Abstract
Cryptosporidium skunk genotype is a zoonotic pathogen commonly identified in surface water. Thus far, no subtyping tool exists for characterizing its transmission in humans and animals and transport in environment. In this study, a subtyping tool based on the 60 kDa glycoprotein (gp60) gene previously developed for Cryptosporidium chipmunk genotype I was used in the characterization of Cryptosporidium skunk genotype in animal and storm runoff samples from a watershed in New York. Altogether, 17 positive samples from this watershed and 5 human and animal specimens from other areas were analyzed. We identified 14 subtypes of Cryptosporidium skunk genotype, 11 of which were seen in the watershed. In phylogenetic analysis, these subtypes belonged to 4 subtype families (XVIa, XVIb, XVIc, and XVId). No host-adapted subtypes were identified and the two subtypes in humans were genetically similar to some in raccoons, otters, and storm runoff samples from the watershed. The characteristics of gp60 protein sequences of the Cryptosporidium skunk genotype are similar to those of other Cryptosporidium species, but only its XVIb subtype family has a putative furin cleavage site. This subtyping tool might be useful in characterizing Cryptosporidium skunk genotype in clinical and environmental samples.
Keywords: Cryptosporidium, gp60, Subtyping, Zoonosis, One health, Water
1. Introduction
Cryptosporidiosis is a major waterborne disease in both industrialized and developing countries (Checkley et al., 2015). Cryptosporidium hominis and C. parvum are the leading causes of cryptosporidiosis in humans globally (Ryan et al., 2014; Xiao, 2010). However, some Cryptosporidium species or genotypes from wildlife, including C. ubiquitum, C. cuniculus, Cryptosporidium chipmunk genotype I, and skunk genotype, have been detected in humans in some areas (Davies et al., 2009; Elwin et al., 2012; Feltus et al., 2006; Lebbad et al., 2013; Robinson et al., 2008). Subtyping these unusual species or genotypes would facilitate the tracking of their environmental contamination.
Cryptosporidium skunk genotype was initially isolated from striped skunks in California, USA and was named this way because of the presumed host specificity of most Cryptosporidium spp. (Xiao et al., 2002). It, however, has been subsequently identified in other wild mammals including raccoons, eastern gray squirrels, American red squirrels, fox squirrels, river otters, Virginia opossums, and southern elephant seals (Feng et al., 2007; Rengifo-Herrera et al., 2011; Stenger et al., 2015b; Zhou et al., 2004). Because of its broad host range, Cryptosporidium skunk genotype is common in surface water in the United States and Canada (Jellison et al., 2009; Jiang et al., 2005; Ruecker et al., 2012; Yang et al., 2008), and has been identified in five human cases in the United States and United Kingdom (Davies et al., 2009; Robinson et al., 2008). Currently, there are no subtyping tools for this zoonotic parasite, which makes it difficult to investigate its transmission in humans (Robinson et al., 2008; Xiao, 2010).
Subtyping tools are widely used in the characterization of transmission and environmental transport of C. parvum and C. hominis (Ryan et al., 2014; Xiao, 2010). One common target used in subtyping of these species is the 60-kDa glycoprotein (gp60) gene, as it is highly polymorphic and divides the two Cryptosporidium species into several major subtype families (Strong et al., 2000). In recent years, subtyping tools based on gp60 sequences have been developed for other Cryptosporidium species and genotypes such as C. meleagridis, C. ubiquitum, C. fayeri, C. viatorum, and Cryptosporidium chipmunk genotype I (Guo et al., 2015; Li et al., 2014; Power et al., 2009; Stensvold et al., 2014; Stensvold et al., 2015). With the availability of whole genome sequences, some of the new tools, such as the one for Cryptosporidium chipmunk genotype I, have used conserved nucleotide sequences in PCR primer design (Guo et al., 2015). The latter was shown to be able to detect C. ubiquitum and possibly Cryptosporidium skunk genotype in water samples in addition to Cryptosporidium chipmunk genotype I (Guo et al., 2015). In this study, we have used this tool in subtyping Cryptosporidium skunk genotype present in wildlife, storm runoff and three humans.
2. Materials and methods
2.1. Specimens
Genomic DNA preparations from 22 samples of Cryptosporidium skunk genotype were used in the study, including six from raccoons, one from eastern gray squirrel, and one from river otter in a watershed in New York, USA, nine from storm runoff collected from creeks in the same watershed, two from striped skunks in California, one from a human in Nebraska, and two from humans in the United Kingdom (Table 1). All three human specimens were from patients who sought medical care because of the occurrence of diarrhea. These specimens were identified as positive for Cryptosporidium skunk genotype by PCR and sequence analysis of the small subunit (SSU) rRNA gene (Xiao et al., 2002).
Table 1.
