Abstract
The genetic code—the language used by cells to translate their genomes into proteins that perform many cellular functions—is highly conserved throughout natural life. Rewriting the genetic code could lead to new biological functions such as expanding protein chemistries with noncanonical amino acids (ncAAs) and genetically isolating synthetic organisms from natural organisms and viruses. It has long been possible to transiently produce proteins bearing ncAAs, but stabilizing an expanded genetic code for sustained function in vivo requires an integrated approach: creating recoded genomes and introducing new translation machinery that function together without compromising viability or clashing with endogenous pathways. In this review, we discuss design considerations and technologies for expanding the genetic code. The knowledge obtained by rewriting the genetic code will deepen our understanding of how genomes are designed and how the canonical genetic code evolved.
Keywords: codon usage, genetic code, orthogonal, synthetic biology, translation engineering
INTRODUCTION
A synthetic organism would revolutionize basic research and biotechnology. Such an entity would have additional protein constituents, noncanonical amino acids (ncAAs) assigned to their own codon in the genetic code. This would be the dream of protein engineers (80); it would allow the design of proteins with novel properties based on the presence of new building blocks in addition to the 20 canonical amino acids (109). Progress along these lines is being made, as codons have been successfully reassigned to encode ncAAs in Escherichia coli (69, 70, 95, 109) and genome synthesis projects aiming at rewriting the genetic codes of E. coli (113, 149), Salmonella typhimurium (71), and yeast (27, 124) are proceeding.
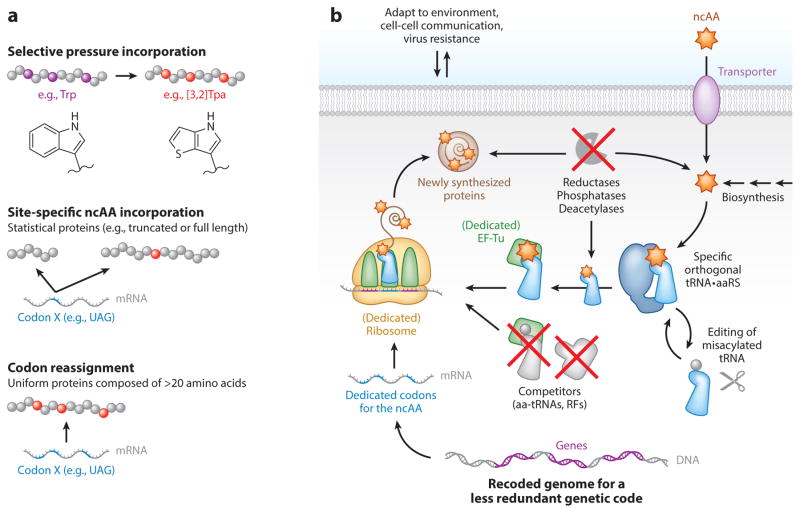
Rewriting the genetic code (Figure 1a, b) involves (a) engineering orthogonal translational components, (b) engineering endogenous translational components, (c) metabolome engineering, (d) massive genome/chromosome engineering for modulating global codon usage, (e) chemical synthesis or biosynthesis of ncAAs, and (f) motivating organisms to maintain the new genetic code and to evolve with it. Traditionally, several methods have been used for incorporating ncAAs into proteins (1) (Figure 1a). The selective pressure incorporation method replaces 1 of the 20 canonical amino acids with its ncAA analog. Expression of a variant of an aminoacyl-tRNA synthetase (aaRS) has facilitated ncAA incorporation into proteins (25). In Bacillus subtilis and E. coli, tryptophan (Trp) has been completely replaced by Trp analogs (such as [3,2]Tpa) in the proteome (1, 47). In contrast, stop or sense codon suppression methods (67, 152) as well as frameshift suppression (151) allow site-specific ncAA incorporation into proteins by using an orthogonal tRNA•aaRS pair specific for the ncAA and the codon (15, 77) (Figure 1b). Thus, the amino acid repertoire of the genetic code is expanded. During stop codon suppression, the stop codon is ambiguously translated as stop or sense (by a ncAA). Similarly, sense codon suppression simultaneously assigns one codon to encode two amino acids, and stochasticity in frameshift suppression results in a mixture of proteins translated in two different frames. All ambiguous decoding methods produce statistical proteins. In contrast, codons can be reassigned (5, 95) to unambiguously encode a ncAA by eliminating native tRNAs or the release factor (RF) originally decoding the codon to be redefined (Figure 1b). To accomplish this, the genomic usage of the codon should be reduced to alleviate the detrimental effects of codon reassignment (70, 112).
Figure 1.
Rewriting the genetic code. (a) Three methods used to augment the genetic code with ncAAs: selective pressure incorporation, site-specific incorporation, and codon reassignment. These methods are not mutually exclusive. (b) A proposed form of an organism having a new genetic code and amino acid repertoire. The ncAA (orange star) is either supplemented in the media and taken up by the cell through a transporter or produced by the cell. Enzymes that degrade the ncAA are inactivated, and an orthogonal aaRS charges the ncAA onto its devoted tRNA. Panel a adapted from Sakamoto (129). Abbreviations: aaRS, aminoacyl-tRNA synthetase; aa-tRNA, aminoacyl-tRNA; mRNA, messenger RNA; ncAA, noncanonical amino acid; RF, release factor; tRNA, transfer RNA.
In this review, we provide an update of recent in vivo genetic code and genome engineering studies in microbes, particularly E. coli, and a comparison with natural cases of noncanonical genetic codes. We focus on ncAAs that resemble canonical amino acids in size and hydrophilicity and may thus be biomolecule-friendly. These ncAAs are, however, often involved in cellular metabolism and posttranslational protein modification, as indicated in Figure 1b. This review does not cover the promising genetic code expansion studies using in vitro protein synthesis (e.g., 126, 137), some of the established applications of in vivo genetic code engineering (15, 57, 77), or the exciting work with supernumerary unnatural base pairs (6, 14).
NATURAL EXPANSION OF THE GENETIC CODE
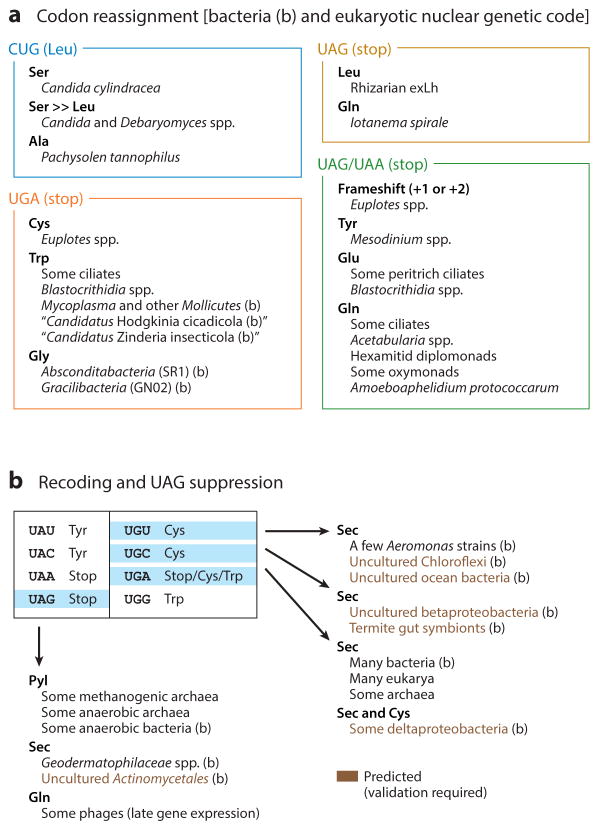
Given the vast amount of new genomic and metagenomic DNA sequence information generated in the past few years, we document here the current knowledge of natural deviations from the standard genetic code (45, 61, 79, 92, 94, 98, 115, 125, 134, 157) (Figure 2) (Supplemental Figure 1). Previously, it was well known that Candida yeast reassigns the CUG codon from leucine (Leu) to serine (Ser) (60) and that many ciliates reassign either the UGA stop codon or both the UAA and the UAG stop codons to a canonical amino acid (12, 61, 88, 119) (Figure 2). A few bacteria reassign the UGA stop codon to either Trp or glycine (11, 76, 155) (Figure 2). The genetic code of mitochondria deviates significantly from the standard genetic code (61, 76) (Supplemental Figure 1), probably because there are only a few genes (encoding membrane proteins) in mitochondrial genomes. It was also known that the UGA stop codon is recoded as selenocysteine (Sec), the twenty-first genetically encoded amino acid, in the three domains of life when guided by Sec-insertion sequence elements (7, 33, 93, 158). In contrast, the UAG stop codon is translated as pyrrolysine (Pyl), the twenty-second genetically encoded amino acid, in some anaerobic archaea and bacteria (35, 93, 158), and as glutamine (Gln) in the late genes of some bacteriophages that kill bacteria whose UGA codon encodes Trp (54) (Figure 2).
Figure 2.
Deviation from the standard genetic code in nature. (a) Codon reassignment occurred in some bacteria and eukaryotes (nuclear genetic code), whereas dual or triple usage of a particular codon, including the assignment of selenocysteine (Sec) and pyrrolysine (Pyl), is found in all three domains of life. (b) Some bacteriophages change the genetic code of their host cells for late gene expression. The full map of codon reassignment (organisms and organelles) can be found in Supplemental Figure 1.
