEGG classification (Fig. 3c). There were no clear differences in the respective gene families and the functional categories among Prototheca and Auxenochlorella species, except for two categories; energy metabolism, and metabolism of cofactors and vitamins that had obvious connection to photosynthesis. Therefore, substantial gene loss and gain for certain functional categories other than that for photosynthesis probably did not occur during the shift from photosynthetic to teh heterotrophic lifestyle. However, Helicosporidium sp. carried more reduced gene families (2,591 gene families) compared to the others. Although Prototheca species are mainly free-living, Helicosporidium is the obligate parasite of insects. Therefore, it is considered that further genome reduction has to be related to the increased dependence on the host21.
Figure 3.
Comparison of nucleus-encoded proteins among Prototheca, Auxenochlorella, and Helicosporidium. (a) Venn diagram of shared gene families among P. cutis, P. stagnora, A. protothecoides, and Helicosporidium sp. (b) The number and size of gene families. Gene families consisting of multiple genes are shown in red, green, and purple according to their family size (two, three, and more than four). (c) The number of genes according to KEGG classification.
The colourless plastids of P. wickerhamii and Helicosporidium sp. were predicted to function in the biosynthesis of starch, fatty acids, tetrapyrrole, terpenoids, and amino acids based on their gene composition for plastid-targeted proteins10,21. We compared the nuclear gene contents related to metabolism in the plastids among P. cutis, P. stagnora, Helicosporidium sp., and A. protothecoides (Fig. 4). The three non-photosynthetic