Cryptosporidium specimens used in this study and their subtype identifications based on sequence analysis of the gp60 gene.
| Source | Sample ID | Year of collection | Location of collection | SSU rRNA genotype | gp60 subtype** | Reference |
|---|---|---|---|---|---|---|
| Human | UKSK1 (42694) | 2010 | East Midlands, UK | Skunk genotype | XVIbA16G2b | This study |
| Human | UKSK2 (42693) | 2013 | South West, UK | Skunk genotype | XVIbA16G2b | This study |
| Human | 44,195 | 2016 | Nebraska, USA | Skunk genotype | XVIcA22 | This study |
| Skunk | 1166 | 1999 | California, USA | Skunk genotype | XVIdA29 | Xiao et al., 2002 |
| Skunk | 1170 | 1999 | California, USA | Skunk genotype | XVIdA29 | Xiao et al., 2002 |
| Eastern gray squirrel | 12,376 | 2006 | New York, USA | Skunk genotype | XVIaA14b | Feng et al., 2007 |
| River otter | 13,641 | 2006 | New York, USA | Skunk genotype | XVIcA19 | Feng et al., 2007 |
| Raccoon | 13,468 | 2006 | New York, USA | Skunk genotype | XVIbA18G2 | Feng et al., 2007 |
| Raccoon | 42,589 | 2015 | New York, USA | Skunk genotype | XVIbA16G2a | This study |
| Raccoon | 42,590 | 2015 | New York, USA | Skunk genotype | XVIbA16G2a | This study |
| Raccoon | 39,654 | 2013 | New York, USA | Skunk genotype | XVIcA10 | This study |
| Raccoon | 39,656 | 2013 | New York, USA | Skunk genotype | XVIcA23 | This study |
| Raccoon | 39,657 | 2013 | New York, USA | Skunk genotype | XVIcA23 | This study |
| Runoff | 8060 | 2003 | New York, USA | Skunk genotype | XVIaA14a | Jiang et al., 2005 |
| Runoff | 6017 | 2002 | New York, USA | Skunk genotype | XVIdA19 | Jiang et al., 2005 |
| Runoff | 8649* | 2003 | New York, USA | Skunk genotype | XVIbA14G2, XVIbA16G2a | Jiang et al., 2005 |
| Runoff | 15,081 | 2007 | New York, USA | Skunk genotype | XVIbA17G1 | This study |
| Runoff | 8519 | 2003 | New York, USA | Skunk genotype | XVIaA15 | Jiang et al., 2005 |
| Runoff | 6316 | 2002 | New York, USA | Skunk genotype | XVIbA16G2a | Jiang et al., 2005 |
| Runoff | 8651* | 2003 | New York | Skunk genotype & C. ubiquitum | XIIb, XIId | Jiang et al., 2005 |
| Runoff | 8514* | 2003 | New York | Skunk genotype & C. ubiquitum | XIIb, XIId | Jiang et al., 2005 |
| Runoff | 6858 | 2002 | New York | Skunk genotype & C. ubiquitum | XIIb | Jiang et al., 2005 |
Multiple PCR products from the storm runoff sample produced different subtypes.
Letters “a” and “b” at the end of subtype names are used to distinguish subtypes that have the same number of trinucleotide repeats but nucleotide substitutions in the downstream non-repeat region of the gp60 gene.
2.2. PCR analysis of gp60 gene
The gp60 gene of Cryptosporidium skunk genotype was amplified from each DNA preparation using nested-PCR. The previously described Chip-F1 (5′ TTTACCCACACATCTGTAACGTCG 3′) and Chip-R1 (5′ CCTGTGAGAATATTCTGGAAATTA 3′) and Chip-F2 (5′ ATAGGTAATAATTACTCAGTATTTAAT 3′) and Chip-R2 (5′ TCATCTTAAAACGCTTAAACTCTTAA 3′) primers for Cryptosporidium chipmunk genotype I were used in primary and secondary PCR, respectively (Guo et al., 2015). Duplicate PCR reactions were used in the analysis of DNA from human and animal specimens whereas quintuplicate PCR reactions were used in the analysis of DNA from storm runoff samples. The PCR condition in this study was identical to the one described previously (Guo et al., 2015), except that the annealing temperature in both primary and secondary PCR was changed from 55 °C to 52 °C. The secondary PCR products were analyzed by 1.5% agarose gel electrophoresis.
2.3. Sequence analysis
Positive PCR products were sequenced using the Chip-F2 and Chip-R2 primers in both directions on an ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, CA). Due to the likely presence of multiple Cryptosporidium species or subtypes (Jiang et al., 2005), all PCR products from storm runoff samples were sequenced. The nucleotide sequences obtained were assembled using ChromasPro 1.7.7 (http://technelysium.com.au/wp/chromaspro/), edited using BioEdit 7.04 (www.mbio.ncsu.edu/BioEdit/bioedit.html), and aligned with each other and reference sequences from the GenBank database using ClustalX2.1 (www.clustal.org/). The subtype families and subtypes were named using the established gp60 subtype nomenclature (Ryan et al., 2014).