Recent eukaryotic genome/transcriptome analyses identified that (a) Pachysolen tannophilus, a yeast species distinct from Candida spp., reassigns CUG to alanine (Ala; 92, 125) (Figure 2); (b) diverse single-celled eukaryotes that are nonciliate also reassign stop codons to amino acids (17, 19, 59, 62, 63, 115, 157) (Figure 2); (c) not only Gln but also Leu, tyrosine (Tyr), and glutamic acid (Glu) are assigned to UAR [R denotes adenosine (A) and guanine (G)] (45, 134) or to UAG (115) (Figure 2); and (d) ciliates Parduczia sp. and Condylostoma magnum and trypanosomatids Blastocrithidia spp. reassign the three stop codons to amino acids, whereas one or three of the codons are still used as a termination signal at the end of open reading frames (45, 134, 157). In the ciliates Euplotes spp., the default function of the UAR stop codons may be frameshifting (79). Therefore, it was proposed that ciliates may have a special mechanism by which the polypeptide release factor eRF1 is tethered to the poly(A) tail of the mRNA, which may facilitate the context-dependent translation termination in ciliates (10, 79, 134). In contrast, rare distributions of UAG/UAA/UGA sense codons in the ribosomal protein genes of the Blastocrithidia spp. indicated that this alternative genetic code may have a young history (157). Interestingly, ciliates having the UGA cysteine (Cys) or Trp codon still assign Sec with UGA (Figure 2), probably in a context-dependent manner (134, 143).
Recent bacterial genome/metagenome/metatranscriptome analyses identified some bacterial species for which Sec is not assigned by UGA but by UAG and the UGU/UGC Cys codons (94) (Figure 2). In particular, all Geodermatophilaceae (actinobacteria) species sequenced so far use UAG for Sec (Figure 2). In contrast, it was predicted that the UGA stop codon would be recoded as both Sec and Cys in a few Deltaproteobacteria species such as Desulfococcus biacutus (98), although experimental validation is required (Figure 2). Furthermore, a group of potential missense and nonsense suppressor tRNA genes was identified in genome/metagenome/metatranscriptome sequences, derived mostly from Acidobacteria species (98), indicating that ambiguous decoding of a particular codon might be a popular mechanism in bacteria.
Taken together, these data make clear that the genetic code has more flexibility than was assumed at the time the code was determined. The basis for many of these code variations derives from the relative simplicity (i.e., one or two mutations in tRNA) by which tRNA identity may be switched (75, 76). Such codon reassignments also make the organism’s DNA refractive to horizontal gene transfer, a property that may be desirable in certain circumstances.
EXPANDING THE GENETIC CODE WITH ORTHOGONAL TRANSLATION SYSTEMS
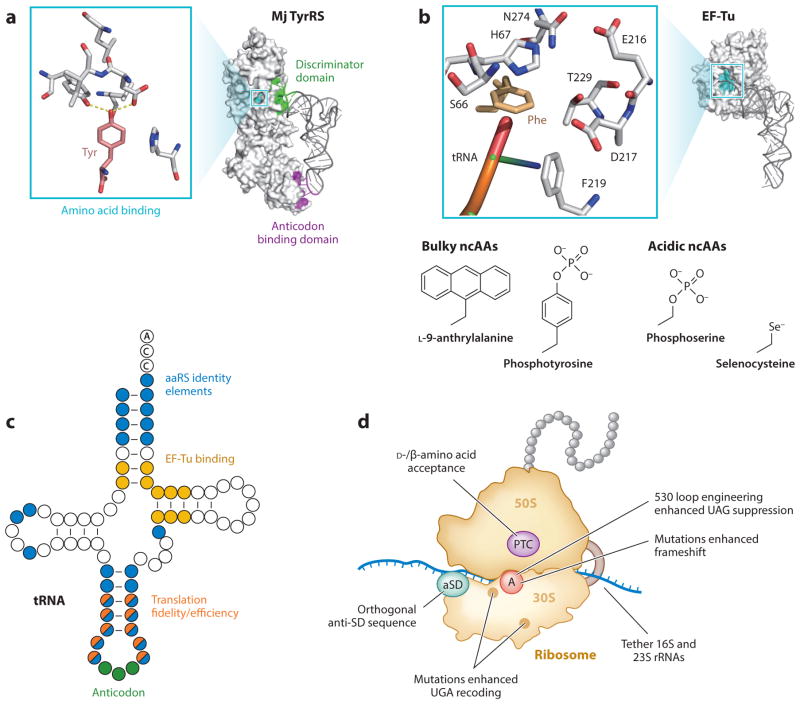
Accurate protein biosynthesis is an immensely complex process involving more than 100 discrete components that must come together to translate proteins with high speed, efficiency, and fidelity. The E. coli ribosome alone is composed of 54 proteins and 3 RNAs, whereas other translation factors include 33 tRNAs, 21 aminoacyl-tRNA synthetases, 3 initiation factors, 3 elongation factors, 2 RFs, and 12 nucleotide-modifying enzymes (34). Efforts to expand the genetic code must introduce new translation components without compromising the function or fidelity of the endogenous translation system or relaxing the endogenous mechanisms of protein quality control. These efforts have predominantly focused on engineering aaRSs, tRNAs, elongation factor EF-Tu, and the ribosome (37) (Figures 1b and 3).
Figure 3.
Engineering the orthogonal translation systems. (a) aaRS engineering with an example of Methanocaldococcus jannaschii (Mj) TyrRS. (b) EF-Tu engineering. The changed residues are shown. Ser66 was modified to alanine to improve azido-phenylalanine recognition (37). (c) tRNA engineering. Colored residues were mutated to change the indicated properties. (d) Ribosome engineering. The PTC, A site, anti-SD sequence, and mutated ribosomal RNA residues are indicated. Abbreviations: aaRS, aminoacyl-tRNA synthetase; anti-SD, anti-Shine-Dalgarno; ncAAs, noncanonical amino acids; PTC, peptidyl transfer center; rRNA, ribosomal RNA; tRNA, transfer RNA; TyrRS, tyrosyl-tRNA synthetase.
Engineering of aaRS•tRNA Pairs
Orthogonal aaRS•tRNA pairs have allowed the introduction—to date—of more than 167 ncAAs into the genetic codes of bacteria, yeast, and animals (26) (Supplemental Table 1). Orthogonality requires that the aaRS•tRNA pair incorporate its cognate amino acid without cross-reacting with other added ncAAs, cellular amino acids, tRNAs, or aaRSs (78, 106). In general, the aaRS specifies an amino acid with its amino acid binding pocket and selects the tRNA species to be charged by recognizing a small number of bases or structural features (identity elements) in the tRNA (39). Although the aaRS enzymes discriminate superbly against any component of the cell’s metabolome, they have no mechanism to reject the many ncAAs that are used by synthetic biologists (31, 106); thus, aaRSs tend to be polyspecific for ncAAs (42). Many aaRS variants used in current work with ncAAs display (in vitro) greater than 100-fold reduced catalytic activity (42, 106). This poor catalytic activity can lead to low ncAA-tRNA levels that are successfully outcompeted by endogenous E. coli tRNAs that engage in near-cognate codon:anticodon interactions (107), with the consequence of inserting an undesired canonical amino acid at the codon of choice. This poor ncAA activation by the aaRS variant needs to be compensated for by overexpression of the orthogonal tRNA•aaRS pair and an elevated presence of the ncAA (42, 122). Clearly, aaRS variants with increased activity and specificity are required for future synthetic biological experiments like the reassignment of multiple codons with different ncAAs. In addition, orthogonal aaRSs equipped with a heterologous editing domain may improve amino acid specificity (110, 123).
Although tRNA identity elements are generally conserved across all three domains of life, several exceptions have been discovered, providing a starting point for evolving orthogonal aaRS•tRNA pairs. Archaeal systems are more likely to be orthogonal in bacteria than eukaryotes, whereas bacterial systems are more likely to be orthogonal in eukaryotes. For example, the tRNATyr•TyrRS pair from the archaeon Methanocaldococcus jannaschii with a few tRNA modifications is orthogonal in E. coli (152), whereas bacterial tRNATyr•TyrRS is orthogonal in eukaryotes (16, 130). The specificity of these aaRS•tRNA pairs can be tuned by generating large libraries of mutations in their amino acid binding pockets or at the residues involved in aaRS•tRNA interactions (Figure 3). Functional variants are identified by performing alternate cycles of positive selections in the presence of a ncAA and negative selections in the absence of a ncAA (77). Recent advances in directed evolution methods (114, 141) such as multiplex automated genome engineering (MAGE; 3, 132) and phage-assisted continuous evolution (29) may accelerate evolution. In addition to the traditional methods that use E. coli and yeast in vivo, positive in vitro selection systems were demonstrated as useful (28, 146), whereas negative selections are still limited to in vivo experiments (83).
Among others, tRNATyr•TyrRS and tRNAPyl•PylRS pairs have been predominantly used for bacterial and mammalian ncAA incorporation (Supplemental Table 1). Likewise, yeast tRNATrp•TrpRS and tRNAPhe•PheRS pairs and archaeal tRNALys•LysRS pairs were used in E. coli, and bacterial tRNALeu•LeuRS and tRNATrp•TrpRS pairs were used in eukaryotes (Supplemental Table 1). tRNATyr•TyrRS, tRNALeu•LeuRS, or tRNAPyl•PylRS systems are also used in gammaproteobacteria including enteropathogenic E. coli, species of Shigella and Salmonella, Yersinia ruckeri, Acinetobacter baylyi, and Pseudomonas syringae; in Synechococcus elongates (cyanobacterium); in gram-positive bacteria, including Mycobacterium tuberculosis, Streptomyces species, and Bacillus cereus; in yeasts, including Saccharomyces cerevisiae, Pichia pastoris, Candida albicans, and Schizosaccharomyces pombe; in the plant Arabidopsis thaliana; and in animals and animal cells (Supplemental Table 1).
