Abstract
Mitochondria and chloroplasts are now called organelles, but they used to be bacteria. As they transitioned from endosymbionts to organelles, they became more and more integrated into the biochemistry and cell biology of their hosts. Work over the last 15 years has shown that other symbioses show striking similarities to mitochondria and chloroplasts. In particular, many sap-feeding insects house intracellular bacteria that have genomes that overlap mitochondria and chloroplasts in terms of size and coding capacity. The massive levels of gene loss in some of these bacteria suggest that they, too, are becoming highly integrated with their host cells. Understanding these bacteria will require inspiration from cell biology, because a traditional microbiological framework is no longer sufficient.
We microbiologists can be a bit defensive about our organisms. We know that microbes are the most genetically and metabolically diverse organisms on Earth [1], that they drive nearly all biogeochemical cycles [2], and that all macroorganisms require the presence of microorganisms for optimal health and survival [3] (but that the opposite is not true). We know all of this, but despite our best efforts to remind our colleagues of these important facts at every possible opportunity, we still suspect that our macroorganism-focused friends really don’t care.
Perhaps, sometimes, this defensiveness can breed zealotry. If a microbial zealot thinks of the eukaryotic cell at all, their focus is usually restricted to the mitochondrion and chloroplast. These structures are clearly the only interesting parts of eukaryotic biology, because they are the only organelles of bacterial origin [4–7]. Although some microbiologists might allow that the morphological diversity of eukaryotes is interesting, to a microbial purist these beautiful forms are really just elaborate ways of moving around mitochondria and displaying chloroplasts.
No matter one’s enthusiasm for the microbial world, the idea that the origin of the mitochondrion was important in the evolution of the eukaryotic cell is not controversial. If true eukaryotes existed before the mitochondrion, none apparently survived; all extant eukaryotes have mitochondria or degenerate mitochondria-like organelles called hydrogenosomes and mitosomes [5,8]. Arguments in the field of eukaryotic cell evolution now concern the timing and effect of the mitochondrial acquisition [9–13]. Did a cell that looked at least a bit like a modern eukaryote become the mitochondrial host, perhaps engulfing it by endocytosis, or did the mitochondrial acquisition happen early, by other mechanisms, and drive the evolution of the eukaryotic cell? These are very difficult questions to answer, because no intermediate forms exist—the last universal eukaryotic ancestor had most of the canonical features seen in eukaryotes today [14].
However, recent results seem to be converging on a partial answer to these questions. Phylogenetic [15,16] and genomic [17] results both point to organisms from the TACK (Thaumarchaeota, Aigarchaeota, Crenarchaeaota, Korarchaeota) superphylum of Archaea as our closest prokaryotic cousins [18,19]. TACK metagenomes encode pathways previously thought to be limited to eukaryotes, including endosomal sorting complexes required for transport (ESCRT) pathways, homologs of actin and tubulin, and the ubiquitin pathway [10,17]. Together these results suggest that eukaryotic organisms are not sister to Archaea as they are generally shown in textbooks [20], but rather they emerged from them. Although these issues are far from resolved [13], several lines of evidence point to the idea that the mitochondrion might have been acquired by a cell with some eukaryotic features, perhaps somewhat late in the evolution of the eukaryotic cell [12,21].
What makes all of these inferences difficult is the fact that the mitochondrial and chloroplast acquisitions each occurred only once, approximately 1.8 billion [22] and 1.5 billion [23] years ago, respectively. Drawing firm conclusions about the timing and nature of the organisms involved in these acquisitions will therefore probably always be difficult [14]. Of course it makes sense to try, because the only way to really understand the evolution of mitochondria and chloroplasts is to study the evolution of mitochondria and chloroplasts. But other, more modern endosymbioses are available that might tell us something about how they came to be, and why they look the way they do. While these symbioses will never recapitulate the evolution of the mitochondria and chloroplasts—nothing could—they can reveal the general outcomes that occur as a result of intimate, long-term endosymbiosis, and thus may have happened during their evolution [24]. Or they may not, but it seems worthwhile to try this approach, too.