To assess the uniqueness of the gp60 protein of Cryptosporidium skunk genotype, the nucleotide sequences obtained were translated into amino acid sequences using the program EditSeq in DNASTAR Lasergene 12.3.1 (http://www.dnastar.com/t-dnastar-lasergene.aspx). Signal peptide was predicted using SignalP 4.1 (www.cbs.dtu.dk/services/SignalP/). The C-terminal glycosylphosphatidylinositol (GPI) anchor and transmembrane domain were predicted using the program Protean in DNASTAR Lasergene 12.3.1, PredGPI (http://gpcr2.biocomp.unibo.it/gpipe/pred.htm) and PSORT II (http://psort.hgc.jp/form2.html), respectively. The predictions of potential N-glycosylation and O-glycosylation sites were performed using NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/) and NetOGlyc 4.0 Server (http://www.cbs.dtu.dk/services/NetOGlyc/), respectively. These sequences were also analyzed for the presence of furin cleavage sites using the ProP 1.0 (http://www.cbs.dtu.dk/services/ProP/).
To assess the genetic relationship among gp60 sequences from the Cryptosporidium skunk genotype and other Cryptosporidium species and genotypes, a maximum likelihood tree was constructed using Tamura-Nei evolutionary distances calculated in MEGA 7.0.7 (http://www.megasoftware.net/). DnaSP 5.10 (www.ub.es/dnasp/) was used to calculate recombination rates among various subtype families of Cryptosporidium skunk genotype and chipmunk genotype I.
2.4. 3.4. Nucleotide sequence accession numbers
Nucleotide sequences of the gp60 gene of Cryptosporidium spp. generated in this study were deposited in GenBank under the accession numbers KX698285 to KX698307.
3. Results
3.1. PCR analysis of gp60 gene in Cryptosporidium skunk genotype
Using the previously described primers for Cryptosporidium chipmunk genotype I (Guo et al., 2015), the gp60 gene was efficiently amplified for all 22 DNA samples. For all human and animal specimens, both replicate PCR were positive, compared with one to five replicates positive for storm runoff samples. DNA sequencing results showed that the gp60 sequences from 19 samples were similar to KP099095 from Cryptosporidium skunk genotype, with sequence lengths of 1035–1109 bp. However, sequences generated from the remaining three storm runoff samples were identical to C. ubiquitum XIIb or XIId subtype family (XIIb in one PCR replicate each from samples 8651, 8514 and 6858; and XIId in four PCR replicates from sample 8651 and one replicate from sample 8514).
3.2. Subtypes of Cryptosporidium skunk genotype in humans, wild animals, and storm runoff
Four major types of nucleotide sequences of the gp60 gene were obtained from the Cryptosporidium skunk genotype in this study. These types of sequences differed significantly among each other in the non-repeat regions of the gene, while within each major type sequences differed from each other mostly in the copy number of the TCA or TCG repeats. According to the established gp60 subtype nomenclature, these four major types of Cryptosporidium skunk genotype sequences were named as XVIa, XVIb, XVIc, and XVId subtype families. There were three subtypes within the subtype family XVIa, five in XVIb, four in XVIc, and two in XVId, resulting in 14 subtypes among these 19 samples of the Cryptosporidium skunk genotype. One human case from Nebraska, USA was identified as subtype XVIcA22, and two human cases from the UK were identified as subtype XVIbA16G2b (Table 1). These two subtypes were not seen in wildlife or storm runoff samples in New York.
There were six subtypes of the Cryptosporidium skunk genotype in storm runoff samples from the New York watershed. Similarly, there were six subtypes of the Cryptosporidium skunk genotype in animal specimens from the same watershed. One of the subtypes, XVIbA16G2a, occurred in both storm runoff and animals from the same watershed, and one water sample (No. 8649) had two XVIb subtypes (Table 1). Therefore, there were a total of 11 subtypes of the Cryptosporidium skunk genotype in the watershed. Subtype XVIbA16G2a had one synonymous substitution (T → C at nucleotide 1029 nt) in the non-repeat region compared with subtype XVIbA16G2b.
