Abstract
Single molecule fluorescent in situ hybridization (smFISH) enables quantitative measurements of gene expression and mRNA localization. The technique is increasingly popular for analysis of cultured cells but is not widely applied to intact organisms. Here we describe a method for labeling and detection of single mRNA molecules in whole embryos of the fruit fly Drosophila melanogaster. This method permits measurements of gene expression in absolute units, enabling new studies of transcriptional mechanisms underlying precision and reproducibility in cell specification.
Keywords: Single molecule, FISH, quantitation, fluorescence imaging, Drosophila
1 Introduction
Single molecule approaches to biology provide unprecedented access to direct observations of many key biological activities [1]. Single molecule mRNA counting has proven invaluable for examining gene expression heterogeneity among otherwise identical cells [2–5]. Quantifying gene expression through single molecule fluorescent in situ hybridization (smFISH) has become increasingly popular as a method for absolute quantification of gene expression in cultured cells [6–9]. Although the use of smFISH in intact organisms has not yet been as widely adopted, interest and demonstrated applications of the method are increasing [10–13].
Here, we present a detailed protocol for single molecule detection of mRNA in Drosophila embryos using multiple short oligonucleotides directly conjugated to fluorophore [14,4,15–17]. We provide details regarding probe preparation, embryo fixation, and in situ hybridization conditions for smFISH. We detail three methods for mounting embryos to maximize signal, minimize noise, and maintain tissue morphology. We discuss confocal scanning conditions for optimal generation of high quality images for quantitative analysis.
2 Materials
2.1 Generating fluorescent probes with DNA-fluorophore coupling
Except where noted, use purified deionized water for preparing solutions.
Oligonucleotides: 48 to 96 DNA oligonucleotides, each 20 nucleotides in length, complementary to a transcript of interest and bearing a 3′ amine modification (see Note 1). Each oligo should be at concentration of 100 μM. Store at −20 °C.
Fluorophore: Dissolve 1 mg of NHS ester-bearing fluorescence dye in 200 μL of N,N-dimethylformamide (DMF, molecular biology grade). Prepare immediately prior to use (see Note 2).
1 M sodium bicarbonate: Weigh 0.84 g NaHCO3 and transfer to a 15 mL conical tube containing 9.2 mL water. Mix well by vortexing until dissolved. Add 5 N NaOH to obtain pH 9.0. Use immediately or aliquot and store at −20 °C (see Note 3).
5 M potassium acetate: Weigh 49.1 g KCH3COO. Add gradually to beaker containing 45 mL water with stirring to dissolve. Use a 100 mL graduated cylinder to bring volume to 100 mL with additional water. Store at room temperature.
DEPC-treated water (see Note 4).
2.2 HPLC purification of labeled oligonucleotides
Buffer A: 0.1M triethylammonium acetate, pH 6.5, filtered and degassed.
Buffer B: Acetonitrile, HPLC grade.
C18 reverse phase HPLC column (10 μm particle size, 300 Å pore size, 4.6 mm i.d. x 250 mm).
Spectrophotometer.
2.3 Embryo collection and crosslinking
Apple juice agar plates, yeast paste, and collection cages (see Note 5).
Wire mesh collection baskets: Use wire-cutting scissors to cut steel gauze (e.g. Alfa Aesar Type 304 stainless steel mesh) into small squares approximately 1.5 x 1.5 cm. Fold the edges upward and inward to create roughly circular-shaped metal baskets.
Scintillation vials, 20 mL volume.
Pasteur pipettes.
Heptane, HPLC grade.
Bleach: 7 % sodium hypochlorite solution.
Distilled or reverse osmosis water.
Phosphate-buffered saline (PBS, 10x): in 1L beaker, add to 800 mL water 80 g NaCl, 2 g KCl, 14.4 g dibasic sodium phosphate (Na2HPO4), and 2.4 g monobasic potassium phosphate (KH2PO4). Stir until dissolved.
4% paraformaldehyde fixation buffer: Combine 1 mL 20% methanol-free paraformaldehyde solution (e.g. Electron Microscopy Sciences 15713) with 0.5 mL 10x PBS in 3.5 mL water in scintillation vial. Vortex briefly to mix.
Methanol, certified ACS reagent grade.
2.4 In situ hybridization
PTw: PBS (phosphate-buffered saline, 1x) + 0.1 v/v % Tween-20. Store at room temperature.