species depicted a similar gene content, in which genes related to carotenoid and chlorophyll biosynthesis were mostly eliminated; however, the genes for other products (e.g. starch, fatty acids, tetrapyrrole, and terpenoids) were retained. Nuclear genes for photosystems, including light-harvesting complexes were not found in the non-photosynthetic species. Therefore, elimination of the genes related to certain biological processes of the plastid has occurred concurrently in both the plastid and nuclear genomes. Exceptionally, P. cutis and Helicosporidium sp. possess a putative gene for chlorophyll b reductase (EC: 1.1.1.294) (Fig. 4), which converts chlorophyll a to b. However, the dN/dS ratios of this gene in P. cutis (0.051) and Helicosporidium sp. (0.050) were clearly higher than that in their photosynthetic relatives, A. protothecoides (0.0065) and C. subellipsoidea (0.0075); the dN/dS ratios were calculated against the gene of C. variabilis. Moreover, the C-terminal domain of the chlorophyll b reductase was truncated in Helicosporidium. Therefore, chlorophyll b reductase genes of the non-photosynthetic species would be under the process of gene disruption.
Figure 4.
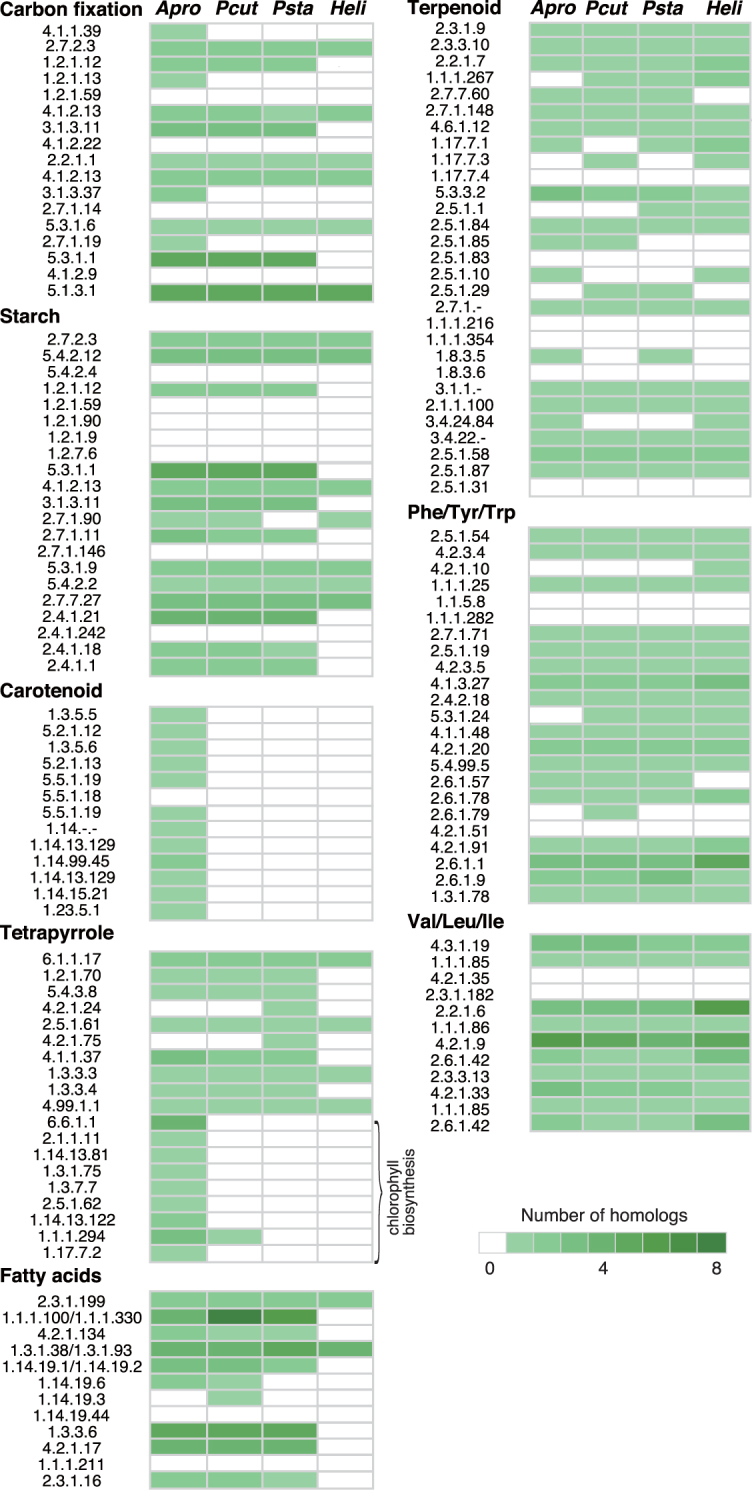
Gene contents related to plastid biosynthesis in Prototheca, Auxenochlorella, and Helicosporidium. Genes for plastid-related proteins were categorized into eight groups according to their functions: carbon fixation, and biosynthesis of starch, carotenoid, tetrapyrrole, fatty acids, terpenoid, Phe/Tyr/Trp, and Val/Leu/Ile. Green coloured boxes indicate the presence of genes for the plastid-related proteins as shown by EC numbers. The colour gradient represents the copy number of genes.