Recent studies provided additional systems for use in E. coli: A part of the cysteinyl-tRNACys synthesis machinery of methanogenic archaea was transplanted into E. coli (and Salmonella enterica), so that the intermediate product phosphoseryl-tRNACys (Sep-tRNACys) produced by SepRS could be used for inserting Sep into proteins (Supplemental Table 1). Although Sec naturally requires the dedicated elongation factor SelB (33), two separate tRNAs have been reengineered to incorporate Sec using the more conventional elongation factor EF-Tu (2, 44, 89, 139), which is not dependent on a Sec insertion sequence in the mRNA. Finally, it would be beneficial to develop orthogonal aaRS•tRNA pairs that can function across all domains, like the tRNAPyl•PylRS pair (97). E. coli strains are being developed that replace the endogenous tRNATyr•TyrRS or tRNATrp•TrpRS pairs with heterologous alternatives, thereby allowing the original pairs to be repurposed for ncAA incorporation (50, 52).
Engineering of Elongation Factor EF-Tu and the Ribosome
AaRS•tRNA pairs are not the only crucial translation components for amino acid incorporation. EF-Tu also has amino acid recognition and tends to reject aminoacyl-tRNAs with bulky (23, 32) or negatively charged (44, 72, 116) amino acids. Directed evolution of the elongation factor’s amino acid binding region (Figure 3) made it possible to cotranslationally incorporate Sep (72, 116), Sec (44), and phosphotyrosine (pTyr; 32) into proteins in E. coli.
Meanwhile, ribosomes reject D-amino acids and define how incoming translation factors affect translation. The ribosome provides an intriguing target for accessing new genetically encoded polymers (30, 126). However, the key challenge that has prevented extensive ribosomal engineering has been the inaccessibility of orthogonal ribosomes. For instance, modifying the peptidyl transfer center of the 50S subunit permitted efficient in vitro translation with D-amino acids (20, 21) and β-amino acids (22, 86), but overexpression of the modified ribosome diminished fitness (20, 21). Several advances in the past decade have now provided a starting point for extensive ribosomal engineering (Figure 3). First, replacing the anti–Shine-Dalgarno (aSD) sequence at the 3′ end of the 16S rRNA produced an orthogonal 16S particle that only translates orthogonal mRNAs bearing the complementary synthetic ribosome binding site (48, 121). This in turn enabled evolution of the ribosomal A site in the orthogonal 30S subunit to improve UAG (150) and frameshift (104) suppression and UGA-to-Sec recoding (138). Subsequently, the orthogonal 16S rRNA was tethered to the circularly permuted 23S rRNA so as to link the 30S and 50S ribosomal subunits, thereby facilitating engineering of the peptidyl transfer center in the 50S subunit of orthogonal ribosomes (36, 111). Furthermore, the 23S rRNA has been engineered to recognize a noncanonical tRNA 3′-terminal tail (CGA or GGA instead of CCA) for in vitro translation (136, 137). Combining these methods will enable extensive engineering of ribosomal function in the near future.
SUSTAINED CODON REASSIGNMENT IN VIVO
In addition to reengineering the biochemical translation machinery, in vivo codon reassignment (Figure 1a) poses additional challenges in genetics and implementation. Synthetic biologists have faced two problems (95): (a) Some endogenous components essential for wild-type cell growth must be eliminated to achieve codon reassignment. (b) The codon to be redefined must be translated predominantly by a new decoding molecule (aa-tRNA) to avoid ambiguous assignment of the codon. Here, we summarize the state-of-the-art genome engineering technologies used to change the codon usage of E. coli.
Conditions for Codon Reassignment
How can an essential gene be knocked out? What if an essential component were depleted from the cytosol? The bacterial prfA gene encoding release factor RF1, responsible for termination at UAG (128), has been extensively studied because a partial RF1-deficiency drastically increases suppression of the UAG codon (128). First, the prfA gene was revealed as nonessential in particular genetic backgrounds (53, 55, 95) (Supplemental Table 2). UAG is the minor stop codon in many bacteria, being present in only 7% of all E. coli genes and 2–3% of essential E. coli genes. Furthermore, E. coli tolerates amber suppression to a significant level. RF1 is essential in E. coli K-12 strains that have RF2(Thr246) (128). However, RF1 is less essential in other E. coli strains and bacteria that contain RF2(Ala/Ser246), RF2 proteins with ten times higher activity than that of the K-12 RF2(Thr246) (55, 56, 69, 91, 108, 145), and is totally dispensable in E. coli expressing Salmonella RF2(L167K) (53) or E. coli RF2(T246A/A293E) (56, 69, 108) variants. These studies proved that RF2 partially substitutes for RF1. Furthermore, RF1 is not essential in E. coli cells expressing amber suppressor tRNA and four essential genes with UAG-to-UAA synonymous changes (95). These studies, together with a supporting theory (131), revealed that an essential component can be depleted from the cellular operating system if another component acts as a substitute. This idea was extended to reassign the rare sense codon AGG in E. coli; homoarginyl-tRNACCU partially substituted for arginyl-tRNACCU (99) (Supplemental Table 2) in accordance with the “similar replaces similar” rule (1).
The next question is more important: How is a new, unambiguous codon assignment achieved? Even in the absence of the original decoders, the new decoding factor must outcompete other cellular factors. Paused translation or ribosome stalling allows enough time for near-cognate tRNAs to mistranslate the codon (58, 107). Alternatively, stalled ribosomes on intact or cleaved mRNAs are rescued by three ribosome rescue pathways (46), resulting in the release of truncated proteins or modified proteins with a C-terminal ssrA degradation tag. In the case of UAG codon reassignment (in which the codon is located at the end of the native gene), the most straightforward way to prevent ribosome stalling is to express a strong amber suppressor tRNA (56, 100, 108). Actually, ribosome stalling was detected in an E. coli ΔRF1 strain expressing a weak UAG-decoding tRNAGln (SupE44) but was resolved by expressing a strong UAG-decoding tRNAGln (SupE3; 108). Similarly, near-cognate UAG decoding was observed in the E. coli ΔRF1 strain JX33 expressing RF2(T246A/A293E) and was resolved by efficient cognate UAG decoding (56). In a later study, UAG was eliminated from 95 genes, including the essential genes in E. coli BL21(DE3) with RF2(Ala246), to produce the E. coli B-95.ΔA strain (96). In B-95.ΔA, both near-cognate UAG decoding and ribosome stalling (or UAG decoding by RF2) were observed in the absence of SupE3 (96).
Recoding the Genome
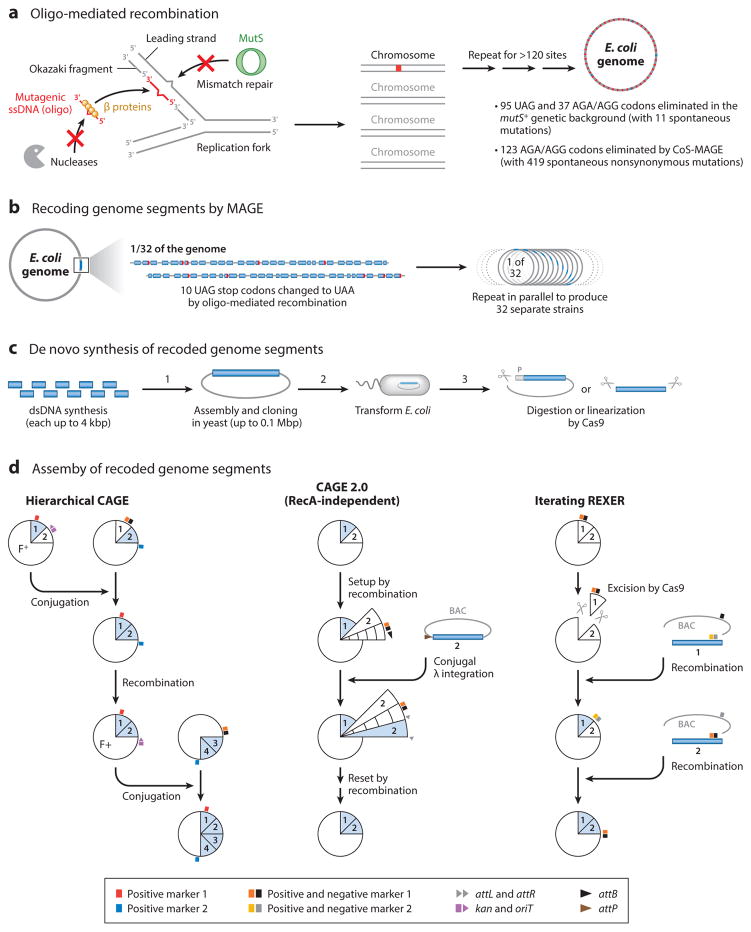
The most promising method of in vivo codon reassignment is to eliminate particular codon assignments throughout the genome (Figure 4a–d). This was achieved in E. coli by using the genome engineering technologies MAGE and conjugative assembly genome engineering (CAGE; 51, 69) (Figure 4a, b, d). In short, the E. coli genome was conceptually split into 32 segments, each containing 10 UAG codons. MAGE was used to produce 32 separate strains, each with all 10 UAG codons in its target segment mutated to synonymous UAA stop codons (Figure 4a, b). CAGE was then used to hierarchically assemble these 32 recoded segments into one fully recoded chromosome (Figure 4d) (51). This genomically recoded organism, or the E. coli strain C321.ΔA (Supplemental Table 2), has no known UAG codon in its genome and lacks RF1 (69). Since the launch of this strain, several derivative strains have been developed for optimized assignments of each ncAA species (Supplemental Table 2). In separate work, coselection-MAGE (CoS-MAGE) was developed for enhancing scarless genome modification (69, 147) and was employed to eliminate all AGA/AGG arginine (Arg) rare codons (123 total) in the essential genes in E. coli (102) (Supplemental Table 2).
