Abstract
Next-generation sequencing (NGS) has enabled comprehensive detection of genomic alterations in lung cancer. Ethnic differences may play a critical role in the efficacy of targeted therapies. The aim of this study was to identify and compare genomic alterations of lung adenocarcinoma between Japanese patients and the Cancer Genome Atlas (TCGA), which majority of patients are from the US. We also aimed to examine prognostic impact of additional genomic alterations in patients harboring EGFR mutations. Genomic alterations were determined in Japanese patients with lung adenocarcinoma (N = 100) using NGS-based sequencing of 415 known cancer genes, and correlated with clinical outcome. EGFR active mutations, i.e., those involving exon 19 deletion or an L858R point mutation, were seen in 43% of patients. Some differences in driver gene mutation prevalence were observed between the Japanese cohort described in the present study and the TCGA. Japanese cohort had significantly more genomic alterations in cell cycle pathway, i.e., CDKN2B and RB1 than TCGA. Concurrent mutations, in genes such as CDKN2B or RB1, were associated with worse clinical outcome in patients with EGFR active mutations. Our data support the utility of comprehensive sequencing to detect concurrent genomic variations that may affect clinical outcomes in this disease.
Introduction
Lung tumors are the most prevalent type of cancer and are one of the leading causes of cancer-related death worldwide1. The discovery that epidermal growth factor receptor (EGFR) mutation is a predictor of clinical response to EGFR tyrosine kinase inhibitors (EGFR-TKIs) has dramatically changed the therapeutic approach to non-small cell lung cancer (NSCLC)2,3. In addition to EGFR, several oncogenic drivers such as anaplastic lymphoma kinase (ALK), ROS proto-oncogene 1, receptor tyrosine kinase (ROS1), and ret proto-oncogene (RET) have been as identified as molecular targets in this disease4,5. The effectiveness of molecularly targeted therapies, and advances in technologies for the detection of genomic alterations in tumors, have driven an increasing interest in such precision medicine approaches.
The advent of next-generation sequencing (NGS) has enabled the comprehensive detection of genomic alterations. Utilizing NGS technology, the Cancer Genome Atlas (TCGA) consortium has reported the molecular profiling of 230 lung adenocarcinoma cases, and the detection of mutations in genes such as neurofibromatosis type 1 (NF1), MET proto-oncogene, receptor tyrosine kinase (MET), and erb-b2 receptor tyrosine kinase 2 gene (ERBB2)6. Although the TCGA represents one of the largest cohorts of NSCLC patients6,7, its limitation is that minor ethnicities are not well represented because samples are derived solely from USA-based institutions. It is well known that cancers differ biologically between ethnicities, but to date, genomic data from east-Asian populations have been scarce8.
We hypothesized that sequencing a panel of cancer-associated genes using NGS technology would identify essentially all actionable genomic driver mutations in lung adenocarcinoma, regardless of ethnic and geographical background. Moreover, we hypothesized that comprehensive sequencing would identify not only the known actionable driver mutations, such as those affecting EGFR, but also other important gene alterations that may impact clinical outcome. To test these hypotheses, we assessed the genomic profile of a Japanese cohort of lung adenocarcinoma patients using NGS-based sequencing of 415 genes, and compared the results with data from TCGA. Finally, it has been reported that the EGFR-mutated patient ratio of east-Asian origin is high compared with Westerners6,9, so we evaluated the clinical benefit of this approach by examining the association between concurrent genomic alterations in patients harboring EGFR active mutations and their subsequent therapeutic outcome.
Results
Patient characteristics
The demographic data for the 100 patients included in this study are shown in Table 1. The median follow-up period after surgery was 32.6 months (range 6.4–104.8). In regard to smoking status, 62 patients identified as never having smoked or as light smokers (pack-years, or PY < 30), and 38 patients were heavy smokers (PY ≥ 30). Overall, the median mutation burden (number of rare SNPs) was 13.5 (range 5–33) and the median number of identified genomic alterations was four (range 1–19), with 98% of patients having one or more actionable mutations, defined as genomic alterations that are either associated with targeted therapy that is FDA/PMDA- approved or would qualify the patient for a clinical trial testing a targeted therapy.
Table 1.
Patient demographics.
| Factor | Category | N = 100 |
|---|---|---|
| Age (Years) | Median | 67 |
| Range | 36–86 | |
| Gender (N) | Male | 67 |
| Female | 33 | |
| Smoking (N) | PY < 30 | 62 |
| PY ≥ 30 | 38 | |
| Stage (N) | I | 31 |
| II | 24 | |
| III | 39 | |
| IV | 6 |
N, number; PY, pack-years.
The frequency of genomic alterations
Tumors were sequenced with a median coverage of 500× , and a total of 281 individual genomic alterations were identified. EGFR was the most commonly mutated gene (48% of patients, n = 48/100), followed by tumor protein p53 gene (TP53) (40%) and cyclin-dependent kinase inhibitor 2B (CDKN2B) (32%) as shown in Table 2. EGFR active mutations, i.e., those involving exon 19 deletion or an L858R point mutation, were detected in 43 patients, with six patients showing more than one mutation in EGFR (see Supplementary Tables S1 and S2). Compared with data from TCGA6,7, there were significantly more genomic alterations in AT-rich interactive domain-containing protein 1A (ARID1A), EGFR, cyclin-dependent kinase inhibitor 1B (CDKN1B), cyclin-dependent kinase inhibitor 2A (CDKN2A), retinoblastoma 1 gene (RB1), phosphatase and tensin homolog (PTEN), activin receptor type 2 (ACVR2A), and F-box/WD repeat-containing protein 7 (FBXW7) (all p < 0.01) and significantly less genomic alterations in erb-b4 receptor tyrosine kinase 4 gene (ERBB4), Kirsten rat sarcoma viral oncogene homolog gene (KRAS), and Kelch-like ECH-associated protein 1 (KEAP1) (p = 0.01, p < 0.01, and p < 0.01, respectively) among our Japanese patients (Table 2).