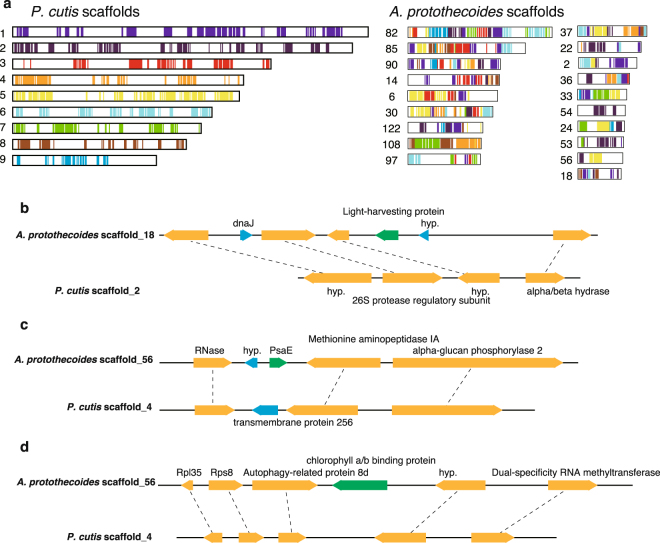
Comparison of the syntenic regions of the plastid genomes of P. cutis, P. wickerhamii, and A. protothecoides revealed that the colourless plastid genomes eliminated the photosynthesis-related genes, while maintaining the gene order, and the remarkable footprints of the missing genes (i.e. truncated pseudogenes) were not found in the syntenic regions (Fig. 1c)19. As described above, the photosynthesis-related nuclear genes have also been lost in Prototheca and Helicosporidium sp.21. To investigate the process of the nuclear gene reduction, comparative analyses of the syntenic regions were performed among P. cutis, P. stagnora, and A. protothecoides. Although the nuclear genomes represented a highly recombinant structure compared to the plastid genomes, a total of 165 syntenic blocks, including 11.9 genes on average was detected between P. cutis and A. protothecoides (Fig. 5a). P. stagnora and A. protothecoides shared 160 syntenic blocks with an average of 6.8 genes, and P. cutis and P. stagnora exhibited 275 syntenic blocks with an average of 5.8 genes (Supplemental Fig. 2). We identified three genes for the photosynthesis-related proteins, light-harvesting protein, PsaE, and chlorophyll a/b binding protein within the syntenic regions (Fig. 5b–d). The junction flanking the psaE gene was substituted by the gene encoding a transmembrane protein in P. cutis (Fig. 5c). The junctions for the other two genes were shortened and did not encode any proteins (Fig. 5b,d). These findings suggest that parts of the photosynthesis-related nuclear genes in P. cutis were omitted from the chromosomes without gene rearrangement similar to the plastid genome during the shift from the photosynthetic to the heterotrophic lifestyle.
Figure 5.
Synteny analysis of the nuclear genomes between P. cutis and A. protothecoides. (a) Syntenic regions between P. cutis and A. protothecoides nuclear genomes are indicated by coloured lines. Numbers beside the scheme represent a scaffold number. (b–d) Syntenic regions including photosynthesis-related genes (green) in A. protothecoides. Homologous genes (orange) between P. cutis and A. protothecoides are connected by dotted lines.
Conclusion
In this study, we report the plastid and nuclear genomes of two Prototheca species, P. cutis and P. stagnora. Our analyses suggest that multiple independent losses of photosynthesis have occurred in the non-photosynthetic trebouxiophytes, which have convergently lost a similar set of genes related to photosynthesis in the plastid and nuclear genomes. Such frequent losses of photosynthesis could possibly imply that some other mixotrophic relative of Prototheca (e.g. some species in genera Chlorella and Auxenochlorella) may eventually give up photosynthesis in future. Long-term monitoring of genome modification in Auxenochlorella under heterotrophic conditions will be an effective approach to investigate the possibility of an irreversible shift from mixotrophy to obligate heterotrophy.
Materials and Methods
Culture and DNA extraction
P. cutis JCM 15793 and P. stagnora JCM 9641 were obtained from the Japan Collection of Microorganisms, RIKEN BioResource Center (RIKEN BRC-JCM), Japan. P. cutis was cultured in 250 mL of YM broth (1% glucose, 0.5% peptone, 0.3% yeast extract, 0.3% malt extract, Difco) for 3 days at 30 °C under constant shaking (150 rpm), and the cells were collected by centrifugation. P. stagnora was cultured on YM agar at 25 °C for 10 days, followed by collecting the cells by scraping. The cell mass was freeze-dried, and ground in a mortar. Total DNA was extracted using phenol/chloroform/isoamyl alcohol, precipitated by adding 2-propanol, and then spooled out with a sterile glass rod. The crude DNA was dissolved in G2 Buffer (Qiagen, Cat. No. 1014636), and purified using a Genomic-tip 100/G (Qiagen, Cat. No. 10243) according to the manufacturer’s instruction. The DNA was further cleaned using PowerClean Pro DNA Clean-Up Kit (MO Bio Laboratories, Cat. No. 12997-50) and used for the library preparation for subsequent sequencing.