Figure 4.
State-of-the-art recombination methods used to engineer the codon usage in the Escherichia coli genome. For serial genome engineering, iterative multiplex oligo-mediated recombination of multiple alleles produced E. coli genomes having more than 120 intended mutations (99, 102) (a). Alternatively, a set of recoded genome segments was prepared by iterating oligo-mediated recombination (MAGE; 51) (b) or by de novo DNA synthesis subjected to assembling in yeast (113, 149) (c). These sets of recoded genome segments were assembled by hierarchical CAGE (51, 69), CAGE 2.0 (105, 113), or iterating REXER (149) (d). The latter two methods are optimized for assembling de novo synthesized segments (50–120 kbp). Hierarchical CAGE was used for assembling 32 segments, each having 10 UAG-to-UAA stop codon changes, into one genome devoid of any UAG stop codon to produce the E. coli C321.ΔA strain (69). Abbreviations: BAC, bacterial artificial chromosome; CAGE, conjugative assembly genome engineering; CoS, coselection; dsDNA, double-stranded DNA; MAGE, multiplex automated genome engineering; REXER, replicon excision for enhanced genome engineering through recombination; ssDNA, single-stranded DNA.
An apparent drawback of MAGE and CoS-MAGE is the accumulation of spontaneous mutations due to deficiency in the methyl-directed mismatch repair (MMR), a mechanism that is necessary to enhance recombination (18). Thus, one to four spontaneous mutations per one intended mutation were detected in the final strains (69, 102). Fortunately, the growth defects caused by the 355 spontaneous mutations and 321 intended mutation in C321.ΔA were largely restored by only six mutations (5 reversion, 1 de novo) to produce C321.ΔA.opt (66) (Supplemental Table 2). In contrast, spontaneous mutation rates dropped significantly in the wild-type MMR-proficient background (to 1 spontaneous per 10 intended; 96, 99) (Figure 4a), whereas recombination efficiencies dropped as well. Transient or local MMR-deficiency can serve as a compromise to boost on-target editing without increasing the spontaneous mutation rate (74, 148). However, the most essential drawback of oligo recombination is that it is currently not practical for generating more than hundreds or thousands of mutations, limiting MAGE to recode only a few rare codons (113).
Two strategies were demonstrated as valid for the de novo synthesis of recoded segments of the E. coli genome (113, 149) (Figure 4c, d). These projects aim to finally create an E. coli genome lacking up to seven codon assignments. In short, de novo synthesized double-stranded DNA fragments were assembled into a 50- to 120-kbp segment in yeast and cloned in an artificial chromosome. After introduction into E. coli, two routes could be used. The first one inserts these chromosomes into the genomic chromosome by λ-integration. In the second approach, the recoded segments are linearized by Cas9 and then inserted by homologous recombination (Figure 4c, d). To avoid aberrant recombination by RecA, λ-integrase–mediated CAGE (CAGE 2.0) was developed (105) (Figure 4d). These recoded segments will then be assembled into a fully recoded genome by iterating these methods. A major challenge will likely be troubleshooting design flaws that cause synthetic lethality as recoded segments are combined. Elimination of the original factors decoding these forbidden codons (68) would make seven codon assignments blank (113). Not only E. coli but also S. typhimurium is remarkably amenable to genome-scale modification (71). Similarly, the synthetic yeast chromosome project is aiming to eliminate the UAG codon (27, 124).
Simple synonymous substitution of a forbidden codon is not always successful, because codon sequence may simultaneously be defined by multiple constraints, including amino acid choice in the gene of interest, amino acid choice in an overlapping gene, DNA motifs affecting replication or transcription, and RNA motifs affecting translation (10, 68, 120) (Supplemental Figure 2). Furthermore, codon bias is involved in the fine-tuning of gene expressions (120). Global change in the codon usage would change the supply and demand of tRNA isoacceptors (24). A comprehensive mutagenesis of the AGA/AGG codons in the essential genes revealed that approximately 10% of these codons reject a simple synonymous replacement with CGU and need troubleshooting (102). Examples of troubleshooting are shown in Supplemental Figure 2. In most cases, these AGA/AGG codons were changed to synonymous Arg codons CGC/CGA/CGG (Supplemental Figure 2a, b). For overlapping genes, insertion of a short sequence allowed safe synonymous replacement by resolving the overlap (99, 113) (Supplemental Figure 2a). However, in a few cases, synonymous substitution was not possible. For example, the AGA codon at position 6 of the repY gene of a ColIb-P9 plasmid (4) was randomized and finally changed to the UUA Leu codon to maintain the plasmid copy number (99) (Supplemental Figure 2c). In another case, the AGG codon of the secE gene comprises the SD sequence for the nusG gene and was instead changed to the GAG Glu codon to maintain the activity of the SD sequence (102). A decrease in the transcription rate of an operon due to multiple codon replacements was compensated for by enhancing the promoter activity (113). Altogether, this knowledge is used to develop algorithms and rules for designing recoded genomes, genome segments, and episomal vectors such as plasmids and phages (81, 99, 113, 149). Oligo-mediated recombination allows one to inspect each case (51, 69, 96, 99, 102), whereas the replacement of genomic regions with recoded segments allows one to explore the feasibility of multiple codon replacements throughout the segments (68, 99, 113, 149). In this way, oligo-mediated recombination and genome synthesis are complementary technologies.
STABILIZING EXPANDED GENETIC CODES
Engineered organisms were traditionally forced to maintain the codon assignment for a ncAA lest the codon assignment be lost (73). Bacterial strains incorporating ncAAs based on the selective pressure incorporation method (Figure 1a) are auxotrophic for the amino acid being substituted (87) and thus are dependent on the substituting ncAA in the absence of the canonical amino acid (47, 65). One interesting example is the B. subtilis HR23 strain that was adapted to use 4-fluorotryptophan and can no longer grow without 4-fluorotryptophan even in the presence of Trp (84, 156), indicating that the functions of some essential proteins became sensitive to this ncAA (156). The same strategy is valid for stabilizing expanded genetic codes (Figure 1).
Reassigning a codon is only half the battle to change the genetic code; the other half is to stabilize the reassigned genetic code. Although tolerance for close analogs of a natural amino acid can be evolved by metabolic supplementation (73), codons can be reassigned to structurally diverse ncAAs only after first replacing all essential instances of the codon with synonymous codons (69, 95). Although incorporation of some amino acids at the remaining original sense codons may be deleterious (100, 107), diverse amino acids tend to be well tolerated (69, 95, 100). This means that the new genetic code remains fragile until cell fitness becomes dependent on the new translation function(s). Although this may occur over time due to natural genetic drift, it was also accomplished by reengineering essential proteins to be dependent on a specific ncAA for proper translation, folding, and function (82, 127). A similar strategy was used to establish ncAA dependence for a conditionally essential β-lactamase gene (antibiotic resistance) in E. coli and other gammaproteobacteria (135).
Aside from codifying the reassigned codon function, this strategy presents a mechanism for bio-containment that could be beneficial for industrial applications of recombinant organisms. First, ncAA dependence prevents escape of the recombinant organism into natural environments where the ncAA is not available. Second, utilizing the reassigned codon throughout the genome presents a firewall against horizontal gene transfer with natural organisms (82), preventing the transfer of functional genes between recombinant and natural populations. This same principle of genetic incompatibility applies to viruses, which depend on their hosts to properly translate the proteins that they need to propagate. Indeed, genomically recoded organism C321.ΔA exhibits increased resistance against multiple natural viruses (69, 81). However, viruses can rapidly evolve to match their host’s genetic code (54). In fact, a T7 bacteriophage mutant has been isolated that exhibits improved fitness as a result of incorporating iodotyrosine in gene 17.5 (43). Achieving true multivirus resistance will require the reassignment of additional codons.
BIOMOLECULE-FRIENDLY NONCANONICAL AMINO ACIDS
Despite the availability of aaRS•tRNA pairs that can incorporate more than 167 ncAAs, only a miniscule fraction of the potential chemical space has been explored. Although these ncAAs offer a diverse selection of functional groups, why are there not more studies that report their utility for improving protein function? The answer is not that the ncAAs lack adequate catalytic functions, as evidenced by Pyl and Sec, which perform functions inaccessible to the canonical 20 amino acids. One barrier may be structural, as the majority of available ncAAs are analogs of Pyl and Tyr (i.e., linked to large, hydrophobic side chains; 26) and do not fit into the functional positions of many natural proteins. If we could incorporate any side chain, what would it be? Smaller amino acids may be more versatile for packing in active sites, and hydrophilic side chains may be beneficial for expanding catalytic functions. However, aaRS design may prove more challenging for such amino acids for two reasons: (a) Smaller amino acids will be more difficult to distinguish from canonical amino acids in an orthogonal aaRS active site. (b) It will be challenging to produce aaRS variants that properly satisfy the hydrogen bonds required to accommodate hydrophilic side chains (99).