Table 2.
Frequency of Gene Alterations in Each Pathway.
| Frequency (%) | Frequency (%) | ||||||
|---|---|---|---|---|---|---|---|
| TCGA | This study | TCGA | This study | ||||
| (N = 216) | (N = 100) | (N = 216) | (N = 100) | ||||
| Transcription factor/regulator | 2.6 | 0.0 | GATA3 | MAPK signaling | 26.3 | 12.0 | KRAS |
| 0.9 | 0.0 | EP300 | 11.8 | 2.0 | NF1 | ||
| 4.0 | 0.0 | TAF1 | 1.8 | 0.0 | MAP3K1 | ||
| 0.4 | 0.0 | RUNX1 | 6.6 | 5.0 | BRAF | ||
| 3.5 | 1.0 | WT1 | 1.8 | 0.0 | NRAS | ||
| 0.4 | 0.0 | FOXA1 | 1.3 | 0.0 | MAP2K4 | ||
| 0.4 | 0.0 | CBFB | PI(3)K signaling | 4.4 | 6.0 | PIK3CA | |
| 1.3 | 0.0 | SOX9 | 2.2 | 17.0 | PTEN | ||
| Histone modifier | 6.1 | 17.0 | ARID1A | 1.3 | 4.0 | PIK3R1 | |
| 1.8 | 4.0 | PBRM1 | 11.4 | 0.0 | TLR4 | ||
| 7.9 | 2.0 | SETD2 | 5.3 | 0.0 | PIK3CG | ||
| 4.8 | 1.0 | KDM5C | 0.0 | 2.0 | AKT1 | ||
| 0.9 | 0.0 | KDM6A | TGF-β signaling | 3.1 | 4.0 | SMAD4 | |
| 1.3 | 0.0 | ASXL1 | 0.9 | 4.0 | TGFBR2 | ||
| 2.2 | 0.0 | EZH2 | 2.2 | 1.0 | ACVR1B | ||
| Genome integrity | 51.8 | 40 | TP53 | 0.9 | 1.0 | SMAD2 | |
| 7.9 | 3.0 | ATM | 0.9 | 12.0 | ACVR2A | ||
| 6.1 | 2.0 | ATRX | Wnt/β-catenin | 9.2 | 16.0 | APC | |
| 5.7 | 10.0 | BRCA2 | 3.5 | 5.0 | CTNNB1 | ||
| 5.7 | 0.0 | ATR | 0.9 | 0.0 | AXIN2 | ||
| 2.6 | 0.0 | STAG2 | Proteolysis | 1.3 | 13.0 | FBXW7 | |
| 1.3 | 1.0 | BAP1 | 17.1 | 2.0 | KEAP1 | ||
| 3.5 | NA | BRCA1 | 0.4 | 0.0 | SPOP | ||
| 1.3 | 0.0 | ERCC2 | Splicing | 2.2 | 0.0 | SF3B1 | |
| RTK signaling | 11.4 | 48.0 | EGFR | HIPPO signaling | 1.3 | 2.0 | CDH1 |
| 4.0 | 1.0 | FLT3 | DNA methylation | 4.0 | 0.0 | DNMT3A | |
| 8.8 | 2.0 | EPHA3 | 3.1 | 0.0 | TET2 | ||
| 7.5 | 1.0 | ERBB4 | Metabolism | 0.9 | 0.0 | IDH1 | |
| 6.6 | 2.0 | PDGFRA | 0.4 | 1.0 | IDH2 | ||
| 9.7 | 0.0 | EPHB6 | NFE2L | 2.2 | 2.0 | NFE2L2 | |
| 3.1 | 0.0 | FGFR2 | Protein phosphatase | 4.6 | 0.0 | PPP2R1A | |
| 1.8 | 3.0 | KIT | TOR signaling | 7.5 | 1.0 | MTOR | |
| 0.4 | 2.0 | FGFR3 | 8.8 | 15.0 | STK11 | ||
| Cell cycle | 6.6 | 17.0 | CDKN2A | Other | 3.1 | 0.0 | NOTCH1 |
| 5.3 | 22.0 | RB1 | 5.3 | 0.0 | USP9X | ||
| 3.1 | 1.0 | CDK12 | 0.9 | 0.0 | NPM1 | ||
| 1.8 | 19.0 | CDKN1B | 10.5 | 1.0 | HGF | ||
| 0.9 | 1.0 | CCND1 | 1.8 | 0.0 | AR | ||
| 0.4 | 0.0 | CDKN1A | |||||
| 0.0 | 0.0 | CDKN2C | |||||
| NA | 32.0 | CDKN2B | |||||
Bolded entries represent those genes for which the frequency of alteration was significantly different between TCGA and the present study; N, number; NA, not available.