DNA sequencing and assembly
A paired-end library with approximate insert size of 240 bp was prepared using TruSeq DNA PCR-free Library Preparation Kit (Illumina, Cat. No. FC-121-3001) according to the manufacturer’s protocol. A mate pair library with approximate insert size of 3 kbp was also prepared using Nextera Mate Pair Sample Preparation Kit (Illumina, Cat. No. FC-132-1001) with some modifications31. Whole genome sequencing was performed using the Illumina HiSeq. 2500 platform to generate 151-base paired-end reads. The mate pair reads were processed with NextClip v.0.832 to trim the adapter sequences. ALLPATHS-LG v.5248833 was used to assemble both paired-end and mate pair reads into scaffolds with default parameters. The number of reads used for the de novo genome assemblies were 35,146,956 paired-end reads (5.3 Gb) and 11,863,706 mate pair reads (1.2 Gb) for P. cutis; and 63,282,152 paired-end reads (9.6 Gb) and 4,759,250 mate pair reads (5.0 Gb) for P. stagnora. The coverage of the paired-end reads of P. cutis and P. stagnora were approximately 265x and 568x, respectively. The N50 values of P. cutis and P. stagnora were 1.4 Mbp and 1.1 Mbp, respectively. For the reconstruction of plastid genomes, 667,790 paired-end reads (101 Mb) and 444,892 mate pair reads (44 Mb) of P. cutis, and 569,540 paired-end reads (86 Mb) and 366,460 mate pair reads (38 Mb) of P. stagnora were randomly sampled and assembled using ALLPATHS-LG with default parameters. Plastid genome sequences were identified using BLAST against the chloroplast genome sequence of P. wickerhamii (accession no. KJ001761).
Gene annotation
For the annotation of plastid genomes, we initially identified the plastid genes using GeneMarkS34, and annotated them using BLASTx35. tRNAscan-SE36 and RNAmmer37 were used to predict tRNA and rRNA, respectively. All the plastid genes were manually curated on the Artemis genome browser38. In the case of nuclear genomes, the coding regions were predicted by MAKER annotation pipeline v.2.31.839, including AUGUSTUS v.3.0.340, SNAP41, and GeneMark-ES v.4.2142, wherein AUGUSTUS and SNAP were trained on A. protothecoides (https://www.ncbi.nlm.nih.gov/assembly/GCF_000733215.1) and C. variabilis (https://www.ncbi.nlm.nih.gov/assembly/GCF_000147415.1/). To estimate assembly completeness, we performed BUSCO analysis22 with the eukaryote dataset using the protein sequences. The estimated completeness of P. cutis and P. stagnora were 92.1% (S: 91.7%, D: 0.7%, F: 4.0%, and M: 3.6%) and 88.4% (S: 87.1%, D: 1.3%, F: 6.3%, and M: 5.3%), respectively. Functional gene annotation was performed according to the sequence homology in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database43 using the KEGG Automatic Annotation Server (KAAS) with BBH method44. Conserved syntenic regions between the two nuclear genomes of P. stagnora, P. cutis, and A. protothecoides were searched using the CHROnicle program of SynChro (January 2015)45. For this analysis, we applied 7, 9, and 19 long scaffolds (>300 kb) of P. stagnora, P. cutis, and A. protothecoides, respectively. Syntenic blocks, including more than two orthologous genes, were identified using reciprocal BLAST hits with a similarity threshold of 40% and a length ratio of 1.3.
Classification of gene families
Annotated nuclear genes of C. variabilis, A. protothecoides46, Helicosporidium sp.21, P. cutis, and P. stagnora were classified into known gene families using TreeFam 930 with an E-value cut-off of 1E-5. Plastid-related proteins were identified using PRIAM (March 2015)47 with an E-value cut-off of 1E-10.
