Abstract
The recent increase in the number of X-ray crystal structures of G-protein coupled receptors (GPCRs) has been enabling for structure-based drug design (SBDD) efforts. These structures have revealed that GPCRs are highly dynamic macromolecules whose function is dependent on their intrinsic flexibility. Unfortunately, the use of static structures to understand ligand binding can potentially be misleading, especially in systems with an inherently high degree of conformational flexibility. Here, we show that docking a set of dopamine D3 receptor compounds into the existing eticlopride-bound dopamine D3 receptor (D3R) X-ray crystal structure resulted in poses that were not consistent with results obtained from site-directed mutagenesis experiments. We overcame the limitations of static docking by using large-scale high-throughput molecular dynamics (MD) simulations and Markov state models (MSMs) to determine an alternative pose consistent with the mutation data. The new pose maintains critical interactions observed in the D3R/eticlopride X-ray crystal structure and suggests that a cryptic pocket forms due to the shift of a highly conserved residue, F6.52. Our study highlights the importance of GPCR dynamics to understand ligand binding and provides new opportunities for drug discovery.
Introduction
G-protein coupled receptors (GPCRs) are one of the largest membrane protein families, encoded by more than 800 genes1. GPCRs recognize a wide range of extracellular signals and transduce them into diverse cellular responses. Blocking or altering these signals provides extraordinary pharmacological opportunities; not surprisingly, ~34% of marketed drugs target GPCRs1. Understanding GPCR binding pocket recognition elements is thus a first step toward designing drugs that modulate these responses.
When first characterized, GPCRs were thought to operate as ‘on-off’ switches through two unique active and inactive states. Recent studies show that GPCRs are more complex than this and have characterized the pathways of entry of different GPCR ligands2, their conformational ensemble landscape3, and many activation pathways4,5. In addition, GPCRs adopt a set of differently populated active, inactive, and intermediate sub-states, and it has been shown that diverse ligands and coupling proteins interact with GPCRs and shift their populations to different extents6.
The most studied GPCR class is the rhodopsin-like class A GPCR family, divided into more than 50 subfamilies7. Some of the rhodopsin-like class A GPCRs recognize biogenic amines as endogenous agonists and are categorized as aminergic receptors. Up to 25% of the currently marketed drugs bind to aminergic receptors, and eleven distinct ligand-bound active and inactive crystal structures have been published7. Aminergic subfamilies include the muscarinic, adrenergic, histamine, trace amine, serotonin and dopamine receptors8.
D3 receptor (D3R) antagonism is a compelling molecular mechanism for its role in drug addiction. D3R modulation has been pursued for more than a decade as a potential target for the treatment of substance use disorders9,10, and several selective D3R antagonists have been reported in the literature11–13. Most of the D3R antagonist structures disclosed to date share a common 4-feature pharmacophore (A1-B-L-A2), where an aromatic (A1) group fills the orthosteric binding site (OBS) followed by a basic cyclic amine (B) hydrogen bonding to the conserved D3.32, connected to a flexible linear chain (L) linked to a second aromatic (A2) group that explores divergent interactions within the secondary binding pocket (SBP)11–14.
We recently published the in vitro and in vivo characterization of PF-4363467 (1, Table 1), a D3R dual antagonist with unusually high D2R receptor occupancy (86%), which translated into unique in-vivo signaling, e.g. lack of locomotor or catalepsy side effects15. Unlike previously reported structures of D3R antagonists, 1 does not share the same pharmacophore feature arrangement (B-A1-L-A2) and presented some challenges in the prediction of its binding mode15. Induced-fit docking of 1 into the published D3R X-ray structure (PDB ID 3PBL)16 suggested the possibility of two binding modes, neither of which was fully predictive of the SAR and selectivity in the series15. The true binding mode of 1 to D3R therefore remains unclear.
Table 1.
Summary of compounds studied in this work.
| ID | Name | 2D structure | Ki (nM) | Mutations |
|---|---|---|---|---|
| 1 | PF-4363467 |
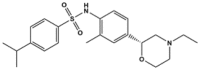
|
3.4 ± 0.4 | I183ECL2F |
| V1895.39A | ||||
| 2 | Eticlopride |
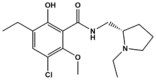
|
0.2416 | V1895.39I |
| Y3737.43F | ||||
| 3 | Haloperidol |
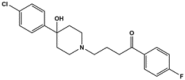
|
6.5 ± 1.0 | C1143.36L, |
| I183ECL2F | ||||
| E902.65Q | ||||
| 4 | GSK598809 |
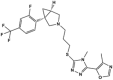
|
2.5 ± 0.4 | Y361.39L |
| E902.65Q | ||||
| Y3737.43F |
Structure, inhibition constant (Ki), and mutations that most affected binding for each compound. Values presented were measured in this work except eticlopride (2), measured in ref.16.