Overall impact of genomic alterations on clinical outcome in lung adenocarcinoma
We next assessed the impact of each genomic alteration on the clinical outcomes of patients in our cohort (Table 3). Univariate analysis indicated that patients with EGFR active mutations had significantly longer disease-free survival (DFS) than patients without these mutations (p = 0.041). Conversely, patients with TP53 mutation or CDKN2B mutation had significantly shorter DFS than patients without those mutations. In multivariate analysis, TP53 mutation and CDKN2B mutation remained independent predictors of DFS (p = 0.037 and p = 0.002, respectively), but EGFR active mutation did not (p = 0.050). In regards to overall survival (OS), however, univariate analysis indicated that patients with any EGFR mutation, not just those with EGFR active mutation, had significantly longer OS (p = 0.020 and p = 0.027, respectively).
Table 3.
Univariate and Multivariate Survival Analysis of DFS and OS in All Patients.
| Genes | Category | N = 100 | 5-year DFS (%) | Univariate Analysis | Multivariate Analysis | 5-year OS (%) | Univariate Analysis | |||
|---|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |||||
| EGFR (All) | WT | 52 | 35.5 | 55.0 | ||||||
| MUT | 48 | 33.4 | 0.610 (0.356–1.045) | 0.072 | 77.3 | 0.356 (0.149–0.847) | 0.020 | |||
| EGFR (active)a | Otherb | 57 | 31.7 | Referencec | 55.6 | |||||
| MUT | 43 | 38.6 | 0.559 (0.320–0.977) | 0.041 | 0.607 (0.335–1.100) | 0.050 | 79.1 | 0.357 (0.143–0.890) | 0.027 | |
| TP53 | WT | 60 | 41.5 | Reference | 76.9 | |||||
| MUT | 40 | 28.9 | 1.795 (1.056–3.051) | 0.031 | 1.727 (1.003–2.975) | 0.037 | 52.4 | 2.042 (0.936–4.458) | 0.073 | |
| CDKN2B | WT | 68 | 51.6 | Reference | 62.9 | |||||
| MUT | 32 | 7.1 | 2.151 (1.253–3.694) | 0.005 | 2.391 (1.376–4.155) | 0.002 | 70.9 | 0.761 (0.317–1.823) | 0.540 | |
| RB1 | WT | 79 | 36.0 | 65.4 | ||||||
| MUT | 21 | 34.3 | 1.459 (0.783–2.722) | 0.234 | 66.2 | 1.208 (0.485–3.011) | 0.685 | |||
| CDKN1B | WT | 81 | 33.8 | 61.8 | ||||||
| MUT | 19 | 40.6 | 0.948 (0.487–1.844) | 0.875 | 75.9 | 0.522 (0.179–1.524) | 0.234 | |||
| CDKN2A | WT | 83 | 44.4 | 64.7 | ||||||
| MUT | 17 | 9.9 | 1.263 (0.665–2.401) | 0.476 | 64.2 | 1.071 (0.401–2.860) | 0.892 | |||
| PTEN | WT | 83 | 33.3 | 63.6 | ||||||
| MUT | 17 | 47.1 | 1.092 (0.533–2.236) | 0.810 | 72.1 | 0.880 (0.303–2.559) | 0.815 | |||
| ARID1A | WT | 83 | 30.9 | 63.6 | ||||||
| MUT | 17 | 58.8 | 0.811 (0.382–1.722) | 0.585 | 71.7 | 1.196 (0.449–3.185) | 0.721 | |||
| APC | WT | 84 | 37.9 | 63.8 | ||||||
| MUT | 16 | 29.5 | 1.061 (0.533–2.110) | 0.866 | 65.6 | 1.295 (0.516–3.254) | 0.582 | |||
| STK11 | WT | 85 | 39.5 | 67.2 | ||||||
| MUT | 15 | 18.8 | 1.707 (0.858–3.395) | 0.128 | 46.7 | 1.793 (0.672–4.782) | 0.243 | |||
| FBXW7 | WT | 87 | 30.3 | 63.6 | ||||||
| MUT | 13 | 65.9 | 0.511 (0.203–1.288) | 0.155 | 72.5 | 1.131 (0.386–3.315) | 0.822 | |||
| KRAS | WT | 88 | 37.0 | 71.9 | ||||||
| MUT | 12 | 25.0 | 1.947 (0.950–3.992) | 0.069 | 44.4 | 2.277 (0.910–5.696) | 0.078 | |||
| ACVR2A | WT | 88 | 33.7 | 63.2 | ||||||
| MUT | 12 | 56.3 | 0.493 (0.178–1.366) | 0.174 | 83.3 | 0.631 (0.149–2.680) | 0.533 | |||
Note: Only those genes mutated in more than 12 patients were analyzed. aActive EGFR mutation refers to exon19 deletion or L858R point mutations. bOther refers to patients with wildtype EGFR or non-active EGFR mutations. cMultivariate analysis was performed for the EGFR active mutation group, since the data for those with active mutations and those with any EGFR mutation overlapped considerably. DFS, disease-free survival; OS, overall survival; N, number; HR, hazard ratio; CI, confidence interval; WT, wildtype; Mut, mutated; Bold values are those with statistical significance of p < 0.05.