Phylogenetic analyses
We performed the phylogenetic analysis using 38 highly conserved plastid encoding proteins (Supplemental Table 2), equivalent to 6,467 amino acids, representing 42 taxa (Supplemental Table 3). Organisms belonging to the core Trebouxiophyceae48 were used as an outgroup. The sequences were aligned using MAFFT 7.164b with the L-INS-i option49, and poorly aligned regions were manually eliminated using MEGA 6.050. Model test was carried out by IQ-TREE multicore v.1.3.251 and maximum likelihood (ML) analyses were performed with the options LG + GAMMA + I + F using RAxML v.8.1.2152. Statistical support was evaluated with the nonparametric bootstrap test using 200 replications. Bayesian analyses were performed using MrBayes v3.2.653 with the same substitutional model. Bayesian inference consisted of 2,000,000 generations with sampling at every 1,000 generations using the four Metropolis-coupled Markov chain Monte Carlo (MCMCMC) simulations. Two separate runs were performed, and the convergence was assessed by the average standard deviation of split frequencies (ASDSF) falling below 0.01. Bayesian posterior probabilities (BPP) were calculated from the majority rule consensus of the trees sampled after the initial 500 burn-in trees.
We also performed phylogenetic analyses using the nucleus-encoded proteins of 7 taxa (P. cutis, P. stagnora, P. wickerhamii, Helicosporidium sp., A. protothecoides, C. variabilis, and C. subellipsoidea). Orthologous sequences among these taxa were searched using the reciprocal best-hit analyses with the cut-off: similarity >70% and HSP coverage >50%. A total of 58 proteins, which were shared by at least six taxa, were used for the analyses (Supplemental Table 6). ML analyses were performed using the same method with the plastid-encoded proteins.
Nucleotide substitution rates of synonymous (dS) and nonsynonymous (dN) sites
The dN/dS ratios of the plastid-encoded ATP synthase genes and chlorophyll b reductase genes were calculated for P. cutis, P. wickerhamii, A. protothecoides, C. variabilis, and C. subellipsoidea. Amino acid sequences were aligned using MAFFT 7.164b with the L-INS-i option. The aligned sequences were converted to nucleotide sequences using PAL2NAL v.1454. Pairwise dN/dS ratios among C. variabilis and the others were calculated using the codeml program of the PAML package v.4.855.
Data deposition
The plastid and nuclear genome sequences of P. cutis JCM 15793 and P. stagnora JCM 9641 were deposited in DDBJ/GenBank/ENA under accession numbers AP018373 (P. cutis plastid), AP018372 (P. stagnora plastid), BCIH01000000 (P. cutis nuclear), and BCJY01000000 (P. stagnora nuclear).
Electronic supplementary material
Acknowledgements
Genome sequencing was supported by the Genome Information Upgrading Program of the National BioResource Project, MEXT, Japan. This study was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers: 23117004, 15K18582, and 14J00572. R.M. was supported by Research Grant to RIKEN Centre for Life Science Technologies, Division of Genomic Technologies from MEXT. S.S. was a recipient of the JSPS Research Fellowships for Young Scientists 26–572.
Author Contributions
Y.H., R.M., and M.O. conceived the study. R.E. and M.O. provided DNA samples, and R.M. performed DNA sequencing, assembly, and annotation. S.S. performed genomic and phylogenetic analyses. Y.H. and S.S. wrote the manuscript. All authors contributed in discussing ideas, and read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18378-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Keeling PJ. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu. Rev. Plant Biol. 2013;64:583–607. doi: 10.1146/annurev-arplant-050312-120144. [DOI] [PubMed] [Google Scholar]
- 2.Burki F, et al. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc. Biol. Sci. 2016;283:20152802. doi: 10.1098/rspb.2015.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krause, K. In Organelle Genetics: Evolution of Organelle Genomes and Gene Expression (ed. Bullerwell, C. E.) 79–103 (Springer-Verlag, 2012), 10.1007/978-3-642-22380-8_4.