SSC, 20x: In 1L beaker, add to 800 mL water 175.3 g NaCl and 88.2 g sodium citrate. Stir until dissolved. Adjust pH to 7.2 with HCl. Tranfer to 1L graduated cylinder and adjust volume to 1L with additional water. Stir again, then autoclave. Store at room temperature.
FISH wash buffer: Combine 7 mL deionized formamide (see Note 6) with 4 mL 20x SSC and 9 mL DEPC-treated water. Add Tween-20 to 0.1 v/v %. Make fresh on the day of use.
Hybridization buffer: 10 w/v % dextran sulfate, 0.01 w/v % salmon sperm ssDNA, 1 v/v % (2mM) vanadyl ribonucleoside, 0.2 mg RNase-free BSA, 4x SSC, 35 v/v % deionized formamide, 0.1 v/v % Tween-20 in DePC-treated water (see Note 7).
DAPI (4′,6-diamidino-2-phenylindole) solution: Make a stock solution by dissolving 10 mg of powder in 10 mL of methanol. The solution can be stored at 4 °C for an indefinite period. Make a working solution by diluting the stock by a factor of 4000 volume:volume in PTw and use immediately.
Nutator rotating platform shaker.
2.5 Mounting embryos on slides
Mounting media: Aqua-Poly/Mount (Polysciences), Prolong Gold Antifade Mountant (Life Technologies), or Vectashield Antifade (Vector Laboratories) mounting medium (see Note 8).
23 gauge needle
Nail polish.
Vacuum filter sterilizer.
Glass slides.
No. 1.5 cover glass.
3 Methods
3.1 Probe-fluorophore chemical coupling
Calculate the volume of each individual oligonucleotide to use for conjugation. 1 mg of fluorophore dissolved in 200 μL DMF provides adequate fluorophore to conjugate 266 μL of combined oligonucleotide at a concentration 100 μM. Therefore, the volume of each individual oligonucleotide will change depending on the number of oligonucleotides per probe set. For example, a set of 48 oligonucleotides uses 266/48 = 5.5 μL of each oligonucleotide. Smaller volumes of combined oligonucleotides may also be used as long as all of the following volumes are scaled appropriately.
Combine all oligos in a single 1.5 mL Eppendorf.
Add 0.25x volumes of fresh 1 M sodium bicarbonate and briefly vortex.
Add fluorophore in DMF at concentration of 5 μg/μL. The volume of fluorophore should be 0.6x the combined volume of oligonucleotides and sodium bicarbonate.
Apply gentle agitation at room temperature for 2 hours protected from light on a rocking platform.
If necessary to contain the combined volume of the reagents added in steps 7 and 8, divide solution into 2 tubes of equal volume.
Add 0.11x volumes of 5M potassium acetate pH ~5.2 and vortex briefly
Add 2.5x volumes of 100% ethanol to each tube and vortex briefly
Immerse tubes in a bath of dry ice and ethanol bath for 2 hours.
Centrifuge for 30 min at full speed in a microfuge at 4 °C. A colored pellet will form.
Completely remove ethanol without disturbing pellet. Allow to air dry briefly.
Add 50 μl DEPC-treated water, or 25 μL per tube if divided into 2 tubes, and resuspend pellet by vigorous tapping. The contents can then be combined into a single tube if divided.
The oligonucleotide mixture in water may be stored at 4 °C for > 1 week.
3.2 HPLC purification of labeled oligonucleotide probes
After conjugation, a fraction of oligonucleotides will remain unlabeled and must be separated from labeled probes by HPLC.
Load material from the conjugation reaction on column equilibrated in 95% Buffer A/5% Buffer B.
Linearly increase percentage of Buffer B to 50% over the first 25 minutes. Unlabeled oligonucleotides will tend to elute from the column prior to minute 14. In our experience, oligonucleotides labeled with Atto 565 will elute between about minute 14 and minute 16. Oligonucleotides labeled with Atto 633 elute between about minutes 16 and 21. Collect eluate from instrument during these times.
At minute 26, increase Buffer B to 95% and maintain through minute 40. Excess unconjugated fluorophore will elute from the column (see Note 9).
Buffer B can then be decreased to 5% at minutes 41 through 50 to re-equilibrate the column. The labeled oligonucleotides may be kept in elution buffer at 4C for up to a week.
Remove solvent from probes using a vacuum concentrator “speed vac.” Place no more than 500 μl of eluate in each tube. With a moderate amount of heating, solvent will evaporate in about 1.5–2 hr. Do not overdry, otherwise it may be difficult to resuspend pellet. Make a stock solution by resuspending in 100 μl DePC treated water (see Note 10).