In order to better understand how 1 potentially binds to the D3R and exerts its therapeutic effect, a comparative study of 1 was performed with three other well-characterized D3R antagonists: eticlopride (2), which has been co-crystallized in complex with D3R, haloperidol (3), the most widely used first-generation antipsychotic, and GSK598809 (4), a dual D2R/D3R antagonist which has been shown to be active for the treatment of drug abuse disorder (Table 1).
Point mutation studies were performed on the D3R to identify key binding interactions for the four antagonists. Using rigid docking the binding poses of 2, 3 and 4 could be rationalized with the point mutation experiments. However, the predicted docking pose for 1 in the D3R was not corroborated by the point mutation studies. This result suggested that 1 may adopt an alternative binding mode, and was further interrogated using unbiased, all-atom, large-scale molecular dynamics (MD) simulations. The use of MD to reconstruct ligand-receptor binding has been widely used in the last five years2,17,18, and proved to be valuable for characterizing binding modes while taking into account protein flexibility, e.g., in serine proteases19,20 and GPCRs21. Recently, it was successfully applied to reconstruct the protein-protein binding process of Barnase-Barstar22. The analysis performed in this work revealed 1) a different binding mode for compound 1 compared to poses generated by rigid docking, and 2) a cryptic pocket induced by compound 1 by displacement of the highly conserved residue F3466.52 between helices V and VI. Our study opens new opportunities for drug design in aminergic GPCRs and highlights the importance of protein dynamics in ligand binding.
Results
Mutagenesis and computational docking studies
The affinities of compounds 1–4 for 12 different D3R mutants were measured using radioligand binding experiments (Table S1). The D3R mutations were located in the OBS or the vicinity of the OBS and were performed on nine different residues (Fig. 1). The contribution of each residue to the binding affinities of 1–4 was determined by measuring their inhibition constants for the wt-D3R and mutant D3R forms. The impact of a mutation was evaluated by computing fold-shifts (Kimutant/Kiwt), where values greater than one are considered indicative of a detrimental effect on the compound’s affinity.
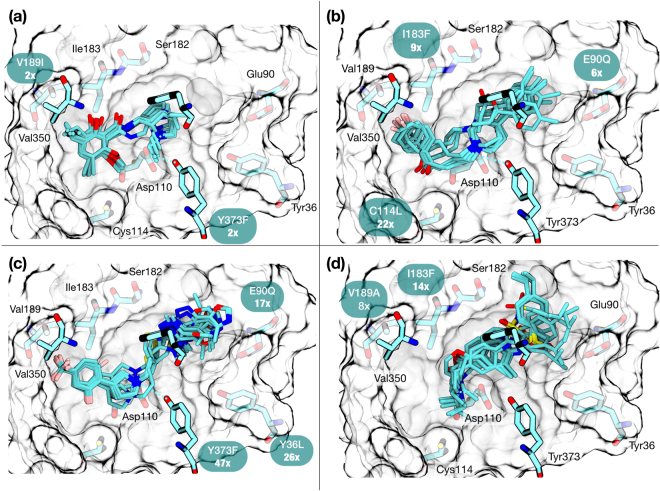
Figure 1.
Binding modes for compounds 1–4 obtained from rigid docking using 3PBL as input coordinates for the receptor. The four structures highlight the same amino acids for direct comparison and the ten highest-scored docked poses. (a) 2 reproduces the crystal binding mode. Compounds 3 (b) and 4 (c) entirely occupy the OBS and extend towards the extracellular vestibule. (d) Compound 1 extends towards the extracellular vestibule.
To interpret the mutation data, compounds 1–4 were docked into the published D3R co-crystal structure (PDB ID: 3PBL) in complex with eticlopride16. The docking grid was created based on the eticlopride binding mode and enclosed the entire orthosteric pocket (see Methods). A priori, the binding mode of the compounds could be inferred from their similarities with eticlopride, as they all share interactions in the OBS. Three highly conserved8 ligand-aminergic receptor interactions were observed: 1) residue D1103.32 forms a salt bridge with the positively charged amino group of the ligands, 2) an aromatic cluster in helix VI W[Y]6.48 - F[Y]6.51 - F6.52 provides a subpocket for aromatic moieties, and 3) residues S5.42 and S5.46 interact with the hydroxyls of catecholamine agonists23. Fig. 1 summarizes the effect of these mutations superimposed with the docking predictions. For the full mutagenesis data set see Table S1 and Fig. S1.
Eticlopride (2), correctly re-docks into the X-ray structure (Fig. 1a). Its tertiary amine in the ethyl-pyrrolidine forms a salt-bridge with D1103.32, while the rest of the structure, kept almost planar due to intramolecular H-bonds, fits tightly in the hydrophobic pocket among helices III, V and the aromatic cluster in helix VI. Two mutations had minor effects on eticlopride affinity, namely, V1895.39I and Y3737.43F. The first mutation is located in the hydrophobic pocket, and the second directly interacts with the tertiary amine.