EGFR genomic alterations and clinical outcome
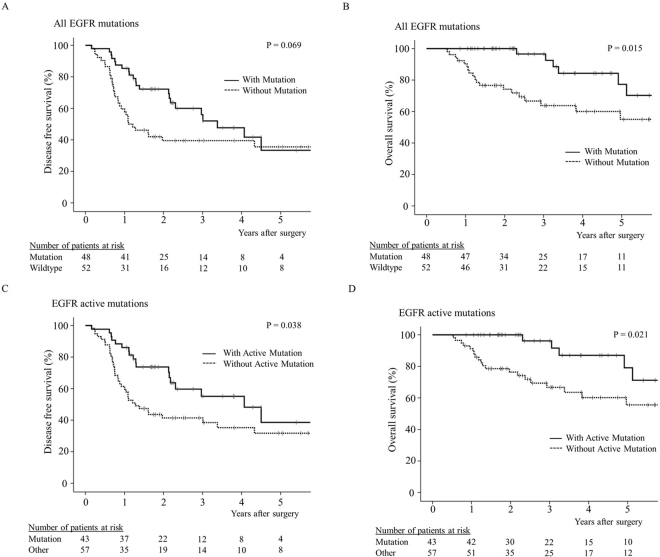
We next examined the clinical outcome of patients with EGFR mutations in more detail. The majority of the patients with EGFR mutations (84%) identified as either having never smoked or as light smokers (see Supplementary Table S1). In terms of clinical outcome, the 48 patients with EGFR mutation showed a trend towards longer DFS and a significantly longer OS, than those without EGFR mutation (Fig. 1A,B; log-rank test, p = 0.069 and p = 0.015, respectively). When only the 43 patients with EGFR active mutations were considered, both DFS and OS were found to be significantly longer than for patients with wildtype EGFR or EGFR non-active mutations (Fig. 1C,D; log-rank test, p = 0.038 and p = 0.021, respectively).
Figure 1.
Effect of EGFR mutation status on patient survival. Postoperative disease-free survival (A) and overall survival (B) curves for patients with or without any type of EGFR mutation, and disease-free survival (C) and overall survival (D) curves for patients with or without EGFR active mutations (i.e., exon19 deletion or L858R point mutation). ‘Other’ indicates patients with either wildtype EGFR or an EGFR non-active mutation.
Concurrent genomic alterations in patients with EGFR active mutation
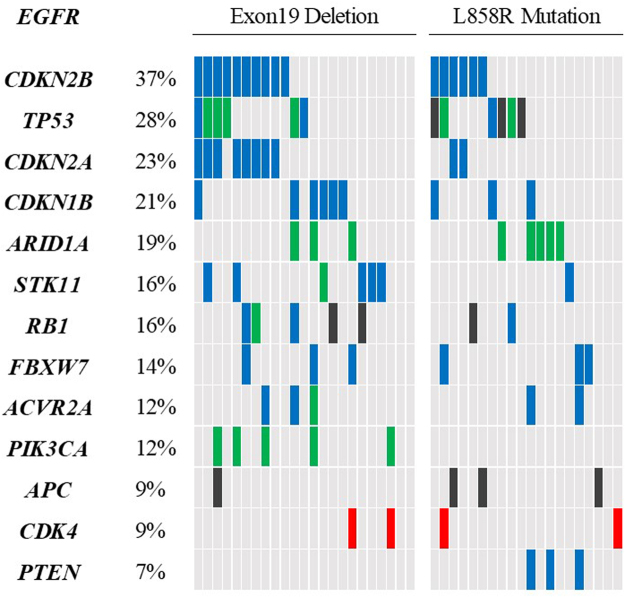
Subsequently, we assessed the incidence of concurrent genomic alteration in the 43 patients with EGFR active mutation. The most frequent concurrent genomic alterations in these patients were CDKN2B (37% of patients), TP53 (28%), CDKN2A (23%), CDKN1B (21%), ARID1A (19%), RB1 (16%), and serine/threonine kinase 11 (STK11) (16%) (Fig. 2). In agreement with previous reports10, there were no concurrent genomic alterations in KRAS, ALK, ROS1, RET, B-Raf proto-oncogene (BRAF) (V600E) or MET. We also examined differences in the pattern of concurrent genomic alteration in patients with EGFR active mutation vs. those with either EGFR wildtype or EGFR non-active mutation. This analysis revealed that patients with EGFR active mutation carried fewer TP53 mutations and PTEN mutations than the others (p = 0.026 and p = 0.018, respectively; see Supplementary Table S3). We also compared the pattern of concurrent genomic alteration in patients with EGFR exon19 deletion and those with EGFR L858R mutation (Fig. 2); however, no significant differences were found between the two groups.
Figure 2.
Concurrent genetic alterations among patients with EGFR active mutation. Percentages indicate the frequency of each mutation in these 43 patients, with the heatmap indicating the presence or absence of the variation in individuals. Green cells, SNPs; black cells, stop-gain mutations; red cells, gene amplification; blue cells, genomic loss.
Impact of concurrent genomic alterations on clinical outcomes in patients with EGFR active mutation
To assess the impact of concurrent genomic alterations on clinical outcomes in patients with EGFR active mutations, survival of these patients was compared to patients with wildtype EGFR or EGFR non-active mutations. Univariate analysis revealed that patients with CDKN2B mutation had significantly shorter DFS than those without CDKN2B mutation (p = 0.001, Table 4), while patients with RB1 mutation had significantly shorter OS than those without RB1 mutation (p = 0.035, see Supplementary Table S4). These findings suggest that concurrent genetic alterations affect clinical outcomes in patients with EGFR active mutations.
Table 4.