Insect endosymbionts are missing (genomic) links between bacteria and organelles
It is now widely appreciated that all animals form symbioses with bacteria [3,25]. Insects are especially interesting in this regard because they form many intracellular symbioses—that is, they allow bacteria to live inside their cells—that are not pathogenic from the host perspective [25–28]. These intracellular bacteria have broad effects on insect biology. They are sometimes reproductive manipulators [29], but often provide an array of protective functions for the host [30,31]. Most relevant here are the special cases where intracellular bacteria provide absolutely critical nutrients for sap-feeding insects [32–34]. Like all animals, sap-feeding insects cannot make 10 of the required 20 amino acids (the essential amino acids), and so must get them from their diet. This is a problem, because essential amino acids are not found at high levels in plant sap [35]. Sap-feeding insects solved this dilemma by establishing intracellular symbioses with bacteria and fungi that function as little essential amino acid producing factories. Symbiosis with these microbes thus allows exploitation of a new food source, but also then requires the presence of these microbes for the host’s continued survival. It’s important to realize that once you go down this ‘rabbit hole,’ it is very difficult to go back [36]. Just as eukaryotic cells are strictly dependent on their mitochondria, sap-feeding insects are strictly dependent on their nutritional bacterial endosymbionts.
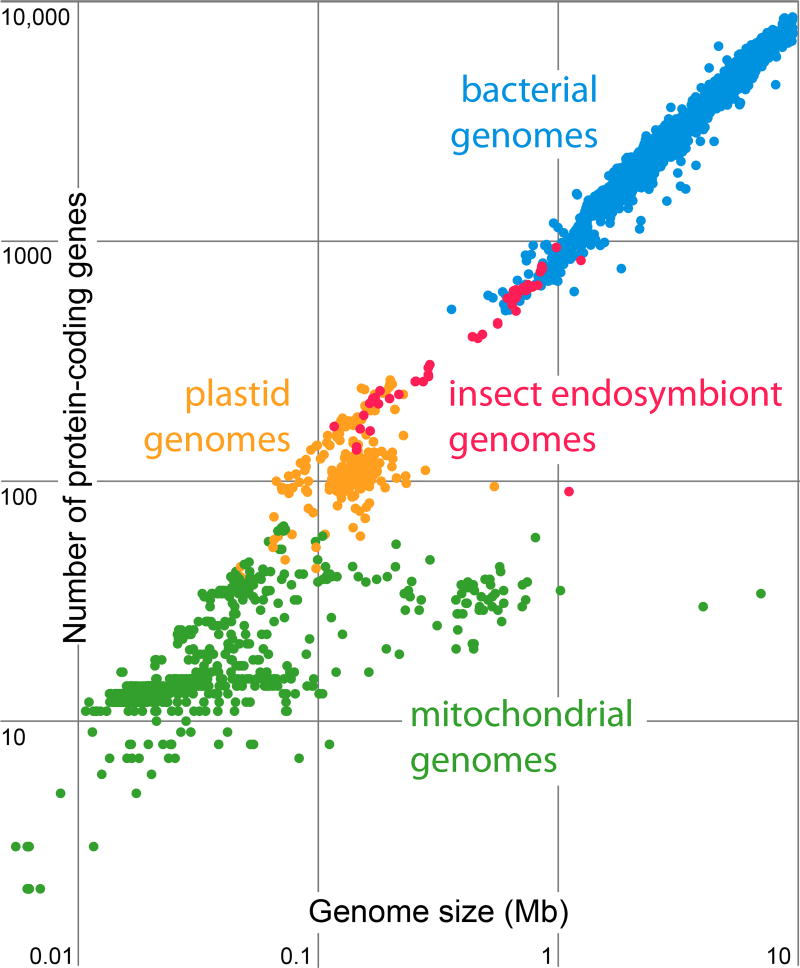
Comparing genome size and coding capacity for bacteria, chloroplasts, and mitochondria reveals three interesting features (Fig. 1). The first is that genome size and coding capacity form an unbroken continuum from free-living bacteria (the largest genomes, typically spanning 3–14 megabase pairs (Mb) in length) to host associated bacteria (typically the smaller genomes centered around 1 Mb), to the endosymbionts of insects (the smallest bacterial genomes, spanning 0.1–1 Mb), followed by the chloroplast and mitochondrial genomes. The second feature, related to the first, is that the genomes of insect endosymbionts are the smallest of all sequenced bacteria and overlap organelle genomes in terms of size and number of genes. The third is that the age of endosymbiosis is reflected in genome size. The oldest endosymbionts, the mitochondria (~1.8 billion years old [22]), have the most reduced genomes encoding the fewest genes. These are followed by the genomes from chloroplasts (~1.5 billion years old [23]), and then the endosymbionts of sap-feeding insects (of various ages, but in the range of 30–300 million years old) [37]. Age recapitulates genome size in endosymbionts.