Measure the probe concentration of a 1:50 or 1:100 dilution of stock solution with a spectrophotometer or nanodrop to determine absorbance at the appropriate wavelength. This value can then be used with the molar extinction coefficient of the fluorophore to calculate the concentration of probe. Typically, the stock concentration is between 0.5 and 5 μM of each individual labeled oligonucleotide, depending on the amount of starting material and the efficiency of labeling.
Prepare a working dilution of probe in hybridization buffer (see Note 11).
3.3 Embryo fixation in paraformaldehyde
Exchange agar plate on cup of adult flies for new plate with a dollop of yeast paste. Leave plates on for desired period of time at 25 °C. Use timed collections to increase fraction of embryos at a given age.
Collect agar plate containing embryos and replace with new plate on cup. Dissolve chorion by adding bleach for 1 min with gentle swirling.
Pour embryos into wire mesh basket. Wash extensively with DI water to remove remaining chorion and excess bleach.
In a scintillation vial, place 5 mL of heptane and 5 mL of fixation buffer. Vortex for 15 sec, then allow phases to separate for 1–2 minutes (see Note 12).
Immerse mesh basket containing embryos. Be sure the basket drops below the interface between the aqueous and organic phases. This is where the embryos will accumulate.
Remove mesh basket from the vial. Check that no embryos remain on the basket. If some are present, the basket can be immersed again. After removing the basket, rinse it in water to ensure no embryos remain.
Cap vial and place onto an orbital shaker set to about 200 rpm. Allow to shake for 20–25 minutes.
Remove vial from shaker. Using a Pasteur pipette, remove all the paraformaldehyde solution (the lower layer) while minimizing the removal of embryos.
When only heptane and embryos remain in the vial, quickly add 5 mL methanol. Recap the vial and vortex vigorously for at least 30 sec and up to 1 min. After vortexing, allow vial to rest for 5 minutes. This step extracts most embryos from the vitelline membrane. Extracted embryos will sink to the bottom of the vial.
With a fresh Pasteur pipette, remove sunken embryos from vial and place into a fresh Eppendorf tube. Allow embryos to settle to bottom of tube. With a fresh Pasteur pipette remove as much methanol from the tube as possible without exposing embryos to air.
Add 1–1.5 mL methanol to tube, ensuring that embryos are mixed and dislodged from the bottom of the tube. This ensures efficient washing. Allow embryos to settle. With a fresh Pasteur pipette, remove as much methanol as possible.
Repeat step 11 at least 2 and up to 5 more times. This ensures that heptane is removed from embryos. The same Pasteur pipette may be used for these methanol washes.
Add 1 mL methanol. Store embryos at −20 °C until use (see Note 13).
3.4 In situ hybridization
Using a Pasteur pipette, transfer a volume containing approximately 25–50 μL of fixed embryos in a fresh Eppendorf tube. Allow embryos to settle and remove as much liquid as possible without exposing embryos to air.
Pipet 1 mL PTw, mix, allow embryos to settle, remove PTw using a Pasteur pipet, and repeat this step 3 more times.
Pipet 1 mL PTw and place embryos on Nutator for 20 minutes. Remove PTw.
Pipet 1 mL FISH wash buffer and mix by inverting tube slowly a few times. Allow embryos to settle. Remove wash buffer. Repeat this step one more time.
Pipet 1 mL FISH wash buffer and mix by inverting. Place tube on Nutator for 20 minutes.
While incubation is underway, preheat diluted probes to 37 °C.
Remove tube from Nutator and allow embryos to settle. Remove nearly all the wash buffer without removing embryos.
Slowly pipet 80–100 μL probe at working dilution. Embryos will rise and float in a layer of wash buffer on top of viscous hybridization buffer. To mix embryos and probe, tap the side of the tube many times and continue until “Schlering lines” are no longer seen (see Note 14).
Place embryos at 37 °C for at least 1.5 hours and up to 16 hours (see Note 15). For short incubation times (less than 6 hours), mix by vigorous tapping every 20–30 minutes. Protect samples from exposure to light.
Remove diluted probe with a pipetman. Used probe can be stored at −20 °C for future use and can be reused at least 2 more times.
Add 1 mL wash buffer preheated to 37 °C. Allow embryos to settle. Remove wash buffer with a Pasteur pipette.
If using long incubation times (>6 hours), add 1 mL preheated wash buffer, place tube at 37 °C for 1 hour, remove buffer, and repeat. For shorter incubation times, repeat step 33 two more times.