Univariate Analysis of DFS in Patients with EGFR Active Mutations.
| Genes | Category | N = 43 | 5-year DFS (%) | Univariate Analysis | |
|---|---|---|---|---|---|
| HR (95% CI) | p-value | ||||
| CDKN2B | WT | 27 | 51.6 | ||
| MUT | 16 | 7.1 | 5.618 (1.995–15.827) | 0.001 | |
| CDKN2A | WT | 33 | 54.7 | ||
| MUT | 10 | 14.6 | 1.887 (0.728–4.892) | 0.191 | |
| CDKN1B | WT | 34 | 28.7 | ||
| MUT | 9 | 62.2 | 0.435 (0.123–1.540) | 0.197 | |
| TP53 | WT | 31 | 47.0 | ||
| MUT | 12 | 31.3 | 2.495 (0.956–6.510) | 0.062 | |
| STK11 | WT | 36 | 44.7 | ||
| MUT | 7 | 33.3 | 0.864 (0.246–3.035) | 0.820 | |
| RB1 | WT | 36 | 41.1 | ||
| MUT | 7 | 28.6 | 1.875 (0.601–5.848) | 0.279 | |
| ARID1A | WT | 35 | 24.1 | ||
| MUT | 8 | 100 | 0.136 (0.030–2.492) | 0.056 | |
| FBXW7 | WT | 37 | 30.3 | ||
| MUT | 6 | 62.5 | 0.715 (0.196–2.621) | 0.613 | |
| EGFR* | WT | 37 | 36.5 | ||
| MUT | 6 | 44.4 | 1.737 (0.566–5.329) | 0.334 | |
Note: Only genes mutated in more than six patients were analyzed. *Refers to EGFR non-active mutations, i.e., excluding Exon19 deletion and L858R. DFS, disease-free survival; N, number; HR, hazard ratio; CI, confidence interval; PY, pack-year; WT, wild type; MUT, mutated; Bold values are those with statistical significance of p < 0.05.
We also examined the impact of concurrent genomic alterations on clinical outcomes in patients with wildtype EGFR or non-active EGFR mutations. In this analysis, patients with STK11 mutation were found to have significantly shorter DFS (p = 0.006, Table 5) and OS (p = 0.046, see Supplementary Table S5) than those without STK11 mutations. Of note, neither CDKN2B nor RB1 were associated with DFS or OS in the patients with wildtype EGFR or non-active EGFR mutations.
Table 5.
Univariate Analysis of DFS in Patients with Wildtype EGFR or Non-active EGFR Mutations.
| Genes | Category | N = 57 | 5-yr DFS (%) | Univariate Analysis | |
|---|---|---|---|---|---|
| HR (95% CI) | p-value | ||||
| TP53 | WT | 29 | 34.4 | ||
| MUT | 28 | 28.3 | 1.386 (0.720–2.668) | 0.329 | |
| CDKN2B | WT | 41 | 38.9 | ||
| MUT | 16 | 0.0 | 1.343 (0.667–2.706) | 0.409 | |
| RB1 | WT | 43 | 30.0 | ||
| MUT | 14 | 12.8 | 1.304 (0.612–2.781) | 0.491 | |
| PTEN | WT | 43 | 28.3 | ||
| MUT | 14 | 35.7 | 1.185 (0.555–2.530) | 0.662 | |
| KRAS | WT | 45 | 33.1 | ||
| MUT | 12 | 25.0 | 1.571 (0.737–3.348) | 0.242 | |
| APC | WT | 45 | 32.1 | ||
| MUT | 12 | 35.0 | 0.762 (0.331–1.752) | 0.522 | |
| CDKN1B | WT | 47 | 33.4 | ||
| MUT | 10 | 20.0 | 1.537 (0.699–3.380) | 0.285 | |
| ARID1A | WT | 48 | 32.4 | ||
| MUT | 9 | 22.2 | 2.004 (0.871–4.614) | 0.102 | |
| STK11 | WT | 49 | 35.3 | ||
| MUT | 8 | 12.5 | 3.268 (1.402–7.618) | 0.006 | |
| BRCA2 | WT | 49 | 27.7 | ||
| MUT | 8 | 50.0 | 0.603 (0.213–1.711) | 0.342 | |
Note: Only genes mutated in more than eight patients were analyzed. DFS, disease-free survival; N, number; HR, hazard ratio; CI, confidence interval; WT, wildtype; MUT, mutated; Bold values are those with statistical significance of p < 0.05.
Clinical outcomes for patients receiving EGFR-TKI therapy
In total, 14 patients received first-generation EGFR-TKI therapy, i.e., gefitinib or erlotinib, for recurrence; however, in one patient an osteosclerotic metastasis in the fourth thoracic vertebra was detected by bone scintigraphy; that lesion was determined to be non-measurable by the RECIST v1.1 guidelines11, and thus the patient was excluded from the analysis. One patient received EGFR-TKI therapy, however, the patient was excluded from this study due to serious adverse event happened early after the administration of EGFR-TKI. The others did not develop recurrence or metastatic disease except two: one patient had a recurrence with brain metastasis and received radiation therapy; one patient developed recurrence but had not started treatment yet. Of the remaining 12 patients, two were designated as non-responders (Fig. 3). Interestingly, the number of genomic alterations in the two non-responders was found to be the highest of any of the patients who received EGFR-TKI treatment (Fig. 3).
Figure 3.
Clinical response of EGFR mutated patients treated with EGFR tyrosine kinase inhibitor (EGFR-TKI). The waterfall plot shows the best percentage change in target lesions from baseline for 13 patients treated with EGFR-TKI. Red boxes indicate those patients with a higher than median mutation burden or number of genomic alterations; *indicates patients without a measurable recurrent lesion prior to EGFR-TKI use.