Figure 1.
Genomes from bacteria, insect endosymbionts, chloroplasts, and mitochondria form an unbroken continuum of size and coding density. The plot is truncated at 10 Mb and 10,000 genes.
Of course, measuring genome size and counting numbers of protein-coding genes tell us little about how organisms function. It’s instructive to dig a bit deeper and compare the genes retained and lost on organelle and endosymbiont genomes.
Genes retained on organelle and insect endosymbiont genomes
First, a disclaimer: The coding capacity of mitochondrial genome varies widely (Fig. 1), making sweeping generalizations about gene content difficult. Some mitochondrial genomes encode 100 genes on a single distinctly bacterium-like chromosome, some retain just a few genes on small linear chromosomes, and some have been completely eliminated [38,39]. So it is only by ignoring this diversity that allows me to say the genes retained on organelle and tiny endosymbiont genomes can be grouped into three broad categories [39–43]. The first relates to translation. Ribosomal RNAs, tRNAs, and ribosomal proteins are universally found on all organelle and endosymbiont genomes. The second are transcription-related genes. The larger mitochondrial, most chloroplast, and endosymbiont genomes encode some fraction of the core bacterial transcriptional machinery. Finally, in what could be considered the ‘signature gene set,’ genes related to the actual function of the organelle or endosymbiont are found. In chloroplasts, these genes are related to generating energy from sunlight [41]. In mitochondria, these genes are related to energy generation from electron transport [40]. In insect nutritional endosymbionts, these genes are related to essential amino acid and vitamin production [34,44]. Genes retained on these genomes thus reflect the core informational processes of translation (mostly), transcription (a bit), and replication (but hardly), as well as the function the endosymbiont or organelle provides to its host.
Genes missing from organelle and endosymbiont genomes
If the genes present on organelle and endosymbiont genomes reveal what they do for their hosts, the genes that are missing hint at what the host must provide to the symbiont. Said another way: How do organelles and endosymbionts survive with such tiny genomes?
The broad outlines of how mitochondria and chloroplasts survive with genomes encoding tens or hundreds of genes are well known. The majority of gene products needed for organelle function are encoded by genes on the nuclear genome, which in many cases have been relocated there by horizontal gene transfer (HGT) during the establishment of the organelles [45,46]. Both mitochondria and chloroplasts have elaborate protein import systems, again encoded on the host genome, which transport these required proteins back into the organelles [47,48]. The targeting and taxonomic affiliation of these proteins can be complex, especially in the photosynthetic eukaryotes [49]. As one example, consider the aminoacyl-tRNA synthetases (aaRSs), the enzymes used to charge tRNAs with the appropriate amino acid. These are essential and ubiquitous enzymes, and most cellular organisms encode genes for twenty enzymes that charge the appropriate tRNA with one of 20 amino acids (or use some defined alternative pathways to perform the same function) [50]. Both mitochondria and chloroplasts have their own translation system requiring their own charged tRNAs, but their genomes encode no aaRSs. How does this work?
The simple expectation is that the nuclear genome of photosynthetic eukaryotes would encode 60 different aaRSs: 20 from the eukaryotic lineage, 20 that have been horizontally transferred from the ancestral mitochondrial genome, and 20 that have been horizontally transferred from the ancestral chloroplast genome. But in Arabidopsis thaliana, only 45 aaRSs are found on the nuclear genome [51]. Twenty-four of these are of mitochondrial or chloroplast origin, and of these 24, 15 are targeted to both mitochondria and chloroplasts [51]. It’s not just aaRSs that are targeted to multiple compartments in the cell—and it’s not just Arabidopsis that shows these complex targeting patterns [52]—many important proteins related to tRNA biogenesis and genome replication, recombination, and repair are also dual targeted [49]. Eukaryotes are mosaic organisms, and have figured out how to build organelles from taxonomically diverse genes sets.