Add 1 mL PTw. Allow embryos to settle, then remove PTw. Repeat this step once.
If desired, stain embryos with DAPI by diluting DAPI stock 1:4000 in PTw. Add 1 mL diluted DAPI to tube. Place on Nutator platform rocker for 30 sec. Remove and allow embryos to settle (usually takes another 30 sec). Remove DAPI solution. Add 1 mL PTw, allow embryos to settle, remove PTw, and repeat 3 more times (see Note 16).
3.5 Mounting embryos for imaging in Auqa-Poly/Mount
Mounting in Aqua-Poly/Mount: Aqua-Poly/Mount is a water-soluble gelling mounting medium. It provides the best option for maintaining structural integrity of embryos and for carefully orienting embryos on slides. However it also yields the lowest signal-to-noise ratio. It is appropriate for experiments that utilize large numbers of probes and thereby generate a high signal.
Using a wide-mouth pipet tip rinse several times in PTw, transfer up to 400 embryos to a glass slide that has been cleaned with 70 % ethanol and dried.
Under stereo dissecting microscope, push embryos into a small pile in the middle of the slide. Use a small piece of paper towel to wick away most of the PTw without allowing embryos to dry.
Deposit directly on the pile of embryos a drop of Aqua-Poly/Mount. Using 23 gauge needle, gently mix embryos into medium until “Schlering lines” are no longer visible.
Use needle to arrange embryos in desired orientation on slide. If medium begins gelling, add a few microliters of water.
When embryos are in desired orientation, allow to dry for a few minutes at room temperature. This minimizes embryos shifting when cover glass is added.
Place a small droplet of Aqua-Poly/Mount on a cover glass and gently spread toward edges using a closed pair of forceps, avoiding introducing bubbles.
Carefully lay cover glass onto mounted embryos. It often works best to hold the cover glass at one edge an angle and slowly tilting the glass down onto the embryos. This helps reduce the introduction of air bubbles.
If desired, embryos can be flattened slightly by placing a small weight on the cover glass (see Note 17).
Allow slide to gel for > 6 hours before imaging (see Note 18). Protect slides from light.
3.6 Mounting embryos in Prolong Gold
Prolong Gold is a hardening mounting medium that offers high signal-to-noise compared to Aqua-Poly/Mount. However, unlike Aqua-Poly/Mount, embryos have a stronger tendency to shrivel and become distorted if left for more than a short period before applying the cover glass. In addition, Prolong Gold is much less pliable than Aqua-Poly/Mount as it begins to cure. The curing process begins within minutes of air exposure. This allows for much less time to arrange or orient embryos on a slide. Nevertheless, if orientation of every embryo is not critical, Prolong Gold is preferred for generating high signal-to-noise data.
Using a wide-mouth pipet tip rinse several times in PTw, transfer up to 400 embryos to a glass slide that has been cleaned with 70% ethanol and dried. Remove the majority of the excess PTw with a piece of paper towel.
Under stereo dissecting microscope, use a 23 gauge needle to separate and arrange embryos on the glass slide. Do not allow embryos to become overly dry, as they can shrink dramatically and introduce artifacts. Add more PTw if too much buffer evaporates.
When embryos are in desired arrangement on the slide, place a droplet (about 25–30 μL) of Prolong Gold on a clean cover glass and set aside.
Use a small folded piece of paper towel to wick away most of the PTw surrounding the embryos. Perform this step in under 1 minute to avoid the start of the curing process of the mountant on the cover glass.
Carefully lay cover glass onto mounted embryos. It often works best to hold the cover glass at one edge an angle and slowly tilting the glass down onto the embryos. This helps reduce the introduction of air bubbles.
Use a paper towel to remove excess Prolong Gold from around the edges of the cover glass.
Allow medium to cure for 48 hours at room temperature before imaging. Protect slides from light during curing process (see Note 19).
3.7 Mounting embryos in Vectashield
Vectashield provides the highest signal-to-noise ratio of the mountants we have tested. However, it is not a hardening medium. It is not possible to orient embryos. It can be challenging to maintain separation between embryos during mounting. Nevertheless, Vectashield is useful for generating usable data from probes/mRNAs that generate low signal, such as in experiments with small numbers of probes. Although a hardening version of Vectashield is available, it is not appropriate for thick specimens such as embryos.
Using a wide-mouth pipet tip rinse several times in PTw, transfer embryos to a glass slide that has been cleaned with 70% ethanol and dried. Remove the majority of the excess PTw with a piece of paper towel.