- 4.Figueroa-Martinez F, Nedelcu AM, Smith DR, Reyes-Prieto A. The plastid genome of Polytoma uvella Is the largest known among colorless algae and plants and reflects contrasting evolutionary paths to nonphotosynthetic lifestyles. Plant Physiol. 2017;173:932–943. doi: 10.1104/pp.16.01628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krause K. From chloroplasts to ‘cryptic’ plastids: evolution of plastid genomes in parasitic plants. Curr. Genet. 2008;54:111–121. doi: 10.1007/s00294-008-0208-8. [DOI] [PubMed] [Google Scholar]
- 6.Smith DR, Lee RW. A plastid without a genome: evidence from the nonphotosynthetic green algal genus. Polytomella. Plant Physiol. 2014;164:1812–1819. doi: 10.1104/pp.113.233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molina J, et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae) Mol. Biol. Evol. 2014;31:793–803. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuzaki M, Kuroiwa H, Kuroiwa T, Kita K, Nozaki H. A cryptic algal group unveiled: a plastid biosynthesis pathway in the oyster parasite Perkinsus marinus. Mol. Biol. Evol. 2008;25:1167–1179. doi: 10.1093/molbev/msn064. [DOI] [PubMed] [Google Scholar]
- 9.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borza T, Popescu CE, Lee RW. Multiple metabolic roles for the nonphotosynthetic plastid of the green alga Prototheca wickerhamii. Eukaryot. Cell. 2005;4:253–261. doi: 10.1128/EC.4.2.253-261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boucias DG, Becnel JJ, White SE, Bott M. In vivo and in vitro development of the protist Helicosporidium sp. J Eukaryot Microbiol. 2001;48:460–470. doi: 10.1111/j.1550-7408.2001.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 12.Boucias DG, Tartar A, Adams BJ, Becnel JJ. Phylogenetic analysis identifies the invertebrate pathogen Helicosporidium sp. as a green alga (Chlorophyta) Int. J. Syst. Evol. Microbiol. 2002;52:273–279. doi: 10.1099/00207713-52-1-273. [DOI] [PubMed] [Google Scholar]
- 13.Tartar A, Boucias DG, Becnel JJ, Adams BJ. Comparison of plastid 16S rRNA (rrn16) genes from Helicosporidium spp.: evidence supporting the reclassification of Helicosporidia as green algae (Chlorophyta) Int. J. Syst. Evol. Microbiol. 2003;53:1719–1723. doi: 10.1099/ijs.0.02559-0. [DOI] [PubMed] [Google Scholar]
- 14.Consuelo Quinet Leimann B, Cezar Fialho Monteiro P, Lazéra M, Ulloa Candanoza E, Wanke B. Protothecosis. Med. Mycol. 2004;42:95–106. doi: 10.1080/13695780310001653653. [DOI] [PubMed] [Google Scholar]
- 15.Lass-Flörl C, Mayr A. Human protothecosis. Clin. Microbiol. Rev. 2007;20:230–42. doi: 10.1128/CMR.00032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tartar A. The non-photosynthetic algae Helicosporidium spp.: emergence of a novel group of insect pathogens. Insects. 2013;4:375–391. doi: 10.3390/insects4030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadakavukaren MJ, McCracken DA. An ultrastructural survey of the genus Prototheca with special reference to plastids. Mycopathologia. 1977;61:117–119. doi: 10.1007/BF00443840. [DOI] [PubMed] [Google Scholar]
- 18.Kiyohara N, et al. Immuno-electron microscopic studies on plastid DNA and photosynthetic proteins in Prototheca wickerhamii. Cytologia (Tokyo). 2006;71:309–314. doi: 10.1508/cytologia.71.309. [DOI] [Google Scholar]
- 19.Yan D, et al. Auxenochlorella protothecoides and Prototheca wickerhamii plastid genome sequences give insight into the origins of non-photosynthetic algae. Sci. Rep. 2015;5:14465. doi: 10.1038/srep14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Koning AP, Keeling PJ. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 2006;4:12. doi: 10.1186/1741-7007-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pombert J-F, Blouin NA, Lane C, Boucias D, Keeling PJ. A lack of parasitic reduction in the obligate parasitic green alga. Helicosporidium. PLoS Genet. 2014;10:e1004355. doi: 10.1371/journal.pgen.1004355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 23.Ueno R, Urano N, Suzuki M. Phylogeny of the non-photosynthetic green micro-algal genus Prototheca (Trebouxiophyceae, Chlorophyta) and related taxa inferred from SSU and LSU ribosomal DNA partial sequence data. FEMS Microbiol. Lett. 2003;223:275–280. doi: 10.1016/S0378-1097(03)00394-X. [DOI] [PubMed] [Google Scholar]