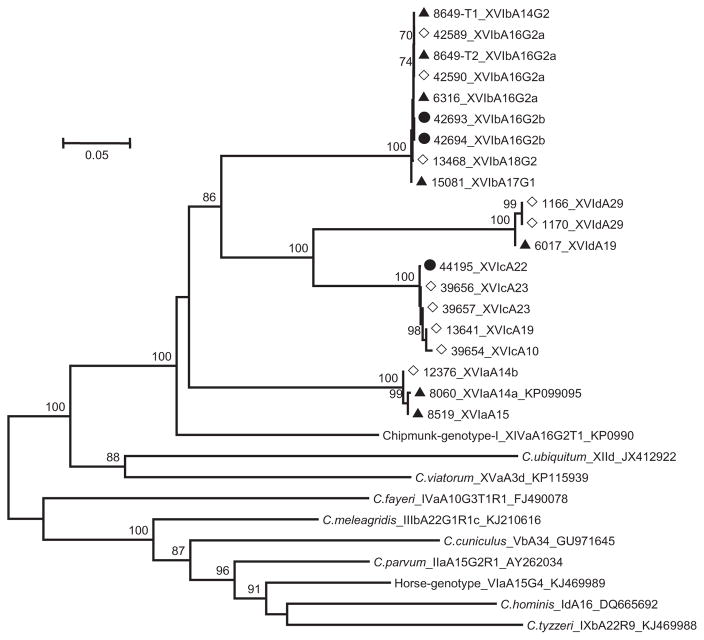
A maximum likelihood tree was constructed with 19 gp60 gene sequences of the Cryptosporidium skunk genotype generated in this study, one Cryptosporidium skunk genotype sequence previously published (KP099095), and GenBank sequences from several other Cryptosporidium species or genotypes (Fig. 1). All 20 sequences of the Cryptosporidium skunk genotype clustered into one large clade that appeared to be a sister to Cryptosporidium chipmunk genotype I. Other Cryptosporidium species or genotypes, in contrast, were more distant (Fig. 1). In agreement with direct sequence comparison, nucleotide sequences of the Cryptosporidium skunk genotype formed four subclades in phylogenetic analysis. The mean genetic distances in nucleotide sequences were 0.232–0.392 substitution/site between subtype families and 0–0.0021 within each subtype family.
Fig. 1.
Phylogenetic relationship among Cryptosporidium skunk genotype and some other published Cryptosporidium species and genotypes in GenBank based on the maximum likelihood analysis of nucleotide sequences of the gp60 gene. Numbers on branches are percent bootstrapping values (> 50) using 1000 replicates. ◇: isolate from wildlife including skunks, raccoons, river otter and eastern gray squirrel; ▲: isolate from storm runoff; ●: isolate from humans.
DnaSP analysis of the gp60 nucleotide sequences revealed the occurrence of 37 recombination events across the gene among four subtype families of Cryptosporidium skunk genotype (XVIa-XVId), but only two recombination events if only XVIa, XVIb, and XVIc sequences were included in the analysis. Pairwise recombination event comparisons revealed the presence of one probable genetic recombination between subtype families XVIb and XVId (Table 2).
Table 2.
Nucleotide sequence similarity (lower triangular matrix) and potential genetic recombination events (upper triangular matrix) among subtype families of Cryptosporidium skunk genotype (XVIa, XVIb, XVIc, and XVId) and chipmunk genotype I (XIVa) at the gp60 locus.
| XVIa | XVIb | XVIc | XVId | |
|---|---|---|---|---|
| XVIa | - | 0 | 0 | 0 |
| XVIb | 77.3% | - | 0 | 1 |
| XVIc | 75.9% | 76.7% | - | 0 |
| XVId | 72.1% | 73.2% | 83.2% | - |
| XIVa | 72.0% | 69.6% | 66.5% | 66.7% |
3.3. Characteristics of gp60 gene of Cryptosporidium skunk genotype
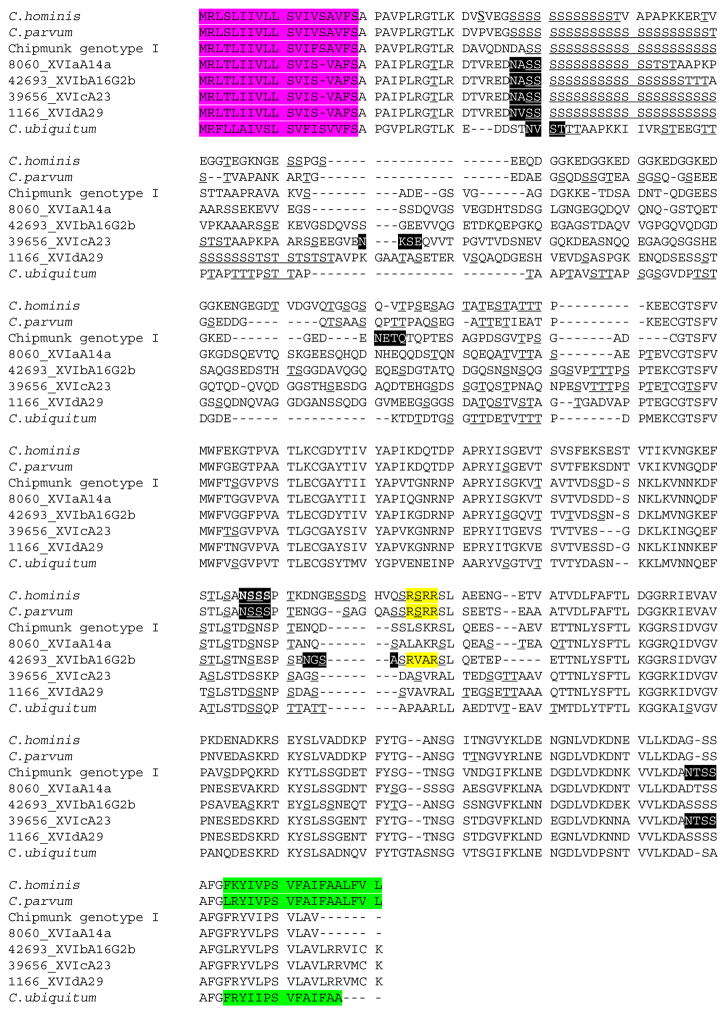
The complete open reading frame (ORF) of the gp60 gene was obtained for 15 of the 19 Cryptosporidium skunk genotype sequences generated in this study. One full sequence of the gp60 from specimen 42,693 is used here to elucidate the characteristics of gp60 gene in Cryptosporidium skunk genotype. The ORF of the gp60 gene consists of 1089 nucleotides, encoding a peptide of 362 amino acids. Except for the 5′ and 3′ regions, the gp60 gene of Cryptosporidium skunk genotype is significantly different from those of C. hominis, C. parvum, C. ubiquitum, chipmunk genotype I and other species or genotypes, with only 40.2% to 72.0% of nucleotide sequence identity. The gp60 