Although many ncAAs function only in protein translation, a subset interacts strongly with cellular metabolism. For example, 4-aminophenylalanine (pAF) (85), Pyl, and pyrroline-carboxy-lysine (Pcl) (13, 38) can be synthesized in E. coli with heterologous biosynthetic pathways (13, 38, 85). O-phosphoserine (Sep), O-phosphotyrosine (pTyr; 32), and Nε-acetyllysine (103) are all common posttranslational modifications. These compatibilities with cellular systems, in turn, facilitate degradation and metabolism of free ncAAs, ncAA residues in proteins, and ncAA moieties of ncAA-tRNAs (41, 49, 116) (Figure 1b). Some ncAAs scramble cellular processes (118). However, ncAAs with a posttranslational modification can be cotranslationally incorporated when deacetylases (103) or phosphatases (32, 116) are deleted from the cell or inhibited with an inhibitor (Figure 1b). Furthermore, elimination of tyrosine/aspartate aminotransferases prevented conversion of p-hydroxy-L-phenyllactic acid to tyrosine (41). Depletion of two reductase genes (trxB and gor) prevented the reduction of azido-tyrosine to amino-tyrosine (49). A mutation in the arginine repressor ArgR(L70P) eliminated the toxicity of homoarginine in the E. coli B strain (96, 118). Future systems biology experiments may reveal additional interactions of ncAAs with the metabolome of an organism.
PREPARING FOR RADICALLY ALTERED GENETIC CODES
Protein Engineering Using More Than 21 Building Blocks
Emerging genome engineering technologies and plummeting DNA synthesis prices are now making it possible to rewrite entire genomes, raising interest in creating radically altered genetic codes (8, 113, 149). But why do we want to reassign more than one codon? It may seem a creative exercise to consider applications requiring more than 21 amino acids. Peptide chemists see expanded genetic codes as an opportunity to explore broader conformational and chemical landscapes (8), and the story need not be any different for proteins (80). For example, incorporating multiple bromo/chlorotyrosine residues into redox enzymes has increased enzyme thermostability due to better side chain packing (109). The above-mentioned T7 bacteriophage mutant uses an iodotyrosine residue for quicker propagation (43). Meanwhile, L-(7-hydroxycoumarin-4-yl)ethylglycine improved the activity of a phosphotriesterase beyond its supposed evolutionary limit with canonical amino acids (144). Furthermore, p-acrylamido-phenylalanine has induced a conformational change of an enzyme in a way that the canonical amino acids cannot (154). Covalent bond formation between a ncAA residue and a proximal Cys residue enables irreversible binding of two proteins (153). Directed evolution (140) and protein design (8, 90, 117) will increasingly integrate expanded genetic codes.
Controlling Translational Fidelity
Elimination of several codon assignments does not imply that these blank codons will be assigned to the same number of ncAAs. First, the number will be reduced to approximately half because of wobble pairing by tRNAs. Second, anticodons corresponding to sense codons are often the most important recognition element for aaRSs (39, 64); therefore, the three recoded genome projects are carefully focusing on Ala, Ser, and Leu codons whose cognate aaRS enzymes do not recognize the anticodon as an identity element (71, 113, 149). In addition, base modification of tRNAs is also important for accurate translation and restricted/extended codon decoding by tRNA (40) in not only a static but also a dynamic manner (142). Modified or unmodified U34 in an anticodon could be paired with any nucleotide at the wobble position of codons in the ribosome. In contrast, modified C34 derivatives can be paired with adenine (101, 133). Studies on modification of orthogonal tRNAs have just started (9), but they could provide opportunities to expand the genetic code by splitting anticodons (70).
OUTLOOK
Given the diverse strategies currently used in genetic code engineering, any firm prediction of future breakthroughs would be folly. However, if codon choice were to become limiting in the future, then efforts with quadruplet codons or with codons containing unusual bases would help build on the strategies described in this review. To engineer eukaryotic cells, context-dependent translation termination might be a practical method of stop codon reassignment. Once a variety of hydrophilic ncAAs and unnatural bases are confirmed to be suitable for cell engineering, they will be used to further expand the genetic code by developing new methods of RNA and amino acid recognition. Certainly, in vitro studies will provide the groundwork for successful in vivo systems.
Supplementary Material
Acknowledgments
Work in the authors’ laboratory was supported by National Institue of General Medical Sciences grants R01 GM22854-42 and R35 GM122560, and by a grant (DE-FG02-98ER20311) from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the Department of Energy. T.M. is a Japan Society for the Promotion of Science postdoctoral fellow for research abroad. M.J.L. is supported by a Cancer Research Institute Irvington Postdoctoral Fellowship and a Washington Research Foundation Innovation Postdoctoral Fellowship.
Footnotes
DISCLOSURE STATEMENT
M.J.L. has a financial interest in GRO Biosciences and is coinventor on patents related to genomically recoded organisms.
LITERATURE CITED
- 1.Acevedo-Rocha CG, Budisa N. Xenomicrobiology: a roadmap for genetic code engineering. Microb Biotechnol. 2016;9:666–76. doi: 10.1111/1751-7915.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldag C, Bröcker MJ, Hohn MJ, Prat L, Hammond G, et al. Rewiring translation for elongation factor Tu-dependent selenocysteine incorporation. Angew Chem Int Ed Engl. 2013;52:1441–45. doi: 10.1002/anie.201207567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amiram M, Haimovich AD, Fan C, Wang Y-S, Aerni H-R, et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat Biotechnol. 2015;33:1272–79. doi: 10.1038/nbt.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano K, Mizobuchi K. Copy number control of IncIαplasmid ColIb-P9 by competition between pseudoknot formation and antisense RNA binding at a specific RNA site. EMBO J. 1998;17:5201–13. doi: 10.1093/emboj/17.17.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkins JF, Baranov PV. The distinction between recoding and codon reassignment. Genetics. 2010;185:1535–36. doi: 10.1534/genetics.110.119016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benner SA, Karalkar NB, Hoshika S, Laos R, Shaw RW, et al. Alternative Watson–Crick synthetic genetic systems. Cold Spring Harb Perspect Biol. 2016;8:a023770. doi: 10.1101/cshperspect.a023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, et al. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–76. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 8.Bhardwaj G, Mulligan VK, Bahl CD, Gilmore JM, Harvey PJ, et al. Accurate de novo design of hyperstable constrained peptides. Nature. 2016;538:329–35. doi: 10.1038/nature19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biddle W, Schmitt MA, Fisk JD. Modification of orthogonal tRNAs: unexpected consequences for sense codon reassignment. Nucleic Acids Res. 2016;44:10042–50. doi: 10.1093/nar/gkw948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brar GA. Beyond the triplet code: Context cues transform translation. Cell. 2016;167:1681–92. doi: 10.1016/j.cell.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell JH, O’Donoghue P, Campbell AG, Schwientek P, Sczyrba A, et al. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. PNAS. 2013;110:5540–45. doi: 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caron F, Meyer E. Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature. 1985;314:185–88. doi: 10.1038/314185a0. [DOI] [PubMed] [Google Scholar]
- 13.Cellitti SE, Ou W, Chiu HP, Grünewald J, Jones DH, et al. D-Ornithine coopts pyrrolysine biosynthesis to make and insert pyrroline-carboxy-lysine. Nat Chem Biol. 2011;7:528–30. doi: 10.1038/nchembio.586. [DOI] [PubMed] [Google Scholar]
- 14.Chen T, Hongdilokkul N, Liu Z, Thirunavukarasu D, Romesberg FE. The expanding world of DNA and RNA. Curr Opin Chem Biol. 2016;34:80–87. doi: 10.1016/j.cbpa.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 16.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–67. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 17.Cocquyt E, Gile GH, Leliaert F, Verbruggen H, Keeling PJ, De Clerck O. Complex phylogenetic distribution of a non-canonical genetic code in green algae. BMC Evol Biol. 2010;10:327. doi: 10.1186/1471-2148-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costantino N, Court DL. Enhanced levels of λ Red-mediated recombinants in mismatch repair mutants. PNAS. 2003;100:15748–53. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Koning AP, Noble GP, Heiss AA, Wong J, Keeling PJ. Environmental PCR survey to determine the distribution of a non-canonical genetic code in uncultivable oxymonads. Environ Microbiol. 2008;10:65–74. doi: 10.1111/j.1462-2920.2007.01430.x. [DOI] [PubMed] [Google Scholar]