The binding mode of 3 has been the subject of intense research. Most studies agree on the region in which haloperidol binds in the OBS; however, there is not consensus on the orientation of the molecule, which diverges by 180°24. The most recent evidence from quantum mechanics25, molecular dynamics24 and experimental studies point to a binding mode with the butyrophenone moiety binding deep in the binding cavity26, in agreement with our docking prediction. The experiments suggest that the affinity of 3 is mainly affected by mutations C1143.36L, I183ECL2F and E902.65Q (Fig. 1b). The effect of C1143.36 mutations on other D3R ligands has been tested in the past. In particular, the affinity of aminotetralin antagonists decreased significantly in the C1143.36S mutant form27. Reported QM-MM docking pose refinements on 3 also revealed the pivotal role of C1143.36 in binding24.
The predicted binding mode of 4 aligns with what has been previously reported28. The basic nitrogen of 4 forms a salt bridge with D1103.32 while the substituted phenyl ring fills the hydrophobic region in the OBS. In addition, the terminal oxazole ring of the aromatic group clearly points toward the SBP in the extracellular opening. Mutations that most affected the binding of compound 4 were Y361.39L, E902.65Q, and Y3737.43F. These residues are located at the cavity between helices I, II and VII where the biaryl system binds (Fig. 1c). Whereas Y361.39 and Y3737.43 do not form specific interactions with 4 and may share a structural role in D3R, the loss of binding affinity observed with the E902.65Q mutation could be explained by the disruption of the favorable electrostatic interaction between the acidic CH in the oxazole moiety and the carboxyl group of E902.65.
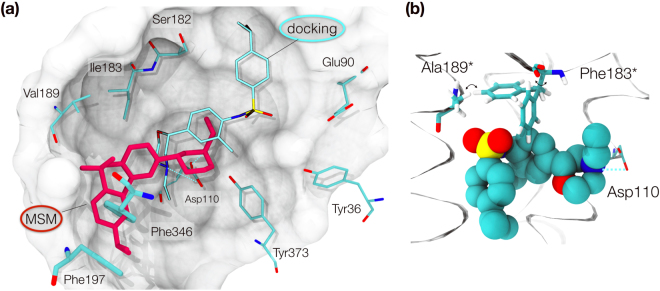
The docked pose for 1 suggests a salt bridge between D1103.32 and the positively charged nitrogen in the morpholine ring (Fig. 1d). The 4-isopropylphenyl group occupies a crevice enclosed by the extracellular loops (also termed the extracellular vestibule), interacting with a cluster of hydrophobic and aromatic residues: L892.64, V95ECL1, F1063.28, C181ECL2, and Y3657.35. Remarkably, this binding mode leaves unoccupied most of the hydrophobic region in the OBS delimited by helix III, V and VI. Mutation of I183ECL2 into Phe resulted in a 14-fold decrease in affinity. A second detrimental mutation located in the same subpocket, V1895.39A, was accompanied by an 8-fold decrease in affinity. These changes in binding affinity could be explained if compound 1 filled the region adjacent to I183ECL2 - V1895.39: the introduction of a larger residue with the I183ECL2F mutation would clash with the compound, and reducing the sidechain size with the V1895.39A mutation would potentially lose favorable hydrophobic interactions. However, in the binding mode predicted by docking, compound 1 is only partially filling this region and positioned more than 6 Å and 9 Å away from the I183ECL2F and V1895.39A mutations, respectively. While most of the mutations seem to agree with our predicted binding mode, these two are exceptions and stand out as incompatible with the docking model for 1.
Molecular dynamics simulations and Markov state models of PF-4363467/D3R complex
To further interrogate the binding mode of 1 with D3R, high-throughput unbiased MD simulations were performed. This approach had initially been validated using 2, for which the binding mode at D3R is known, as a control experiment (details in Text S1 and Fig. S2). In total, 700 μs of aggregated simulation time were produced, and 1 was observed to spontaneously bind to D3R in multiple different poses in this timeframe. The simulations were run on the distributed computing project GPUGRID.net29 using ACEMD30 directed sampling and the Markov state model (MSM) analysis with HTMD31. We used an unsupervised adaptive sampling protocol32 that allows for efficient exploration of conformational space without biasing the dynamics of the system by performing simulations in successive epochs. At each epoch, the cumulative ensemble of simulations is automatically analyzed using MSMs from which the starting conformations for the following epoch of simulations are identified (see Methods).