protein of Cryptosporidium skunk genotype has a signal peptide in the first 18 amino acids with a cleavage site between amino acids Ser18 and Ala19. Following the signal peptide, the deduced amino acid sequence contains one polyserine track including 18 Ser residues, which were predicted to be O-glycosylation sites. Several other Ser and Thr residues throughout the sequence were predicted to be O-glycosylation sites. Two potential N-glycosylation sites were identified in the gp60 protein sequence. A hydrophobic region is present at the C-terminus of the gp60 protein, and is linked to a GPI anchor. However, no transmembrane domain is present in the gp60 protein sequence. In addition, the predicted furin cleavage site sequence RVAR in the XVIb subtype family is different from the conserved sequence RSRR in C. hominis and C. parvum (Guo et al., 2015; Stensvold et al., 2015). No furin cleavage site was found in the other three subtype families (XVIa, XVIc and XVId) of Cryptosporidium skunk genotype (Fig. 2).
Fig. 2.
Alignment of gp60 protein sequences of Cryptosporidium skunk genotype compared with C. hominis (ACQ82748), C. parvum (AF022929), Cryptosporidium chipmunk genotype I (AJW72309), and C. ubiquitum (AJW72317). The signal peptides in the amino acid sequences are shaded in pink, the transmembrane domains in green, the furin cleavage sites in yellow, potential N-glycosylation sites in black, and O-glycosylation sites are underlined. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In this study, we have shown that gp60 PCR primers we previously designed for Cryptosporidium chipmunk genotype I can efficiently amplify the gp60 gene of Cryptosporidium skunk genotype in water samples and stool specimens. We hypothesized that because Cryptosporidium skunk genotype is genetically related to Cryptosporidium chipmunk genotype I at the SSU rRNA and 70 kDa heat shock protein loci (Feng et al., 2007; Lv et al., 2009; Stenger et al., 2015a), gp60 PCR primers designed for Cryptosporidium chipmunk genotype I could amplify the gp60 gene of Cryptosporidium skunk genotype. In addition, the nested-PCR primers for subtyping Cryptosporidium chipmunk genotype I were designed based on conserved nucleotide sequences flanking the gp60 gene of C. parvum, C. hominis, C. ubiquitum, and Cryptosporidium chipmunk genotype I (Guo et al., 2015). It is well known that because of the extensive sequence differences in the gp60 gene among various Cryptosporidium species and genotypes, PCR primers based on C. parvum and C. hominis sequences alone generally fail to amplify the gp60 gene of other Cryptosporidium species and genotypes that are genetically distant from these two species (Feng et al., 2011; Xiao, 2010). Phylogenetic analysis of gp60 sequences obtained in this study has shown a close genetic relatedness between Cryptosporidium skunk genotype and Cryptosporidium chipmunk genotype I, reinforcing the usefulness of the Cryptosporidium chipmunk genotype I primers in PCR analysis of the gp60 gene of Cryptosporidium skunk genotype.