- 20.Dedkova LM, Fahmi NE, Golovine SY, Hecht SM. Enhanced D-amino acid incorporation into protein by modified ribosomes. J Am Chem Soc. 2003;125:6616–17. doi: 10.1021/ja035141q. [DOI] [PubMed] [Google Scholar]
- 21.Dedkova LM, Fahmi NE, Golovine SY, Hecht SM. Construction of modified ribosomes for incorporation of D-amino acids into proteins. Biochemistry. 2006;45:15541–51. doi: 10.1021/bi060986a. [DOI] [PubMed] [Google Scholar]
- 22.Dedkova LM, Fahmi NE, Paul R, del Rosario M, Zhang L, et al. β-Puromycin selection of modified ribosomes for in vitro incorporation of β-amino acids. Biochemistry. 2012;51:401–15. doi: 10.1021/bi2016124. [DOI] [PubMed] [Google Scholar]
- 23.Doi Y, Ohtsuki T, Shimizu Y, Ueda T, Sisido M. Elongation factor Tu mutants expand amino acid tolerance of protein biosynthesis system. J Am Chem Soc. 2007;129:14458–62. doi: 10.1021/ja075557u. [DOI] [PubMed] [Google Scholar]
- 24.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–63. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 25.Döring V, Mootz HD, Nangle LA, Hendrickson TL, de Crécy-Lagard V, et al. Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science. 2001;292:501–4. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- 26.Dumas A, Lercher L, Spicer CD, Davis BG. Designing logical codon reassignment—expanding the chemistry in biology. Chem Sci. 2015;6:50–69. doi: 10.1039/c4sc01534g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dymond JS, Richardson SM, Coombes CE, Babatz T, Müller H, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–76. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellefson JW, Meyer AJ, Hughes RA, Cannon JR, Brodbelt JS, Ellington AD. Directed evolution of genetic parts and circuits by compartmentalized partnered replication. Nat Biotechnol. 2014;32:97–101. doi: 10.1038/nbt.2714. [DOI] [PubMed] [Google Scholar]
- 29.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fahnestock S, Rich A. Ribosome-catalyzed polyester formation. Science. 1971;173:340–43. doi: 10.1126/science.173.3994.340. [DOI] [PubMed] [Google Scholar]
- 31.Fan C, Ho JM, Chirathivat N, Söll D, Wang YS. Exploring the substrate range of wild-type aminoacyl-tRNA synthetases. ChemBioChem. 2014;15:1805–9. doi: 10.1002/cbic.201402083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan C, Ip K, Söll D. Expanding the genetic code of Escherichia coli with phosphotyrosine. FEBS Lett. 2016;590:3040–47. doi: 10.1002/1873-3468.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer N, Neumann P, Bock LV, Maracci C, Wang Z, et al. The pathway to GTPase activation of elongation factor SelB on the ribosome. Nature. 2016;540:80–85. doi: 10.1038/nature20560. [DOI] [PubMed] [Google Scholar]
- 34.Forster AC, Church GM. Towards synthesis of a minimal cell. Mol Syst Biol. 2006;2:45. doi: 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fournier GP, Andam CP, Gogarten JP. Ancient horizontal gene transfer and the last common ancestors. BMC Evol Biol. 2015;15:70. doi: 10.1186/s12862-015-0350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fried SD, Schmied WH, Uttamapinant C, Chin JW. Ribosome subunit stapling for orthogonal translation in E. coli. Angew Chem Int Ed Engl. 2015;54:12791–94. doi: 10.1002/anie.201506311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gan R, Perez JG, Carlson ED, Ntai I, Isaacs FJ, et al. Translation system engineering in Escherichia coli enhances non-canonical amino acid incorporation into proteins. Biotechnol Bioeng. 2016;114:1074–86. doi: 10.1002/bit.26239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaston MA, Zhang L, Green-Church KB, Krzycki JA. The complete biosynthesis of the genetically encoded amino acid pyrrolysine from lysine. Nature. 2011;471:647–50. doi: 10.1038/nature09918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–35. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grosjean H, Westhof E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016;44:8020–40. doi: 10.1093/nar/gkw608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo J, Wang J, Anderson JC, Schultz PG. Addition of an α-hydroxy acid to the genetic code of bacteria. Angew Chem Int Ed Engl. 2008;47:722–25. doi: 10.1002/anie.200704074. [DOI] [PubMed] [Google Scholar]
- 42.Guo LT, Wang YS, Nakamura A, Eiler D, Kavran JM, et al. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. PNAS. 2014;111:16724–29. doi: 10.1073/pnas.1419737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammerling MJ, Ellefson JW, Boutz DR, Marcotte EM, Ellington AD, Barrick JE. Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat Chem Biol. 2014;10:178–80. doi: 10.1038/nchembio.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haruna K, Alkazemi MH, Liu Y, Söll D, Englert M. Engineering the elongation factor Tu for efficient selenoprotein synthesis. Nucleic Acids Res. 2014;42:9976–83. doi: 10.1093/nar/gku691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heaphy SM, Mariotti M, Gladyshev VN, Atkins JF, Baranov PV. Novel ciliate genetic code variants including the reassignment of all three stop codons to sense codons in Condylostoma magnum. Mol Biol Evol. 2016;33:2885–89. doi: 10.1093/molbev/msw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Himeno H, Nameki N, Kurita D, Muto A, Abo T. Ribosome rescue systems in bacteria. Biochimie. 2015;114:102–12. doi: 10.1016/j.biochi.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 47.Hoesl MG, Oehm S, Durkin P, Darmon E, Peil L, et al. Chemical evolution of a bacterial proteome. Angew Chem Int Ed Engl. 2015;54:10030–34. doi: 10.1002/anie.201502868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hui AS, Eaton DH, de Boer HA. Mutagenesis at the mRNA decoding site in the 16S ribosomal RNA using the specialized ribosome system in Escherichia coli. EMBO J. 1988;7:4383–88. doi: 10.1002/j.1460-2075.1988.tb03337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikeda-Boku A, Ohno S, Hibino Y, Yokogawa T, Hayashi N, Nishikawa K. A simple system for expression of proteins containing 3-azidotyrosine at a pre-determined site in Escherichia coli. J Biochem. 2013;153:317–26. doi: 10.1093/jb/mvs153. [DOI] [PubMed] [Google Scholar]
- 50.Iraha F, Oki K, Kobayashi T, Ohno S, Yokogawa T, et al. Functional replacement of the endogenous tyrosyl-tRNA synthetase-tRNATyr pair by the archaeal tyrosine pair in Escherichia coli for genetic code expansion. Nucleic Acids Res. 2010;38:3682–91. doi: 10.1093/nar/gkq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–53. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Italia JS, Addy PS, Wrobel CJJ, Crawford CC, Lajoie MJ, et al. An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nat Chem Biol. 2017;13:446–50. doi: 10.1038/nchembio.2312. [DOI] [PubMed] [Google Scholar]
- 53.Ito K, Uno M, Nakamura Y. Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. PNAS. 1998;95:8165–69. doi: 10.1073/pnas.95.14.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivanova NN, Schwientek P, Tripp HJ, Rinke C, Pati A, et al. Stop codon reassignments in the wild. Science. 2014;344:909–13. doi: 10.1126/science.1250691. [DOI] [PubMed] [Google Scholar]
- 55.Johnson DB, Wang C, Xu J, Schultz MD, Schmitz RJ, et al. Release factor one is nonessential in Escherichia coli. ACS Chem Biol. 2012;7:1337–44. doi: 10.1021/cb300229q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson DB, Xu J, Shen Z, Takimoto JK, Schultz MD, et al. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol. 2011;7:779–86. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson JA, Lu YY, Van Deventer JA, Tirrell DA. Residue-specific incorporation of non-canonical amino acids into proteins: recent developments and applications. Curr Opin Chem Biol. 2010;14:774–80. doi: 10.1016/j.cbpa.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 59.Karpov SA, Mikhailov KV, Mirzaeva GS, Mirabdullaev IM, Mamkaeva KA, et al. Obligately phagotrophic aphelids turned out to branch with the earliest-diverging fungi. Protist. 2013;164:195–205. doi: 10.1016/j.protis.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 60.Kawaguchi Y, Honda H, Taniguchi-Morimura J, Iwasaki S. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature. 1989;341:164–66. doi: 10.1038/341164a0. [DOI] [PubMed] [Google Scholar]
- 61.Keeling PJ. Genomics: evolution of the genetic code. Curr Biol. 2016;26:R851–53. doi: 10.1016/j.cub.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 62.Keeling PJ, Doolittle WF. Widespread and ancient distribution of a noncanonical genetic code in diplomonads. Mol Biol Evol. 1997;14:895–901. doi: 10.1093/oxfordjournals.molbev.a025832. [DOI] [PubMed] [Google Scholar]