To gain a clearer understanding of factors that may have contributed to the poor clinical response of these two patients, we examined their medical records in more detail. Briefly, Case 1 initially received right middle lobectomy with lymph node dissection. Two months following surgery, chest CT revealed the presence of mediastinal lymph nodes and supraclavicular lymph node metastases. After the failure of concurrent platinum-based chemotherapy and thoracic radiotherapy, new multiple small (<10 mm) brain and osteosclerotic bone metastases were found to have developed. At this point the patient received EGFR-TKI therapy; however, multiple bone metastases were detected approximately 2 months later. Case 2 underwent right lower lobectomy with lymph node dissection. Seven months after surgery, bone scintigraphy revealed osteosclerotic metastases to the cervical vertebrae. Following irradiation of the cervical vertebrae lesion the patient received EGFR-TKI therapy, but malignant pleural effusion and lymphangiosis carcinomatosa developed approximately 1 month later.
Notably, we investigated the association between the number of genomic alterations and progression-free survival (PFS) of the patients with EGFR-TKI therapy. We found the patients with 4 or more genomic alterations had significantly poorer PFS than those with less than 4 (p = 0.006) (Supplementary Fig. S6).
Discussion
NGS technologies enable us to identify not only potentially targetable driver mutations but also other important genomic alterations that are associated with clinical outcome. In the present study, we identified actionable genomic driver mutations in 98% of patients in a Japanese lung adenocarcinoma cohort by comprehensive NGS-based sequencing of a panel of 415 genes with relevance for cancer, with an average of 500× depth. In contrast, TCGA performed whole-exome sequencing on tumor, with a mean coverage of 97.6× depth. Although the sequence techniques had been not the same, of note, there were differences in the observed prevalence for several driver gene mutations between the Japanese and TCGA cohorts, consistent with previous reports12,13. Furthermore, we found that concurrent mutations, in genes such as CDKN2B and RB1, may impact the survival of patients with EGFR mutations.
The CDKN2B gene lies adjacent to the tumor suppressor gene CDKN2A in a genomic region that is frequently mutated and/or deleted in various tumor types. CDKN2B encodes a cyclin-dependent kinase (CDK) inhibitor known as p15Ink4b, which forms a complex with CDK4 or CDK6 and prevents their ability inactivate the RB1 gene product (Rb) and other Rb-family proteins during the G1 phase of the cell cycle. The p15Ink4b protein thus functions as a cell growth regulator that inhibits cell cycle progression14. Zhao et al. reported that CDKN2B loss is associated with poor overall survival of patients with lung squamous cell carcinoma15, and in NSCLC patients, loss of chromosome 9p (encompassing the 9p21.3 locus where CDKN2A and CDKN2B are located) has also been associated with poor survival outcome16,17. Moreover, CDKN2A and CDKN2B are known to be frequently inactivated by allelic loss and promotor methylation in NSCLC, resulting in the deregulation of cell proliferation through the loss of G1 arrest control18.
Like CDKN2B, the RB1 gene is also an important regulator of cell cycle progression. Together with other Rb family members (such as p107 and p130), Rb is phosphorylated by CDK4/6 and other cyclin-CDK complexes, inducing the release of transcription factors of the E2F family and the consequent transcription of genes required for S-phase entry19. Molecular alterations involving the CyclinD1-CDK4/6-Rb pathway occur in a variety of malignancies such as breast cancer20, prostate cancer21, and osteosarcoma22, and have been associated with poor prognosis. In regard to lung cancer, recent whole-genome sequencing has revealed that the RB1 gene is altered in almost all cases of small cell carcinoma23. It remains premature to determine the clinical outcome on this population due to the small sample size of this study. However, considering that the CyclinD1-CDK4/6-Rb pathway is downstream of the EGFR signaling pathway, it is likely that this cell cycle pathway, which involves CDKN2B and RB1, plays an important role in lung cancer progression and/or therapeutic resistance in patients with EGFR active mutation.
A previous study has reported that lung adenocarcinoma patients with relapse have a significantly larger proportion of private non-trunk mutations in their primary tumor than those without relapse, indicating not only that subclonal mutations are crucial for tumor progression, but that they increase postoperative risk of relapse24. Notably, the two lung adenocarcinoma patients in our cohort that carried the highest number of oncogenic gene alterations, showed no clinical response to EGFR-TKI therapy. This observation supports the concept that it is not just major driver mutations that affect therapeutic response and clinical outcome, and highlights the importance of conducting comprehensive genome sequencing in patients with lung adenocarcinoma.
In regard to the EGFR gene, we identified differences in DFS and OS between patients with EGFR active mutation and those with wildtype EGFR or non-active EGFR mutations. Although the prognostic impact of EGFR mutation in lung adenocarcinoma remains controversial, several retrospective studies have reported that patients with EGFR mutation survived for longer periods than those without mutations, irrespective of therapy25,26. Such results may in part be a reflection of cohort composition in regard to gender and smoker status, with several investigators having reported that among patients with NSCLC, those that have never smoked have a better OS than smokers24,27.
It is important to point out that the current study has some limitations. Firstly, this is a retrospective study with a relatively small number of patients. Secondly, we noted an imbalance between the proportion of Stage I/II and Stage III/IV surgically resected cases in comparison with reported data for other Japanese cohorts28. Since only patients from whom sufficient amounts of tumor DNA could be extracted from FFPE specimens were included in this study, the proportion of advanced stage cases in our study was understandably higher. Third, when assessing the impact of each genomic alteration on DFS and OS, we did not perform multiple testing correction. This is because we analyzed only about 10 genomic alterations on survival and the sample size of this study was limited due to the cost of NGS analyses. However, to our knowledge, the present study represents the largest cohort of Japanese lung adenocarcinoma patients to have been genomically characterized using a comprehensive gene panel, and thus contributes substantially to our understanding of the clinical progression of lung adenocarcinoma in this population.