The detection of C. ubiquitum gp60 sequences in some storm runoff samples in addition to those from Cryptosporidium skunk genotype supports the broad specificity of the gp60 PCR primers used in this study. As storm runoff samples frequently contain mixed Cryptosporidium species/genotypes (Xiao et al., 2006), three of the nine samples analyzed produced C. ubiquitum gp60 sequences, in agreement with the result of SSU rRNA-based PCR-sequencing analysis of the samples. These three runoff samples had 1 and 2, 1 and 4, and 2 and 2 replicates positive for Cryptosporidium skunk genotype and C. ubiquitum in quintuplicate PCR, respectively. Previously, using these Cryptosporidium chipmunk genotype I primers, we were able to amplify the gp60 gene of C. ubiquitum in some storm runoff samples (Guo et al., 2015). In one of the samples analyzed (KP099095), a nucleotide sequence representing a new gp60 subtype family was obtained. As the sample had concurrent presence of Cryptosporidium skunk genotype, it was suggested that the gp60 sequence could be from this parasite (Guo et al., 2015). As a result, a new subtype family name XVIa was designated. Here, we detected similar XVIa sequences in one eastern squirrel specimen and one storm runoff sample that are known to have Cryptosporidium skunk genotype based on SSU rRNA-based PCR-RFLP and sequence analyses (Table 1).
A significant genetic heterogeneity is apparently present in Cryptosporidium skunk genotype. In this study, in addition to XVIa, three other subtype families of gp60 sequences were detected in humans, animals, and storm runoff samples. According to the established gp60 subtype nomenclature, they were named as XVIb, XVIc, and XVId. Genetic recombination, as suggested in other Cryptosporidium species (Li et al., 2014), could play a potential role in the generation of high genetic heterogeneity in Cryptosporidium skunk genotype, as potential genetic events were detected among some of the subtype families in this study. This is especially the case in subtype family XVId, which has very high nucleotide sequence similarity to XVIc (Fig. 1 and Table 2). Recombinant event analysis indicates that it could be a recombinant between XVIc and XVIb subtype families.
Unlike in some Cryptosporidium species such as C. parvum, C. tyzzeri, and C. ubiquitum in which host adaptation exists among subtype families (Kvac et al., 2013; Li et al., 2014; Xiao, 2010), there might be no apparent host-adaptation among subtype families of Cryptosporidium skunk genotype based on the characterization of specimens from several animal species. In fact, all four subtype families described in this study were found in animals and storm runoff in the New York watershed. Altogether, 14 subtypes in these four subtype families were seen in humans, raccoons, skunks, river otter, east gray squirrel and storm runoff. However, most of the animal specimens were from carnivores, which has prevented full assessment of host adaptation. Among the 14 subtypes described in this study, 11 were detected in animal and storm runoff samples from the New York watershed examined. Two subtypes (XVIbA16G2b and XVIcA22) were detected in humans. The XVIb subtype family appears to be the most common one, being detected in humans, various animals, and storm runoff. In particular, its XVIbA16G2b subtype detected in two human cases in the United Kingdom is very similar to the XVIbA16G2a subtype, which was found in raccoons and storm runoff samples in the United States and has only one synonymous nucleotide substitution (T → C) in the gp60 gene. Therefore, at least XVIb and XVIc subtype families of Cryptosporidium skunk genotype could be human pathogens. More extensive characterizations of human and animal specimens are needed to understand host specificity and cross-species transmission of Cryptosporidium skunk genotype.
The close genetic relatedness between Cryptosporidium skunk genotype and Cryptosporidium chipmunk genotype I is interesting. Data obtained thus far indicate that the former is mostly a pathogen of Carnivora (Table 1) whereas the latter is mostly a parasite of Rodentia and Eulipotyphla (Feng et al., 2007; Guo et al., 2015; Kvac et al., 2008; Lv et al., 2009; Song et al., 2015). The occurrence of both in humans as pathogens indicates that both Cryptosporidium genotypes have much broad host ranges than the names have indicated. The current practice in naming new Cryptosporidium genotypes after their initial hosts has some obvious drawbacks. Although there is clear host adaptation among most Cryptosporidium spp., no Cryptosporidium species or genotypes are known to have absolute host specificity and some of them, such as C. parvum and C. ubiquitum, are known to have a broad host range. Designating species names to these Cryptosporidium genotypes upon extensive biologic and genetic characterizations would alleviate some of the confusions.
The gp60 gene of Cryptosporidium skunk genotype has typical characteristics of the gene seen in other Cryptosporidium species, including a signal peptide in the first 18 amino acids, a polyserine track, various O-glycosylation sites and two N-glycosylation sites, and a GPI anchor at the C-terminus. Interestingly, only the XVIb subtype family of this parasite has a furin cleavage site, which is needed for the cleavage of the gp60 precursor protein into gp15 and gp40 by a subtilisin-like protease during the invasion process (Wanyiri et al., 2009). Cryptosporidium chipmunk genotype I also does not have the classic furin cleavage sequence RSRR in its gp60 gene. Instead, the sequence was replaced by LSKR, which may or may not be cleaved by the subtilisin protease (Guo et al., 2015). Neither RSRR nor LSKR sequences are present in the gp60 gene of XVIa, XVIc, and XVId subtype families of Cryptosporidium skunk genotype, indicating that the gp60 protein may be processed differently in these parasites. The gp60 gene of C. ubiquitum also does not appear to have any furin cleavage site (Li et al., 2014). Further studies are needed to examine the significance of the lack of a furin cleavage site in gp60 protein in Cryptosporidium invasion.