When the first insect endosymbiont genome was sequenced, the lack of genes required to produce components of the cell envelope such as lipopolysaccharides and phospholipids was surprising, and it suggested that these bacteria required extensive cooperation from the host [32]. Looking more broadly across the many tiny endosymbiont genomes now available reveals profound levels of gene loss in fundamental cellular processes [42–44]. The tiniest genomes such as those from Nasuia, Tremblaya, and Hodgkinia encode few or no genes involved in constructing a cell envelope, and are missing many aaRSs and genes involved in genome replication and repair [42–44]. These genomes therefore not only show parallels to organelle genomes in terms of size and genes retained, but also in genes lost.
From microbiology to eukaryotic cell biology
The similarities between insect endosymbionts and organelles are clear. Both are of bacterial origin and have experienced large levels of gene loss. Both encode their tiny gene sets on genomes that are often, but not always [53–56], small in size. Both have lost genes to autonomously construct cell envelopes, along with essential translation-related genes such as tRNAs and aaRSs. Both rely on bacterial genes that have been horizontally transferred from other bacteria to the host genomes [45,57–61]. (There is now evidence that protein made from at least one of these horizontally transferred genes is transported back into insect endosymbiont cells [62].) Both endosymbionts and organelles are absolutely required for normal host function and host survival. The question is: What do these parallels tell us about how endosymbionts actually work?
There are many more questions than answers, especially when considering these symbionts as I have for most of my career—that is, as a microbiological zealot. How do you make a bacterium without the ability to make a cell envelope, and without all of the required aaRSs? The cell biological and organelle literature at least provide a bit of focus. The endoplasmic reticulum (ER) contains most of the enzymes eukaryotic cells use to build membranes, and mitochondrial membrane systems are likely constructed via special ER interaction sites [63,64] (yet they somehow retain some bacterial lipid signatures [65]). What kind of interactions might endosymbionts have with the ER, and could this be how membranes are built in endosymbionts that lack the ability to do it themselves? If so, did they coopt the mitochondrial system, or did they find another way to do it? Mitochondria and chloroplasts retain the ability to translate some proteins but must import aaRSs and tRNAs of diverse taxonomic origins to do so. Some endosymbionts with tiny genomes such as Hodgkinia have incomplete sets of aaRSs and tRNAs, but shotgun proteomics shows that Hodgkinia protein synthesis still occurs [66]. Has Hodgkinia coopted the preexisting aaRS targeting system to become another cellular compartment that requires import of cytoplasmic or mitochondrial aaRS, or has it found another way to assure charged tRNAs are available for translation? These are important questions. They are also, alas, not microbiological questions. Answering them will require a jump into cell biology from a field dominated by microbe-loving scientists.
Acknowledgments
I was supported by the National Science Foundation (IOS-1256680 and IOS-1553529) and the National Aeronautics and Space Administration Astrobiology Institute award NNA15BB04A during the writing of this review. I thank James Van Leuven for help in drawing Fig. 1.