In conclusion, using an NGS sequencing approach we have identified actionable genomic driver mutations in 98% of Japanese lung adenocarcinoma patients, and discovered that the mutation frequencies of several key genes in this population appear to differ from those described in the TCGA database. Notably, concurrent loss of CDKN2B or RB1 was associated with poor prognosis in patients with EGFR active mutation. In addition, our data indicate that lung adenocarcinoma patients with high numbers of oncogenic gene alterations may show the worst responses to EGFR-TKI targeted therapy. Although further studies are needed to verify these findings, our data improve our understanding of the relationship between genomic alteration and prognosis in lung adenocarcinoma.
Material and Methods
Patients and tissues
This study was approved by the Institutional Review Boards of Niigata University, and Kyusyu University Hospital. At Niigata University, patients were recruited from January 2008 to December 2014, and at Kyushu University from October 2013 to August 2015. All methods were performed in accordance with the relevant guidelines and regulations, and written informed consent was obtained from all patients. Patients were selected using the following three criteria: firstly, a tumor content of >20% based on pathological review of hematoxylin and eosin (H&E) stained slides. Secondly, radiological confirmation of lung adenocarcinoma with a consolidation/tumor ratio (C/T ratio) >0.5 using thin-section computed tomography (CT). Thirdly, the successful extraction of ≥150 ng DNA from each sample. One hundred patients who underwent surgery for primary lung adenocarcinoma were finally enrolled into the study, with all clinicopathology data, including smoking history, being retrieved from medical records. Six patients with Stage IV disease were included in our cohort. Preoperative work up revealed no metastasis and their diseases were clinically diagnosed as Stage I in 3 patients, Stage II in 2 patients, and Stage III in one preoperatively, however, they were found to have dissemination to visceral pleura by postoperative pathological examination (Table 1).
Sequencing library preparation
Formalin-fixed, paraffin embedded (FFPE) cancer tissue from surgical specimens was used for analysis. An independent pathologist evaluated the tumor content using H&E slides. Where applicable, unstained slides were macro-dissected to enrich for tumor content and genomic DNA was extracted using the BiOstic FFPE Tissue DNA Isolation Kit (Mo Bio Laboratories, Inc; Carlsbad, CA). Sample preparation, genomic sequencing and subsequent analyses were all performed in a CLIA/CAP-accredited laboratory (KEW Inc.; Cambridge, MA).
Comprehensive genomic sequencing
FFPE genomic DNA (150 ng) was converted into libraries and enriched for a 415 gene panel with CANCERPLEX (KEW Inc.; Cambridge, MA). CANCERPLEX is a clinically validated gene panel enriched for the coding regions and selected introns of 415 genes with known relevance for cancer. Sequencing was performed on the Illumina MiSeq and NextSeq platforms with an average of 500× sequencing depth. Genomic data were then processed through a proprietary bioinformatics platform and knowledge base to identify multiple classes of genomic abnormalities, including single-nucleotide substitutions (SNPs), small insertions/deletions (indels), copy number variations (CNVs), and translocations in ALK, RET, and ROS1. A threshold of 10% allelic fraction was used for SNPs and indels, and thresholds of >2.5-fold (gains) and 0.5-fold (losses) were used for CNVs. To assess the somatic status of mutations in the absence of constitutive samples, we employed a filtering strategy similar to one recently published, but with minor differences29,30. Based on both published evidence and our own experience, this approach allows the correct discrimination between germline and somatic variants in >99% of cases. Mutation burden was determined by the number of nonsynonymous SNPs present in the tumor that had population frequencies of <1% in dbSNP and the 1000 genomes databases. Actionable mutations were defined as known oncogenic alterations in key driver genes that are associated with response to approved targeted therapies (e.g., exon 19 deletions in EGFR). The number of genomic alterations was calculated in the context of the total number of genes represented on the CANCERPLEX panel. EGFR active mutations were defined as those involving EGFR exon 19 deletion or an L858R point mutation.
Statistical analysis
Associations between each genotype and clinical characteristics were analyzed using two-tailed Student’s t tests and Fisher’s exact tests. Disease-free survival (DFS) was defined as the time from surgery to documented clinical progression or death. Overall survival (OS) was defined as the time from surgery until death. Progression-free survival (PFS) was defined as the time from the treatment to disease progression or death from any cause. As for PFS, we have established the cutoff value (4 or more vs. less than 4), which was based on the median number of identified genomic alterations in all patients. Survival curves were constructed using the Kaplan-Meier method, with statistical significance determined by log-rank test. Univariate and multivariate Cox proportional hazards models were developed to determine which factors had a significant impact on survival and to assess independent prognostic significance. All factors that attained a significance level of p < 0.05 in univariate analysis were included in the multivariate analysis. Statistical analysis was performed using SPSS for Windows Version 23.0 (SPSS, Inc., Chicago, IL, USA), with p < 0.05 being considered statistically significant.