- 24.Ueno R, Hanagata N, Urano N, Suzuki M. Molecular phylogeny and phenotypic variation in the heterotrophic green algal genus Prototheca (Trebouxiophyceae, Chlorophyta) J. Phycol. 2005;41:1268–1280. doi: 10.1111/j.1529-8817.2005.00142.x. [DOI] [Google Scholar]
- 25.Kuhsel MG, Strickland R, Palmer JD. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990;250:1570–3. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- 26.Besendahl A, Qiu YL, Lee J, Palmer JD, Bhattacharya D. The cyanobacterial origin and vertical transmission of the plastid tRNA(Leu) group-I intron. Curr. Genet. 2000;37:12–23. doi: 10.1007/s002940050002. [DOI] [PubMed] [Google Scholar]
- 27.Knauf U, Hachtel W. The genes encoding subunits of ATP synthase are conserved in the reduced plastid genome of the heterotrophic alga Prototheca wickerhamii. Mol. Genet. Genomics. 2002;267:492–497. doi: 10.1007/s00438-002-0681-6. [DOI] [PubMed] [Google Scholar]
- 28.Donaher N, et al. The complete plastid genome sequence of the secondarily nonphotosynthetic alga Cryptomonas paramecium: reduction, compaction, and accelerated evolutionary rate. Genome Biol. Evol. 2009;1:439–48. doi: 10.1093/gbe/evp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamikawa R, et al. Proposal of a twin arginine translocator system-mediated constraint against loss of ATP synthase genes from nonphotosynthetic plastid genomes. Mol. Biol. Evol. 2015;32:2598–2604. doi: 10.1093/molbev/msv134. [DOI] [PubMed] [Google Scholar]
- 30.Ruan J, et al. TreeFam: 2008 update. Nucleic Acids Res. 2008;36:D735–740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park N, et al. An improved approach to mate-paired library preparation for Illumina sequencing. Methods Next Gener. Seq. 2013;1:10–20. [Google Scholar]
- 32.Leggett RM, Clavijo BJ, Clissold L, Clark MD, Caccamo M. NextClip: an analysis and read preparation tool for Nextera Long Mate Pair libraries. Bioinformatics. 2014;30:566–568. doi: 10.1093/bioinformatics/btt702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–18. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagesen K, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 39.Cantarel BL, et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2007;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson AD, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drillon G, Carbone A, Fischer G. SynChro: A fast and easy tool to reconstruct and visualize synteny blocks along eukaryotic chromosomes. PLoS One. 2014;9:e92621. doi: 10.1371/journal.pone.0092621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao C, et al. Oil accumulation mechanisms of the oleaginous microalga Chlorella protothecoides revealed through its genome, transcriptomes, and proteomes. BMC Genomics. 2014;15:582. doi: 10.1186/1471-2164-15-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Claudel-Renard C, Chevalet C, Faraut T, Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–9. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemieux C, Otis C, Turmel M. Chloroplast phylogenomic analysis resolves deep-level relationships within the green algal class Trebouxiophyceae. BMC Evol. Biol. 2014;14:211. doi: 10.1186/s12862-014-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:286–98. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 50.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronquist F, et al. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.