Under stereo dissecting microscope, use a 23 gauge needle to separate and arrange embryos on the glass slide. Do not allow embryos to become overly dry, as they can shrink dramatically and introduce artifacts. Add more PTw if too much buffer evaporates.
When embryos are in desired arrangement on the slide, place a droplet (about 25–30 μL) of Vectashield on a clean cover glass and set aside.
Use a small folded piece of paper towel to wick away most of the PTw surrounding the embryos.
Carefully lay cover glass onto mounted embryos. It works best to keep the droplet of Vectashield in the middle of the cover glass and drop the glass directly onto the embryos.
Use a paper towel to remove excess Vectashield from around the edges of the cover glass. This will also help to flatten the embryos. Do not over-flatten.
Seal the cover glass to the slide with nail polish and allow to dry. Slide may be imaged immediately (see Note 20).
3.8 Confocal microscopy
Acquire 3D stacks by laser scanning confocal microscopy using a high magnification, high NA objective (e.g. 63x NA 1.4). Example images are shown in Fig. 1. We have principally used a Leica SP5 equipped with GaAsP “HyD” avalanche photodiode detectors in photon counting mode (see Note 21). For thick specimens such as embryos, confocal microscopy is essential to visualize diffraction-limited objects. Wide-field fluorescence microscopy, used in many studies of cultured cells, is inappropriate because of the overwhelming signal from out-of-focus planes.
Voxel size: For most applications, diffraction-limited objects are most effectively detected by our custom MATLAB software in images obtained with pixels of 76 x 76 nm and a z increment of between 250 and 420 nm (see Note 22).
Total section thickness: We typically collect image stacks spanning 10 to 20 μm in z (see Note 23).
Scanner settings and image acquisition: To improve our ability to locate and measure the signal of diffraction-limited objects, we sum the fluorescence of multiple scans, usually between 4 and 8, of the same volume. This is usually best accomplished by setting “line accumulation” to between 4 and 8 iterations. For dual-color imaging, we use sequential scans, alternating channels with every line, in order to minimize cross-talk between channels. Because of the small numbers of fluorophores, the scanning speed often must be 2 to 8 times slower than that typically used for most confocal imaging applications.
Laser power: As with all fluorescent imaging, an important goal is to obtain images of high quality while minimizing photobleaching. Therefore it is desirable to use as little laser power as necessary to provide sufficient excitation to lift signal above background or autofluorescence. The laser power necessary to achieve this goal is experiment-specific and depends on the number of probes employed, the efficiency of the hybridization, the amount of background and autofluorescence, and the sensitivity of the photodetectors. The necessary laser power must therefore be determined empirically (see Note 24).
As controls for single molecule counting, it is advisable when possible to perform the labeling and imaging on embryos that are heterozygous for mRNA-null mutations in a gene of interest. Counts and particle densities can be compared between similarly-staged embryos of different genotypes. In the absence of compensation mechanisms, heterozygotes will produce mRNAs at half the rate of wild-type embryos. In addition, mRNA-null homozygous embryos can be used as controls for background fluorescence.
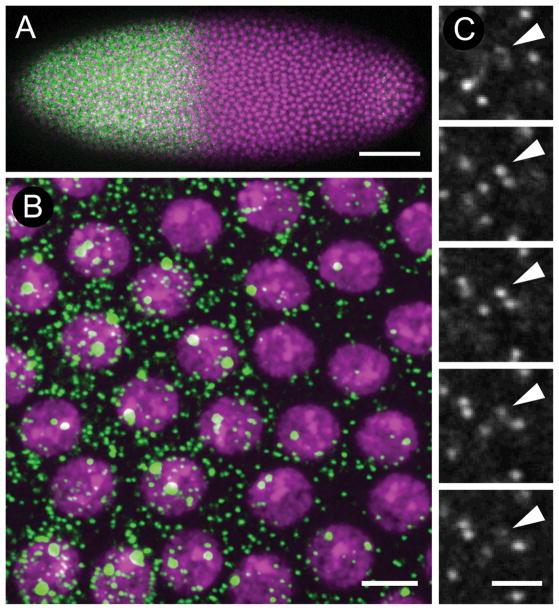
Figure 1.