- 63.Kollmar M, Mühlhausen S. Nuclear codon reassignments in the genomics era and mechanisms behind their evolution. BioEssays. 2017;39:1600221. doi: 10.1002/bies.201600221. [DOI] [PubMed] [Google Scholar]
- 64.Krishnakumar R, Prat L, Aerni HR, Ling J, Merryman C, et al. Transfer RNA misidentification scrambles sense codon recoding. ChemBioChem. 2013;14:1967–72. doi: 10.1002/cbic.201300444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuthning A, Durkin P, Oehm S, Hoesl MG, Budisa N, Sussmuth RD. Towards biocontained cell factories: An evolutionarily adapted Escherichia coli strain produces a new-to-nature bioactive lantibiotic containing thienopyrrole-alanine. Sci Rep. 2016;6:33447. doi: 10.1038/srep33447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuznetsov G, Goodman DB, Filsinger GT, Landon M, Rohland N, et al. Optimizing complex phenotypes through model-guided multiplex genome engineering. Genome Biol. 2017;18:100. doi: 10.1186/s13059-017-1217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwon I, Kirshenbaum K, Tirrell DA. Breaking the degeneracy of the genetic code. J Am Chem Soc. 2003;125:7512–13. doi: 10.1021/ja0350076. [DOI] [PubMed] [Google Scholar]
- 68.Lajoie MJ, Kosuri S, Mosberg JA, Gregg CJ, Zhang D, Church GM. Probing the limits of genetic recoding in essential genes. Science. 2013;342:361–63. doi: 10.1126/science.1241460. [DOI] [PubMed] [Google Scholar]
- 69.Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–60. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lajoie MJ, Söll D, Church GM. Overcoming challenges in engineering the genetic code. J Mol Biol. 2016;428:1004–21. doi: 10.1016/j.jmb.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lau YH, Stirling F, Kuo J, Karrenbelt MAP, Chan YA, et al. Large-scale recoding of a bacterial genome by iterative recombineering of synthetic DNA. Nucleic Acids Res. 2017;45:6971–80. doi: 10.1093/nar/gkx415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee S, Oh S, Yang A, Kim J, Söll D, et al. A facile strategy for selective incorporation of phosphoserine into histones. Angew Chem Int Ed Engl. 2013;52:5771–75. doi: 10.1002/anie.201300531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lemeignan B, Sonigo P, Marlière P. Phenotypic suppression by incorporation of an alien amino acid. J Mol Biol. 1993;231:161–66. doi: 10.1006/jmbi.1993.1269. [DOI] [PubMed] [Google Scholar]
- 74.Lennen RM, Wallin AI, Pedersen M, Bonde M, Luo H, et al. Transient overexpression of DNA adenine methylase enables efficient and mobile genome engineering with reduced off-target effects. Nucleic Acids Res. 2016;44:e36. doi: 10.1093/nar/gkv1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ling J, Daoud R, Lajoie MJ, Church GM, Söll D, Lang BF. Natural reassignment of CUU and CUA sense codons to alanine in Ashbya mitochondria. Nucleic Acids Res. 2014;42:499–508. doi: 10.1093/nar/gkt842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ling J, O’Donoghue P, Söll D. Genetic code flexibility in microorganisms: novel mechanisms and impact on physiology. Nat Rev Microbiol. 2015;13:707–21. doi: 10.1038/nrmicro3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–44. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 78.Liu DR, Magliery TJ, Pastrnak M, Schultz PG. Engineering a tRNA and aminoacyl-tRNA synthetase for the site-specific incorporation of unnatural amino acids into proteins in vivo. PNAS. 1997;94:10092–97. doi: 10.1073/pnas.94.19.10092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lobanov AV, Heaphy SM, Turanov AA, Gerashchenko MV, Pucciarelli S, et al. Position-dependent termination and widespread obligatory frameshifting in Euplotes translation. Nat Struct Mol Biol. 2017;24:61–68. doi: 10.1038/nsmb.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu C. Imaginary proteins made real. Cell. 2016;164:7. [Google Scholar]
- 81.Ma NJ, Isaacs FJ. Genomic recoding broadly obstructs the propagation of horizontally transferred genetic elements. Cell Syst. 2016;3:199–207. doi: 10.1016/j.cels.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maranhao AC, Ellington AD. Evolving orthogonal suppressor tRNAs to incorporate modified amino acids. ACS Synth Biol. 2016;6:108–19. doi: 10.1021/acssynbio.6b00145. [DOI] [PubMed] [Google Scholar]
- 84.Mat WK, Xue H, Wong JT. Genetic code mutations: the breaking of a three billion year invariance. PLOS ONE. 2010;5:e12206. doi: 10.1371/journal.pone.0012206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, et al. Generation of a bacterium with a 21 amino acid genetic code. J Am Chem Soc. 2003;125:935–39. doi: 10.1021/ja0284153. [DOI] [PubMed] [Google Scholar]
- 86.Melo Czekster C, Robertson WE, Walker AS, Söll D, Schepartz A. In vivo biosynthesis of a β-amino acid-containing protein. J Am Chem Soc. 2016;138:5194–97. doi: 10.1021/jacs.6b01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Merkel L, Schauer M, Antranikian G, Budisa N. Parallel incorporation of different fluorinated amino acids: on the way to “Teflon” proteins. ChemBioChem. 2010;11:1505–7. doi: 10.1002/cbic.201000295. [DOI] [PubMed] [Google Scholar]
- 88.Meyer F, Schmidt HJ, Plümper E, Hasilik A, Mersmann G, et al. UGA is translated as cysteine in pheromone 3 of Euplotes octocarinatus. PNAS. 1991;88:3758–61. doi: 10.1073/pnas.88.9.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miller C, Bröcker MJ, Prat L, Ip K, Chirathivat N, et al. A synthetic tRNA for EF-Tu mediated selenocysteine incorporation in vivo and in vitro. FEBS Lett. 2015;589:2194–99. doi: 10.1016/j.febslet.2015.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mills JH, Khare SD, Bolduc JM, Forouhar F, Mulligan VK, et al. Computational design of an unnatural amino acid dependent metalloprotein with atomic level accuracy. J Am Chem Soc. 2013;135:13393–99. doi: 10.1021/ja403503m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mora L, Heurgué-Hamard V, de Zamaroczy M, Kervestin S, Buckingham RH. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J Biol Chem. 2007;282:35638–45. doi: 10.1074/jbc.M706076200. [DOI] [PubMed] [Google Scholar]
- 92.Mühlhausen S, Findeisen P, Plessmann U, Urlaub H, Kollmar M. A novel nuclear genetic code alteration in yeasts and the evolution of codon reassignment in eukaryotes. Genome Res. 2016;26:945–55. doi: 10.1101/gr.200931.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mukai T, Crnković A, Umehara T, Ivanova NN, Kyrpides NC, Söll D. RNA-dependent cysteine biosynthesis in bacteria and archaea. mBio. 2017;8:e00561–17. doi: 10.1128/mBio.00561-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mukai T, Englert M, Tripp HJ, Miller C, Ivanova NN, et al. Facile recoding of selenocysteine in nature. Angew Chem Int Ed Engl. 2016;55:5337–41. doi: 10.1002/anie.201511657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mukai T, Hayashi A, Iraha F, Sato A, Ohtake K, et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010;38:8188–95. doi: 10.1093/nar/gkq707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mukai T, Hoshi H, Ohtake K, Takahashi M, Yamaguchi A, et al. Highly reproductive Escherichia coli cells with no specific assignment to the UAG codon. Sci Rep. 2015;5:9699. doi: 10.1038/srep09699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Adding L-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem Biophys Res Commun. 2008;371:818–22. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 98.Mukai T, Vargas-Rodriguez O, Englert M, Tripp HJ, Ivanova NN, et al. Transfer RNAs with novel cloverleaf structures. Nucleic Acids Res. 2016;45:2776–85. doi: 10.1093/nar/gkw898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mukai T, Yamaguchi A, Ohtake K, Takahashi M, Hayashi A, et al. Reassignment of a rare sense codon to a non-canonical amino acid in Escherichia coli. Nucleic Acids Res. 2015;43:8111–22. doi: 10.1093/nar/gkv787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mukai T, Yanagisawa T, Ohtake K, Wakamori M, Adachi J, et al. Genetic-code evolution for protein synthesis with non-natural amino acids. Biochem Biophys Res Commun. 2011;411:757–61. doi: 10.1016/j.bbrc.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 101.Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNAMet. Nat Chem Biol. 2016;12:546–51. doi: 10.1038/nchembio.2099. [DOI] [PubMed] [Google Scholar]
- 102.Napolitano MG, Landon M, Gregg CJ, Lajoie MJ, Govindarajan L, et al. Emergent rules for codon choice elucidated by editing rare arginine codons in Escherichia coli. PNAS. 2016;113:E5588–97. doi: 10.1073/pnas.1605856113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding Nε-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–34. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 104.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–44. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 105.Norville JE, Gardner CL, Aponte E, Camplisson CK, Gonzales A, et al. Assembly of radically recoded E. coli genome segments. 2016 bioRxiv 070417 https://doi.org/10.1101/070417.