Electronic supplementary material
Supplementary Table S1, Table S2, Table S3, Table S4, Table S5, and Figure S6
Acknowledgements
This project was supported by funding from Denka Co. Ltd. S. Sato is supported by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research Grant Number 15K19931. M. Nagahashi is supported by the JSPS Grants-in-Aid for Scientific Research Grant Numbers 15H05676 and 15K15471, and the Takeda Science Foundation. T. Wakai is supported by the JSPS Grants-in-Aid for Scientific Research Grant Numbers 15H04927 and 16K15610. M. Tsuchida is supported by the JSPS Grant-in-Aid for Scientific Research Grant Number 15K10235. S. Lyle is supported by a grant from the Massachusetts Life Sciences Center. The remaining authors declare no conflict of interest. We are grateful to our colleagues Dr. Akihiko Kitahara and Dr. Tatsuya Goto for patient data collection, and to Mr. T Hatano and Ms. A. Kimoto for their technical support.
Author Contributions
S.S. and M.N. contributed to the concept, design, data acquisition, analysis, interpretation, and writing. S.W., T.O., S.L., T.W., and M.T. contributed to the concept, design, analysis, and interpretation. T.K., H.I., Y.S., T.K., K.T., R.N., E.O., and K.T. contributed to data acquisition and interpretation. K.A. and Y.L., and S.O. contributed to data analysis. S.L., W.T., and M.T. supervised the study. All authors have read and approved the submitted manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Seijiro Sato, Masayuki Nagahashi, Toshifumi Wakai and Masanori Tsuchida contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18560-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Toshifumi Wakai, Email: wakait@med.niigata-u.ac.jp.
Masanori Tsuchida, Email: masatsu@med.niigata-u.ac.jp.
References
- 1.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oxnard GR, Binder A, Janne PA. New targetable oncogenes in non-small-cell lung cancer. J Clin Oncol. 2013;31:1097–1104. doi: 10.1200/JCO.2012.42.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature511, 543–550, 10.1038/nature13385 (2014). [DOI] [PMC free article] [PubMed]
- 7.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda M, et al. Clinical application of amplicon-based next-generation sequencing to therapeutic decision making in lung cancer. Ann Oncol. 2015;26:2477–2482. doi: 10.1093/annonc/mdv475. [DOI] [PubMed] [Google Scholar]
- 9.Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII) Am J Cancer Res. 2015;5:2892–2911. [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, et al. Comprehensive Characterization of Oncogenic Drivers in Asian Lung Adenocarcinoma. J Thorac Oncol. 2016;11:2129–2140. doi: 10.1016/j.jtho.2016.08.142. [DOI] [PubMed] [Google Scholar]
- 11.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Kohno T, et al. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res. 2015;4:156–164. doi: 10.3978/j.issn.2218-6751.2014.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers–a different disease. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 14.Matsushime H, et al. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell. 1992;71:323–334. doi: 10.1016/0092-8674(92)90360-O. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, et al. Association of copy number loss of CDKN2B and PTCH1 with poor overall survival in patients with pulmonary squamous cell carcinoma. Clin Lung Cancer. 2011;12:328–334. doi: 10.1016/j.cllc.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Kim TM, et al. Genome-wide screening of genomic alterations and their clinicopathologic implications in non-small cell lung cancers. Clin Cancer Res. 2005;11:8235–8242. doi: 10.1158/1078-0432.CCR-05-1157. [DOI] [PubMed] [Google Scholar]
- 17.Tomizawa Y, et al. Prognostic significance of allelic imbalances on chromosome 9p in stage I non-small cell lung carcinoma. Clin Cancer Res. 1999;5:1139–1146. [PubMed] [Google Scholar]
- 18.Gazzeri S, Gouyer V, Vour’ch C, Brambilla C, Brambilla E. Mechanisms of p16INK4A inactivation in non small-cell lung cancers. Oncogene. 1998;16:497–504. doi: 10.1038/sj.onc.1201559. [DOI] [PubMed] [Google Scholar]
- 19.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 20.Malorni L, et al. A gene expression signature of Retinoblastoma loss-of-function is a predictive biomarker of resistance to palbociclib in breast cancer cell lines and is prognostic in patients with ER positive early breast cancer. Oncotarget. 2016;7:68012–68022. doi: 10.18632/oncotarget.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamoah K, et al. Novel Biomarker Signature That May Predict Aggressive Disease in African American Men With Prostate Cancer. J Clin Oncol. 2015;33:2789–2796. doi: 10.1200/JCO.2014.59.8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren, W. & Gu, G. Prognostic implications of RB1 tumour suppressor gene alterations in the clinical outcome of human osteosarcoma: a meta-analysis. Eur J Cancer Care (Engl)26, 10.1111/ecc.12401 (2017). [DOI] [PubMed]
- 23.George J, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshino I, et al. Smoking status as a prognostic factor in patients with stage I pulmonary adenocarcinoma. Ann Thorac Surg. 2006;81:1189–1193. doi: 10.1016/j.athoracsur.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 25.Bell DW, et al. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol. 2005;23:8081–8092. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- 26.Eberhard DA, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 27.Sardari Nia P, et al. Prognostic value of smoking status in operated non-small cell lung cancer. Lung Cancer. 2005;47:351–359. doi: 10.1016/j.lungcan.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 28.Committee for Scientific Affairs TJAFTS, et al. Thoracic and cardiovascular surgery in Japan during 2014: Annual report by The Japanese Association for Thoracic Surgery. Gen Thorac Cardiovasc Surg. 2016;64:665–697. doi: 10.1007/s11748-016-0695-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garofalo A, et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med. 2016;8:79. doi: 10.1186/s13073-016-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagahashi M, et al. Genomic landscape of colorectal cancer in Japan: clinical implications of comprehensive genomic sequencing for precision medicine. Genome Med. 2016;8:136. doi: 10.1186/s13073-016-0387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1, Table S2, Table S3, Table S4, Table S5, and Figure S6