Single molecule mRNA detection in Drosophila embryos. A: Low magnification image of an embryo approximately 2 hours after fertilization following the 12th nuclear division cycle and labeled with probes complementary to the gene hunchback (hb). Embryo is oriented with anterior to the left. Green: hb expression; magenta: DAPI stain to highlight nuclei. B: Single confocal slice through the nuclear layer illustrating nascent transcription sites and single mRNAs. C: Series of adjacent confocal slices surrounding a single mRNA illustrating detection on multiple z planes. Scale bars: 50 μm (A), 5 μm (B), 2 μm (C).
3.9 Analysis
Upon request, custom MATLAB analysis software is available which provides semi-automated threshold detection and which outputs spatial coordinates and estimates of fluorescence intensity from TIFF image stacks.
Footnotes
For a given mRNA target, we usually utilize at least 48 oligonucleotides designed using the oligo RNA FISH probe design tool provided online by Biosearch Technologies (http://biosearchtech.com/products/rna-fish/custom-stellaris-probe-sets). Using this tool, we design 20-mers complementary to a transcript of interest with a minimum spacing of 2 nucleotides between oligonucleotides. This ensures that presence of fluorophore does not hinder binding of neighboring probe. Larger numbers of probes increases signal of single transcripts. To maximize utilization of unique sequences, implement repeat masking. CG content of each oligonucleotide should be between 40–60%. Avoid short repeats and continuous stretches of A/T longer than 5 nt. We obtain 3′-amine-modified oligonucleotides from Biosearch Technologies (Petaluma, CA) in 96-well format. Each oligo bears a single free amine at its 3′ end. We order oligos bearing the mdC(TEG-Amino) 3′ modification at the 10 nmol delivered scale. Other modifications that add additional free amines may also be used, although we have not tested them in our protocol. Oligos from Biosearch Technologies may be ordered in solution at a concentration of 100 μM. This is essentially a lifetime supply for most genes we have studied. Biosearch Technologies states that the amine group will remain reactive for 6 months when stored at −20 °C with minimal freeze-thaw cycles. We have successfully conjugated fluorophore to oligos over the course of more than three years even with several freeze-thaw cycles.
We have had excellent success with the “Atto” family of fluorophores. We typically use Atto 565 NHS ester and Atto 633 NHS ester. The Atto dyes exhibit photostability and minimal photobleaching even under prolonged excitation. We have also successfully used Alexa 514 NHS ester and Alexa 594 NHS ester. The NHS reactive group is unstable. For best results, use dyes in the coupling reaction immediately upon arrival. If necessary, store unopened package at −20 °C. Prolonged storage is not recommended. Condensation of water in the air onto the fluorophore will tend to inactivate reagent. Therefore, if stored at −20 °C, allow package to reach room temperature before opening. During the coupling reaction, protect the dye from light as much as possible. It is usually convenient to order 1 mg of fluorophore. Immediately before performing the coupling, open the packaging and dissolve the 1 mg of powder in 200 μL DMF.
Freshly made sodium bicarbonate solution is the most effective. Alternately, small aliquots can be made and stored at −20C. After thawing, discard any unused portion.
Add 0.1 mL diethylpyrocarbonate (DEPC) to 100 mL of water in 250 mL beaker. Stir for > 12 hours. Autoclave for >15 minutes to remove DEPC.
Reference 18 provides a concise description of a simple method for obtaining large numbers of embryos [18].
Formamide is often deionized prior to shipment from supplier. However, in our experience, superior results are obtained by deionizing formamide in-house, even when using material marked by the manufacturer as deionized. To deionize formamide, place a large volume (100–500 mL) of formamide in a 500 mL beaker with a stir bar and add 10 w/v % deionizing resin, e.g. mixed bed resin for deionizing (Sigma-Aldrich M8032). Stir for 1 hour at low speed using the lowest stirring speed that does not allow the resin to settle at the bottom of the beaker. After 1 hour, filter out the resin using a vacuum-driven filter sterilizer. Aliquot and store at −20 °C until needed. With our FISH wash buffer recipe with 35 % formamide, it is convenient to make either 3.5 or 7.0 mL aliquots for 10 or 20 mL of wash buffer.
The solution of dextran sulfate, SSC, and water can be heated to ~60 °C and vortexed repeatedly until the dextran has dissolved. Allow the solution to cool to room temperature before adding the formamide and other components.
Different mountants are useful for different applications. If orientation and morphology of embryos is crucial, Aqua-Poly/Mount is preferred. For small numbers of probes where the signal-to-noise is too low in Aqua-Poly/Mount, VectaShield is preferred, although morphology is often distorted. For a balance between orientation/morphology and signal-to-noise, Prolong Gold offers a reasonable compromise.