In summary, the gp60 primers described previously for Cryptosporidium chipmunk genotype I can be used efficiently in subtyping Cryptosporidium skunk genotype in animal and storm runoff samples from watershed. A preliminary application of this tool in analysis of stool specimens and water samples indicates the presence of heterogeneous populations of the parasite in a small geographic area. Further studies are needed to improve our knowledge on the environmental ecology and transmission of this zoonotic pathogen.
Acknowledgments
We thank the Nebraska Public Health Laboratory for providing one clinical specimen. This study was supported by China Scholarship Council (No. 201208410177), National Natural Science Foundation of China (No. 31001058 and 31425025), and U.S Centers for Disease Control and Prevention.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the U.S. Centers for Disease Control and Prevention.
References
- Checkley W, White AC, Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA, Jr, Priest JW, Roos DS, Striepen B, Thompson RC, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AP, Campbell B, Evans MR, Bone A, Roche A, Chalmers RM. Asymptomatic carriage of protozoan parasites in children in day care centers in the United Kingdom. Pediatr Infect Dis J. 2009;28:838–840. doi: 10.1097/INF.0b013e31819d646d. [DOI] [PubMed] [Google Scholar]
- Elwin K, Hadfield SJ, Robinson G, Chalmers RM. The epidemiology of sporadic human infections with unusual cryptosporidia detected during routine typing in England and Wales, 2000–2008. Epidemiol Infect. 2012;140:673–683. doi: 10.1017/S0950268811000860. [DOI] [PubMed] [Google Scholar]
- Feltus DC, Giddings CW, Schneck BL, Monson T, Warshauer D, McEvoy JM. Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. J Clin Microbiol. 2006;44:4303–4308. doi: 10.1128/JCM.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Alderisio KA, Yang W, Blancero LA, Kuhne WG, Nadareski CA, Reid M, Xiao L. Cryptosporidium genotypes in wildlife from a New York watershed. Appl Environ Microbiol. 2007;73:6475–6483. doi: 10.1128/AEM.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Lal AA, Li N, Xiao L. Subtypes of Cryptosporidium spp. in mice and other small mammals. Exp Parasitol. 2011;127:238–242. doi: 10.1016/j.exppara.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Guo Y, Cebelinski E, Matusevich C, Alderisio KA, Lebbad M, McEvoy J, Roellig DM, Yang C, Feng Y, Xiao L. Subtyping novel zoonotic pathogen Cryptosporidium chipmunk genotype I. J. Clin. Microbiol. 2015;53:1648–1654. doi: 10.1128/JCM.03436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellison KL, Lynch AE, Ziemann JM. Source tracking identifies deer and geese as vectors of human-infectious Cryptosporidium genotypes in an urban/suburban watershed. Environ Sci Technol. 2009;43:4267–4272. doi: 10.1021/es900081m. [DOI] [PubMed] [Google Scholar]
- Jiang J, Alderisio KA, Xiao L. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol. 2005;71:4446–4454. doi: 10.1128/AEM.71.8.4446-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvac M, Hofmannova L, Bertolino S, Wauters L, Tosi G, Modry D. Natural infection with two genotypes of Cryptosporidium in red squirrels (Sciurus vulgaris) in Italy. Folia Parasitol. 2008;55:95–99. [PubMed] [Google Scholar]
- Kvac M, McEvoy J, Loudova M, Stenger B, Sak B, Kvetonova D, Ditrich O, Raskova V, Moriarty E, Rost M, Macholan M, Pialek J. Coevolution of Cryptosporidium tyzzeri and the house mouse (Mus musculus) Int J Parasitol. 2013;43:805–817. doi: 10.1016/j.ijpara.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebbad M, Beser J, Insulander M, Karlsson L, Mattsson JG, Svenungsson B, Axen C. Unusual cryptosporidiosis cases in Swedish patients: extended molecular characterization of Cryptosporidium viatorum and Cryptosporidium chipmunk genotype I. Parasitology. 2013;140:1735–1740. doi: 10.1017/S003118201300084X. [DOI] [PubMed] [Google Scholar]
- Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, Santin M, Fayer R, Kvac M, Ryan U, Sak B, Stanko M, Guo Y, Wang L, Zhang L, Cai J, Roellig D, Feng Y. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg Infect Dis. 2014;20:217–224. doi: 10.3201/eid2002.121797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv C, Zhang L, Wang R, Jian F, Zhang S, Ning C, Wang H, Feng C, Wang X, Ren X, Qi M, Xiao L. Cryptosporidium spp. in wild, laboratory, and pet rodents in China: prevalence and molecular characterization. Appl Environ Microbiol. 2009;75:7692–7699. doi: 10.1128/AEM.01386-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power ML, Cheung-Kwok-Sang C, Slade M, Williamson S. Cryptosporidium fayeri: diversity within the GP60 locus of isolates from different marsupial hosts. Exp Parasitol. 2009;121:219–223. doi: 10.1016/j.exppara.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Rengifo-Herrera C, Ortega-Mora LM, Gomez-Bautista M, Garcia-Moreno FT, Garcia-Parraga D, Castro-Urda J, Pedraza-Diaz S. Detection and characterization of a Cryptosporidium isolate from a southern elephant seal (Mirounga leonina) from the Antarctic peninsula. Appl Environ Microbiol. 2011;77:1524–1527. doi: 10.1128/AEM.01422-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Elwin K, Chalmers RM. Unusual Cryptosporidium genotypes in human cases of diarrhea. Emerg Infect Dis. 2008;14:1800–1802. doi: 10.3201/eid1411.080239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruecker NJ, Matsune JC, Wilkes G, Lapen DR, Topp E, Edge TA, Sensen CW, Xiao L, Neumann NF. Molecular and phylogenetic approaches for assessing sources of Cryptosporidium contamination in water. Water Res. 2012;46:5135–5150. doi: 10.1016/j.watres.2012.06.045. [DOI] [PubMed] [Google Scholar]
- Ryan U, Fayer R, Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014;141:1667–1685. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- Song J, Kim CY, Chang SN, Abdelkader TS, Han J, Kim TH, Oh H, Lee JM, Kim DS, Kim JT, Oh HS, Hur M, Suh JH, Park JH. Detection and molecular characterization of Cryptosporidium spp. from wild rodents and insectivores in South Korea. Korean J Parasitol. 2015;53:737–743. doi: 10.3347/kjp.2015.53.6.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger BL, Clark ME, Kvac M, Khan E, Giddings CW, Dyer NW, Schultz JL, McEvoy JM. Highly divergent 18S rRNA gene paralogs in a Cryptosporidium genotype from eastern chipmunks (Tamias striatus) Infect Genet Evol. 2015a;32:113–123. doi: 10.1016/j.meegid.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger BL, Clark ME, Kvac M, Khan E, Giddings CW, Prediger J, McEvoy JM. North American tree squirrels and ground squirrels with overlapping ranges host different Cryptosporidium species and genotypes. Infect Genet Evol. 2015b;36:287–293. doi: 10.1016/j.meegid.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Stensvold CR, Beser J, Axen C, Lebbad M. High applicability of a novel method for gp60-based subtyping of Cryptosporidium meleagridis. J Clin Microbiol. 2014;52:2311–2319. doi: 10.1128/JCM.00598-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensvold CR, Elwin K, Winiecka-Krusnell J, Chalmers RM, Xiao L, Lebbad M. Development and application of a gp60-based typing assay for Cryptosporidium viatorum. J Clin Microbiol. 2015;53:1891–1897. doi: 10.1128/JCM.00313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong WB, Gut J, Nelson RG. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect Immun. 2000;68:4117–4134. doi: 10.1128/iai.68.7.4117-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanyiri JW, Techasintana P, O’Connor RM, Blackman MJ, Kim K, Ward HD. Role of CpSUB1, a subtilisin-like protease, in Cryptosporidium parvum infection in vitro. Eukaryot Cell. 2009;8:470–477. doi: 10.1128/EC.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Xiao L, Sulaiman IM, Ryan UM, Zhou L, Atwill ER, Tischler ML, Zhang X, Fayer R, Lal AA. Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int J Parasitol. 2002;32:1773–1785. doi: 10.1016/s0020-7519(02)00197-2. [DOI] [PubMed] [Google Scholar]
- Xiao L, Alderisio KA, Jiang J. Detection of Cryptosporidium oocysts in water: effect of the number of samples and analytic replicates on test results. Appl Environ Microbiol. 2006;72:5942–5947. doi: 10.1128/AEM.00927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chen P, Villegas EN, Landy RB, Kanetsky C, Cama V, Dearen T, Schultz CL, Orndorff KG, Prelewicz GJ, Brown MH, Young KR, Xiao L. Cryptosporidium source tracking in the Potomac River watershed. Appl Environ Microbiol. 2008;74:6495–6504. doi: 10.1128/AEM.01345-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Fayer R, Trout JM, Ryan UM, Schaefer FW, 3rd, Xiao L. Genotypes of Cryptosporidium species infecting fur-bearing mammals differ from those of species infecting humans. Appl Environ Microbiol. 2004;70:7574–7577. doi: 10.1128/AEM.70.12.7574-7577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]