- 106.O’Donoghue P, Ling J, Wang YS, Söll D. Upgrading protein synthesis for synthetic biology. Nat Chem Biol. 2013;9:594–98. doi: 10.1038/nchembio.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.O’Donoghue P, Prat L, Heinemann IU, Ling J, Odoi K, et al. Near-cognate suppression of amber, opal and quadruplet codons competes with aminoacyl-tRNAPyl for genetic code expansion. FEBS Lett. 2012;586:3931–37. doi: 10.1016/j.febslet.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ohtake K, Sato A, Mukai T, Hino N, Yokoyama S, Sakamoto K. Efficient decoding of the UAG triplet as a full-fledged sense codon enhances the growth of a prfA-deficient strain of Escherichia coli. J Bacteriol. 2012;194:2606–13. doi: 10.1128/JB.00195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ohtake K, Yamaguchi A, Mukai T, Kashimura H, Hirano N, et al. Protein stabilization utilizing a redefined codon. Sci Rep. 2015;5:9762. doi: 10.1038/srep09762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oki K, Sakamoto K, Kobayashi T, Sasaki HM, Yokoyama S. Transplantation of a tyrosine editing domain into a tyrosyl-tRNA synthetase variant enhances its specificity for a tyrosine analog. PNAS. 2008;105:13298–303. doi: 10.1073/pnas.0803531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Orelle C, Carlson ED, Szal T, Florin T, Jewett MC, Mankin AS. Protein synthesis by ribosomes with tethered subunits. Nature. 2015;524:119–24. doi: 10.1038/nature14862. [DOI] [PubMed] [Google Scholar]
- 112.Osawa S, Jukes TH, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992;56:229–64. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ostrov N, Landon M, Guell M, Kuznetsov G, Teramoto J, et al. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819–22. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- 114.Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16:379–94. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 115.Pánek T, Žihala D, Sokol M, Derelle R, Klimeš V, et al. Nuclear genetic codes with a different meaning of the UAG and the UAA codon. BMC Biol. 2017;15:8. doi: 10.1186/s12915-017-0353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–54. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pearson AD, Mills JH, Song Y, Nasertorabi F, Han GW, et al. Trapping a transition state in a computationally designed protein bottle. Science. 2015;347:863–67. doi: 10.1126/science.aaa2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Peyru GM, Maas WK. Inhibition of Escherichia coli B by homoarginine. J Bacteriol. 1967;94:712–18. doi: 10.1128/jb.94.3.712-718.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Preer JR, Jr, Preer LB, Rudman BM, Barnett AJ. Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature. 1985;314:188–90. doi: 10.1038/314188a0. [DOI] [PubMed] [Google Scholar]
- 120.Quax TE, Claassens NJ, Söll D, van der Oost J. Codon bias as a means to fine-tune gene expression. Mol Cell. 2015;59:149–61. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rackham O, Chin JW. A network of orthogonal ribosome•mRNA pairs. Nat Chem Biol. 2005;1:159–66. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- 122.Rauch BJ, Porter JJ, Mehl RA, Perona JJ. Improved incorporation of noncanonical amino acids by an engineered tRNATyr suppressor. Biochemistry. 2016;55:618–28. doi: 10.1021/acs.biochem.5b01185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Richardson CJ, First EA. Hyperactive editing domain variants switch the stereospecificity of tyrosyl-tRNA synthetase. Biochemistry. 2016;55:2526–37. doi: 10.1021/acs.biochem.6b00157. [DOI] [PubMed] [Google Scholar]
- 124.Richardson SM, Mitchell LA, Stracquadanio G, Yang K, Dymond JS, et al. Design of a synthetic yeast genome. Science. 2017;355:1040–44. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- 125.Riley R, Haridas S, Wolfe KH, Lopes MR, Hittinger CT, et al. Comparative genomics of biotechnologically important yeasts. PNAS. 2016;113:9882–87. doi: 10.1073/pnas.1603941113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rogers JM, Suga H. Discovering functional, non-proteinogenic amino acid containing, peptides using genetic code reprogramming. Org Biomol Chem. 2015;13:9353–63. doi: 10.1039/c5ob01336d. [DOI] [PubMed] [Google Scholar]
- 127.Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, et al. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rydén SM, Isaksson LA. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol Gen Genet. 1984;193:38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- 129.Sakamoto K. Innovative technology for recombinant protein production using engineered E. coli genetic codes. Kagaku Seibutsu. 2016;54:343–50. [Google Scholar]
- 130.Sakamoto K, Hayashi A, Sakamoto A, Kiga D, Nakayama H, et al. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–99. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schultz DW, Yarus M. Transfer RNA mutation and the malleability of the genetic code. J Mol Biol. 1994;235:1377–80. doi: 10.1006/jmbi.1994.1094. [DOI] [PubMed] [Google Scholar]
- 132.Singh V, Braddick D. Recent advances and versatility of MAGE towards industrial applications. Syst Synth Biol. 2015;9(Suppl 1):1–9. doi: 10.1007/s11693-015-9184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Suzuki T, Numata T. Convergent evolution of AUA decoding in bacteria and archaea. RNA Biol. 2014;11:1586–96. doi: 10.4161/15476286.2014.992281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Swart EC, Serra V, Petroni G, Nowacki M. Genetic codes with no dedicated stop codon: context-dependent translation termination. Cell. 2016;166:691–702. doi: 10.1016/j.cell.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tack DS, Ellefson JW, Thyer R, Wang B, Gollihar J, et al. Addicting diverse bacteria to a noncanonical amino acid. Nat Chem Biol. 2016;12:138–40. doi: 10.1038/nchembio.2002. [DOI] [PubMed] [Google Scholar]
- 136.Terasaka N, Hayashi G, Katoh T, Suga H. An orthogonal ribosome-tRNA pair via engineering of the peptidyl transferase center. Nat Chem Biol. 2014;10:555–57. doi: 10.1038/nchembio.1549. [DOI] [PubMed] [Google Scholar]
- 137.Terasaka N, Iwane Y, Geiermann AS, Goto Y, Suga H. Recent developments of engineered translational machineries for the incorporation of non-canonical amino acids into polypeptides. Int J Mol Sci. 2015;16:6513–31. doi: 10.3390/ijms16036513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Thyer R, Filipovska A, Rackham O. Engineered rRNA enhances the efficiency of selenocysteine incorporation during translation. J Am Chem Soc. 2013;135:2–5. doi: 10.1021/ja3069177. [DOI] [PubMed] [Google Scholar]
- 139.Thyer R, Robotham SA, Brodbelt JS, Ellington AD. Evolving tRNASec for efficient canonical incorporation of selenocysteine. J Am Chem Soc. 2015;137:46–49. doi: 10.1021/ja510695g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tian F, Tsao ML, Schultz PG. A phage display system with unnatural amino acids. J Am Chem Soc. 2004;126:15962–63. doi: 10.1021/ja045673m. [DOI] [PubMed] [Google Scholar]
- 141.Tizei PA, Csibra E, Torres L, Pinheiro VB. Selection platforms for directed evolution in synthetic biology. Biochem Soc Trans. 2016;44:1165–75. doi: 10.1042/BST20160076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tuorto F, Lyko F. Genome recoding by tRNA modifications. Open Biol. 2016;6:160287. doi: 10.1098/rsob.160287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Turanov AA, Lobanov AV, Fomenko DE, Morrison HG, Sogin ML, et al. Genetic code supports targeted insertion of two amino acids by one codon. Science. 2009;323:259–61. doi: 10.1126/science.1164748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ugwumba IN, Ozawa K, Xu ZQ, Ely F, Foo JL, et al. Improving a natural enzyme activity through incorporation of unnatural amino acids. J Am Chem Soc. 2011;133:326–33. doi: 10.1021/ja106416g. [DOI] [PubMed] [Google Scholar]
- 145.Uno M, Ito K, Nakamura Y. Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie. 1996;78:935–43. doi: 10.1016/s0300-9084(97)86715-6. [DOI] [PubMed] [Google Scholar]
- 146.Uyeda A, Watanabe T, Kato Y, Watanabe H, Yomo T, et al. Liposome-based in vitro evolution of aminoacyl-tRNA synthetase for enhanced pyrrolysine derivative incorporation. ChemBioChem. 2015;16:1797–802. doi: 10.1002/cbic.201500174. [DOI] [PubMed] [Google Scholar]
- 147.Wang HH, Kim H, Cong L, Jeong J, Bang D, Church GM. Genome-scale promoter engineering by coselection MAGE. Nat Methods. 2012;9:591–93. doi: 10.1038/nmeth.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang HH, Xu G, Vonner AJ, Church G. Modified bases enable high-efficiency oligonucleotide-mediated allelic replacement via mismatch repair evasion. Nucleic Acids Res. 2011;39:7336–47. doi: 10.1093/nar/gkr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang K, Fredens J, Brunner SF, Kim SH, Chia T, Chin JW. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539:59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol. 2007;25:770–77. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 151.Wang K, Schmied WH, Chin JW. Reprogramming the genetic code: from triplet to quadruplet codes. Angew Chem Int Ed Engl. 2012;51:2288–97. doi: 10.1002/anie.201105016. [DOI] [PubMed] [Google Scholar]
- 152.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 153.Xiang Z, Ren H, Hu YS, Coin I, Wei J, et al. Adding an unnatural covalent bond to proteins through proximity-enhanced bioreactivity. Nat Methods. 2013;10:885–88. doi: 10.1038/nmeth.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xiao H, Nasertorabi F, Choi SH, Han GW, Reed SA, et al. Exploring the potential impact of an expanded genetic code on protein function. PNAS. 2015;112:6961–66. doi: 10.1073/pnas.1507741112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yamao F, Muto A, Kawauchi Y, Iwami M, Iwagami S, et al. UGA is read as tryptophan in Mycoplasma capricolum. PNAS. 1985;82:2306–9. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yu AC, Yim AK, Mat WK, Tong AH, Lok S, et al. Mutations enabling displacement of tryptophan by 4-fluorotryptophan as a canonical amino acid of the genetic code. Genome Biol Evol. 2014;6:629–41. doi: 10.1093/gbe/evu044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Záhonová K, Kostygov AY, Ševčíková T, Yurchenko V, Eliáš M. An unprecedented non-canonical nuclear genetic code with all three termination codons reassigned as sense codons. Curr Biol. 2016;26:2364–69. doi: 10.1016/j.cub.2016.06.064. [DOI] [PubMed] [Google Scholar]
- 158.Zhang Y, Baranov PV, Atkins JF, Gladyshev VN. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J Biol Chem. 2005;280:20740–51. doi: 10.1074/jbc.M501458200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.