There can be unconjugated free fluorophore after the reaction. Do not collect the free fluorophore. Atto 633 elutes at around minute 24–25 and shows no absorption at 260 nm. Atto 565 elutes around minute 18 and shows weak absorption at 260 nm.
Probes may be stored for years at −20 °C until ready for use. We have found that repeated freeze-thaw cycles have no discernable impact on probe performance.
We have obtained best results with a working concentration of around 1–2 nM for each individual labeled oligonucleotide. Note that the hybridization buffer is viscous compared to the stock solution. Vigorous vortexing for at least 1 minute is recommended for thorough mixing of probe stock in hybridization buffer. Diluted probe can be stored indefinitely at −20 °C. Diluted probe tolerates many freeze-thaw cycles without loss of performance. If using previously diluted probe stored at −20 °C, warm to 37 °C prior to adding to embryos.
The mixing and vortexing can be performed during the chorion removal (step 2).
Superior results tend to be obtained from embryos stored <1 week at −20 °C.
Mixing of embryos in all solutions is essential. When mixing embryos and probe, embryos that contact with a surface that has not been first coated with hybridization buffer will tend to stick to the tube. Thus, gentle tapping is advised at first. This will begin mixing embryos and hybridization buffer while also splashing the sides of the tube in hybridization buffer. As mixing continues, the tapping can become more and more vigorous. Embryos will also tend to become more and more transparent as mixing continues into hybridization buffer.
For some well-behaved probe sets, a minimal incubation time will produce high quality images. Others probe sets or mRNAs require longer incubation times. Optimal incubation time is best determined empirically.
Stained embryos may be stored at 4 °C for at least a week, but best results are obtained by proceeding directly to mounting.
A small amount of weight generated by, for example, a stack of small coins, can be placed onto a piece of Kimwipe folded over onto the cover glass. This will slightly flatten the embryos, which can be advantageous for placing many nuclei within the same imaging plane. However, caution is warranted: too much weight or too long application will distort the tissue. These distortions will alter the apparent density of mRNA molecules and will lead to increased measurement error in assessing mRNA density.
Embryos mounted in Aqua-Poly/Mount may be stored at 4 °C for up to a month prior to imaging. However, in our experience, superior results are obtained when imaged within a few days of mounting.
Embryos mounted in Prolong Gold may be stored at room temperature for up to a month prior to imaging. In our experience, superior results are obtained when imaged within a few days of mounting.
Embryos mounted in Vectashield may be stored at 4 °C for up to a month after mounting. In our experience, superior results are obtained when imaged within 1 or 2 weeks of mounting.
Low noise, high sensitivity detectors are essential for imaging dim objects labeled with few fluorophores. The “hybrid detectors” found on most current Leica scanning confocal systems offer high performance. These detectors can be operated in either “standard” mode or photon counting mode. Standard mode offers the option to apply gain and offset settings to amplify signal and reduce putative background. This can be useful for counting objects, but can also be misleading for making quantitative measurements. For quantitative measurements of fluorescence, photon counting mode is strongly preferred.
Efficient detection of diffraction limited objects requires high-resolution images. A simple calculation of Nyquist sampling for most commercial confocal setups can be found at https://svi.nl/NyquistCalculator. This provides an initial estimate for adequate sampling density, that is, the number of pixels per unit of distance. In practice, for the fluorophores we employ on a Leica SP5, we have had good success with pixels of dimension 76 x 76 nm in xy. In addition, we deliberately sample in the axial (z) direction more frequently than strictly required, in order to ensure that all true objects appear on multiple adjacent imaging planes. We use this feature as a powerful means of separating true objects from background signal. This procedure is described in detail in reference 4. We have used z sampling intervals as small as 250 nm [4] and as large as 420 nm [15].
Total section thickness depends on the particular experiment or application. For example, to count most or all zygotically expressed mRNAs typically requires a total imaging thickness of at least 15 μm. Alternatively, to estimate the total number of all transcripts in whole embryos, then it is required to image a very thick section spanning tens of microns. At the other extreme, if objects of interest are found in roughly the same imaging plane, this shortens data collection time. Because the quantification software requires multiple z planes, it is imperative to image at least 3 z slices, and probably more are advised in most cases.