References
- 1.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 2.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 3.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray MW, Doolittle WF. Has the endosymbiont hypothesis been proven? Microbiol. Rev. 1982;46:1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 6.Margulis L. Origin of Eukaryotic Cells. Yale University Press; 1970. [Google Scholar]
- 7.Keeling PJ. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol. 2013;64:583–607. doi: 10.1146/annurev-arplant-050312-120144. [DOI] [PubMed] [Google Scholar]
- 8.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 9*.Koumandou VL, Wickstead B, Ginger ML, van der Giezen M, Dacks JB, Field MC. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol. 2013;48:373–396. doi: 10.3109/10409238.2013.821444. An excellent overview of what the last eukaryotic ancestor might have looked like, from a cell biological perspective. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koonin EV. Archaeal ancestors of eukaryotes: not so elusive any more. BMC Biol. 2015 doi: 10.1186/s12915-015-0194-5. doi:10.1186–s12915–015–0194–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ku C, Nelson-Sathi S, Roettger M, Garg S, Hazkani-Covo E, Martin WF. Endosymbiotic gene transfer from prokaryotic pangenomes: Inherited chimerism in eukaryotes. Proc Natl Acad Sci USA. 2015;112:10139–10146. doi: 10.1073/pnas.1421385112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pittis AA, Gabaldón T. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature. 2016 doi: 10.1038/nature16941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin WF, Garg S, Zimorski V. Endosymbiotic theories for eukaryote origin. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015;370:20140330. doi: 10.1098/rstb.2014.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Poole AM, Gribaldo S. Eukaryotic Origins: How and When Was the Mitochondrion Acquired? Cold Spring Harbor Perspectives in Biology. 2014;6:a015990–a015990. doi: 10.1101/cshperspect.a015990. A tightly argued piece on the difficultly of inferring the steps that lead to the mitochondria. Actually, the entire collection, “The Origin and Evolution of Eukaryotes,” edited by Patrick Keeling and Eugene Koonin, is fantastic. Also included in this book are citations 18, 21, and 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams TA, Foster PG, Nye TMW, Cox CJ, Embley TM. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc R Soc B. 2012;279:4870–4879. doi: 10.1098/rspb.2012.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–179. doi: 10.1038/nature14447. The most recent genomic evidence that some archaea encode a large number of pathways thought once to have been restricted to eukaryotes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guy L, Saw JH, Ettema TJG. The archaeal legacy of eukaryotes: a phylogenomic perspective. Cold Spring Harbor Perspectives in Biology. 2014;6:a016022–a016022. doi: 10.1101/cshperspect.a016022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Williams TA, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2014;504:231–236. doi: 10.1038/nature12779. A terrific review that outlines the data and ideas supporting the concept that eukaryotes are not sister to Archaea, but rather arose out of them. [DOI] [PubMed] [Google Scholar]
- 20.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin EV, Yutin N. The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harbor Perspectives in Biology. 2014;6:a016188–a016188. doi: 10.1101/cshperspect.a016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eme L, Sharpe SC, Brown MW. On the age of eukaryotes: evaluating evidence from fossils and molecular clocks. Cold Spring Harbor Perspectives in Biology. 2014 doi: 10.1101/cshperspect.a016139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parfrey LW, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Keeling PJ, McCutcheon JP, Doolittle WF. Symbiosis becoming permanent: survival of the luckiest. Proc Natl Acad Sci USA. 2015;112:10101–10103. doi: 10.1073/pnas.1513346112. It’s not really this paper that’s special, but the National Academy of Sciences Sackler Colloquium titled “Symbioses Becoming Permanent: The Origins and Evolutionary Trajectories of Organelles” it introduces. Also included in this collection are citations 11, 36, 55, and 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douglas AE. The Symbiotic Habit. 2014 [Google Scholar]
- 26.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 27.Moya A, Pereto J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 2008;9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- 28.Toft C, Andersson SGE. Evolutionary microbial genomics: insights into bacterial host adaptation [Internet] Nature Rev Genet. 2010;11:465–475. doi: 10.1038/nrg2798. [DOI] [PubMed] [Google Scholar]
- 29.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 30.Oliver KM, Degnan PH, Burke GR, Moran NA. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 2010;55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- 31.Nakabachi A, Ueoka R, Oshima K, Teta R, Mangoni A, Gurgui M, Oldham NJ, van Echten-Deckert G, Okamura K, Yamamoto K, et al. Defensive bacteriome symbiont with a drastically reduced genome. Current Biology. 2013;23:1478–1484. doi: 10.1016/j.cub.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 32.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 33.McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 million years of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett GM, Moran NA. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol. 2013;5:1675–1688. doi: 10.1093/gbe/evt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandstrom J, Pettersson J. Amino acid composition of phloem sap and the relation to intraspecific variation in pea aphid (Acyrthosiphon pisum) performance. J. Insect Physiol. 1994;40:947–955. [Google Scholar]