Our experiments typically require us to image both relatively dim and relatively bright objects, for example, single mRNAs that produce a low level of signal and nascent transcription sites which can contain the equivalent of 50–100 mRNAs. To simultaneously image both single mRNAs and bright sites of transcription, we employ Leica’s HyD detectors in photon counting mode. The low noise of these detectors allows us to easily discern single mRNAs from background and simultaneously to measure the fluorescence of transcription sites without saturating the detector. We can thus apply a single scan to capture all data simultaneously. We determine the magnitude of laser power empirically for every probe set with the goal of minimizing photobleaching during prolonged scanning while still providing adequate signal to separate true objects from noise. As a general rule, objects that are clearly discernable by eye in confocal stacks will be most readily separated from background noise during the analysis steps. We have found that traditional photomultiplier tubes (PMTs) do not offer sufficient dynamic range for both reliable detection of single mRNAs and non-saturated transcription sites in the same scan. However, traditional PMTs may still be used by performing two scans of the same sample at two different laser intensities. A low-power scan is first taken for measuring the intensities of transcription sites, followed by a high power stack that saturates the signal from transcription sites but provides high signal from individual mRNAs so that they may be separated from imaging noise. We have not extensively tested other microscopes or imaging configurations. Overall, to determine optimal parameter settings, a simple rule of thumb is that an image that looks good by eye will always generate more reliable results; the more distinct and the brighter objects appear compared to the surrounding background, the more easily and reliably those objects will be detected by the software.
Contributor Information
Shawn C. Little, Department of Cell and Developmental Biology, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA
Thomas Gregor, Department of Physics; Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ, USA.
References
- 1.Chen H, Larson DR. What have single-molecule studies taught us about gene expression? Genes Dev. 2016;30(16):1796–1810. doi: 10.1101/gad.281725.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chong S, Chen C, Ge H, Xie XS. Mechanism of transcriptional bursting in bacteria. Cell. 2014;158(2):314–326. doi: 10.1016/j.cell.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gandhi SJ, Zenklusen D, Lionnet T, Singer RH. Transcription of functionally related constitutive genes is not coordinated. Nat Struct Mol Biol. 2011;18(1):27–34. doi: 10.1038/nsmb.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Little SC, Tikhonov M, Gregor T. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell. 2013;154(4):789–800. doi: 10.1016/j.cell.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4(10):e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348(6233):aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwabe A, Bruggeman FJ. Single yeast cells vary in transcription activity not in delay time after a metabolic shift. Nat Commun. 2014;5:4798. doi: 10.1038/ncomms5798. [DOI] [PubMed] [Google Scholar]
- 8.Skinner SO, Xu H, Nagarkar-Jaiswal S, Freire PR, Zwaka TP, Golding I. Single-cell analysis of transcription kinetics across the cell cycle. Elife. 2016;5:e12175. doi: 10.7554/eLife.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329(5991):533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bahar Halpern K, Tanami S, Landen S, Chapal M, Szlak L, Hutzler A, Nizhberg A, Itzkovitz S. Bursty gene expression in the intact mammalian liver. Mol Cell. 2015;58(1):147–156. doi: 10.1016/j.molcel.2015.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayer LV, Batish M, Formel SK, Bratu DP. Single-Molecule RNA In Situ Hybridization (smFISH) and Immunofluorescence (IF) in the Drosophila Egg Chamber. Methods Mol Biol. 2015;1328:125–136. doi: 10.1007/978-1-4939-2851-4_9. [DOI] [PubMed] [Google Scholar]
- 12.Bolkova J, Lanctot C. Quantitative gene expression analysis in Caenorhabditis elegans using single molecule RNA FISH. Methods. 2016;98:42–49. doi: 10.1016/j.ymeth.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Stapel LC, Lombardot B, Broaddus C, Kainmueller D, Jug F, Myers EW, Vastenhouw NL. Automated detection and quantification of single RNAs at cellular resolution in zebrafish embryos. Development. 2016;143(3):540–546. doi: 10.1242/dev.128918. [DOI] [PubMed] [Google Scholar]
- 14.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280(5363):585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 15.Little SC, Tkacik G, Kneeland TB, Wieschaus EF, Gregor T. The formation of the Bicoid morphogen gradient requires protein movement from anteriorly localized mRNA. PLoS Biol. 2011;9(3):e1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petkova MD, Little SC, Liu F, Gregor T. Maternal origins of developmental reproducibility. Curr Biol. 2014;24(11):1283–1288. doi: 10.1016/j.cub.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachmann A, Knust E. The use of P-element transposons to generate transgenic flies. Methods Mol Biol. 2008;420:61–77. doi: 10.1007/978-1-59745-583-1_4. [DOI] [PubMed] [Google Scholar]