- 36.Bennett GM, Moran NA. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 2015;112:10169–10176. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the Bacterial phylum Bacteroidetes. Appl. Environ. Microbiol. 2005;71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Proc R Soc B. 2010;365:713–727. doi: 10.1098/rstb.2009.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burger G, Gray MW, Forget L, Lang BF. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 2013;5:418–438. doi: 10.1093/gbe/evt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 41.Barbrook AC, Howe CJ, Kurniawan DP, Tarr SJ. Organization and expression of organellar genomes. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2010;365:785–797. doi: 10.1098/rstb.2009.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCutcheon JP. The bacterial essence of tiny symbiont genomes. Curr. Opin. Microbiol. 2010;13:73–78. doi: 10.1016/j.mib.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moran NA, Bennett GM. The Tiniest Tiny Genomes. Annu. Rev. Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 44.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 45.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nature Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 46.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 47.Truscott KN, Brandner K, Pfanner N. Mechanisms of protein import into mitochondria. Current Biology. 2003;13:R326–37. doi: 10.1016/s0960-9822(03)00239-2. [DOI] [PubMed] [Google Scholar]
- 48.Schleiff E, Soll J. Travelling of proteins through membranes: translocation into chloroplasts. Planta. 2000;211:449–456. doi: 10.1007/s004250000357. [DOI] [PubMed] [Google Scholar]
- 49.Carrie C, Small I. A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2013;1833:253–259. doi: 10.1016/j.bbamcr.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 50.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duchene AM, Giritch A, Hoffmann B, Cognat V, Lancelin D, Peeters NM, Zaepfel M, Marechal-Drouard L, Small ID. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gile GH, Moog D, Slamovits CH, Maier UG, Archibald JM. Dual organellar targeting of aminoacyl-tRNA synthetases in diatoms and cryptophytes. Genome Biol Evol. 2015;7:1728–1742. doi: 10.1093/gbe/evv095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rice DW, Alverson AJ, Richardson AO, Young GJ, Sanchez-Puerta MV, Munzinger J, Barry K, Boore JL, Zhang Y, dePamphilis CW, et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342:1468–1473. doi: 10.1126/science.1246275. [DOI] [PubMed] [Google Scholar]
- 54.Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, Taylor DR. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10:e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell MA, Van Leuven JT, Meister RC, Carey KM, Simon C, McCutcheon JP. Genome expansion via lineage splitting and genome reduction in the cicada endosymbiont Hodgkinia. Proc. Natl. Acad. Sci. U.S.A. 2015;112:10192–10199. doi: 10.1073/pnas.1421386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci USA. 2015;112:10177–10184. doi: 10.1073/pnas.1422049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nature Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 58*.Nikoh N, Nakabachi A. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 2009;7:12. doi: 10.1186/1741-7007-7-12. Together with citations 59–61, these papers describe the diversity of bacterial genes that exist on sap-feeding insect genomes, and how many of these genes are overexpressed in the insect tissue housing these bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Husnik F, Nikoh N, Koga R, Ross L, Duncan RP, Fujie M, Tanaka M, Satoh N, Bachtrog D, Wilson ACC, et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153:1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 60.Sloan DB, Nakabachi A, Richards S, Qu J, Murali SC, Gibbs RA, Moran NA. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol. Biol. Evol. 2014;31:857–871. doi: 10.1093/molbev/msu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luan J-B, Chen W, Hasegawa DK, Simmons AM, Wintermantel WM, Ling K-S, Fei Z, Liu S-S, Douglas AE. Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol Evol. 2015;7:2635–2647. doi: 10.1093/gbe/evv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62*.Nakabachi A, Ishida K, Hongoh Y, Ohkuma M, Miyagishima S-Y. Aphid gene of bacterial origin encodes a protein transported to an obligate endosymbiont. Current Biology. 2014;24:R640–1. doi: 10.1016/j.cub.2014.06.038. This paper shows, for the first time, that protein from at least one of the bacterial genes described in references 58–61 is transported back into the endosymbiont. [DOI] [PubMed] [Google Scholar]
- 63.Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat. Rev. Mol. Cell Biol. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64*.Vance JE. MAM (mitochondria-associated membranes) in mammalian cells: Lipids and beyond. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2014;1841:595–609. doi: 10.1016/j.bbalip.2013.11.014. Along with references 63 and 65, a great review of the fascinating interactions between the ER and mitochondria. [DOI] [PubMed] [Google Scholar]
- 65.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PloS Genet. 2009;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]