Initially, a set of exploratory simulations composed of short trajectories of 25 ns was run, accumulating an aggregate of 259 μs. During the simulations, the ligand freely diffused from bulk and initiated interactions at different locations in the receptor. Over the course of the successive simulation epochs, the MSM analysis provided intermediate metastable binding modes (Fig. S3). Subsequently, a second set of 100 ns trajectories was run. In this case, the ligand was located in bulk in 50% of the starting points and was in contact with the protein following the previous analysis’ equilibrium distribution in the other 50% (Text S2).
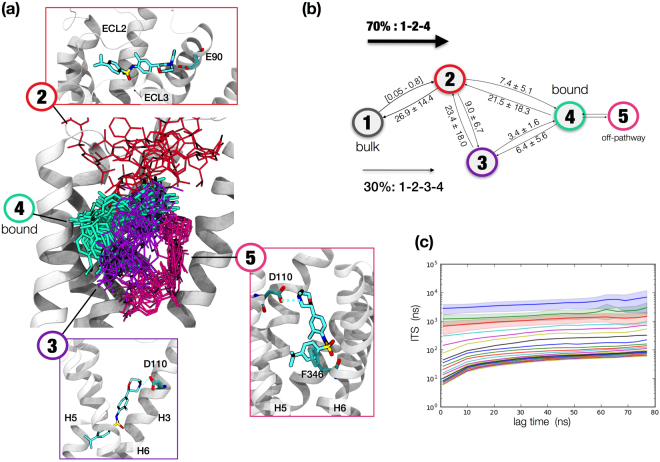
In total, 220 μs from the second set of trajectories were used for this analysis. The data was separated into 800 clusters that were then kinetically lumped together into five states33. These states corresponded to the basins of the complex energy landscape, i.e. to bulk, intermediate and bound poses in the ligand binding pathway (Fig. 2a). State 1 corresponds to bulk, or the initial state in the reaction pathway, when the ligand is free in solution. State 2 is located at the extracellular vestibule and corresponds to the first state when the ligand initiates contacts with the protein. It recognizes E902.65, in agreement with a recent study on the binding pathways of clozapine and haloperidol to D2R and D3R24. State 3 is a broad state in the OBS that encompasses several meta-stable poses before reordering in the final bound pose, state 4. Finally, state 5 is an off-pathway state that does not convert to other states without reverting to bound.
Figure 2.
Details of the MSM analysis. (a) The MSM produced 5 states. State 1, bulk, is not shown for clarity. State 2 binds to the extracellular vestibule while poses 3–5 occupy the OBS. Bound state is shown in Fig. 3 in detail. (b) Transitions between states before reaching bound, in microseconds. (c) The implied timescales of the MSM.
The whole process of binding occurred with a mean first passage time (MFPTon) of 7.0 ± 5.0 μs, which corresponded to a kon of (12 ± 8) 106 M−1 s−1, in line with previous works on drug binding to GPCRs2. Compound 1 can reach the bound state through two different interconnected pathways of binding (Fig. 2b). In the fastest and most common route of binding, compound 1 must traverse two energetic barriers. The first step consists of a diffusion-limited process where 1 recognizes the extracellular vestibule in the nanosecond timescale, ranging from around 50 to 800 ns. The second, rate-limiting step occurs in 7.4 ± 5.1 μs and comprises the recognition between the charged amine in 1 and D1103.32, followed by a rearrangement of the complex to reach the bound state (Fig. 2b). The second, less populated pathway occurs in 30% of the binding processes. It shares the first step with the previous pathway, but includes an additional intermediate state of binding between the extracellular pose and the bound state. Conversion between extracellular state 2 and the alternate bound state 3 occurs in an average time of 9.0 ± 6.7 µs, which is then followed by a reordering in the pocket toward the bound state, crossing a lower energetic barrier (3.4 ± 1.6 µs). An example of a binding trajectory is shown in Movie S1. The three states in complex with the receptor present delta free energies of binding of the order of 4.8–5.4 kcal/mol. The MSM is therefore underestimating the binding free energy (ΔGexp = −11.6 kcal/mol). State 4 is the most probable, and although states 2 and 3 have free energies comparable within the error bars, state 4 provides better alignment with the mutation data.
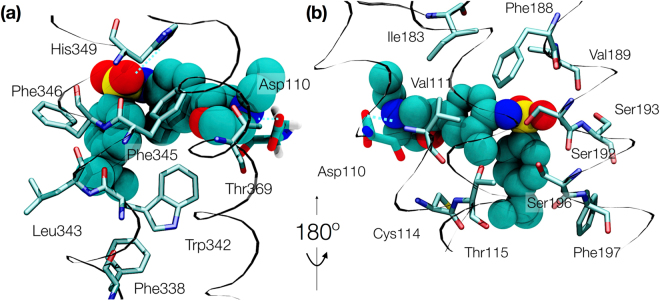
From the almost 12,000 frames belonging to state 4, the pose with the closest RMSD to the pocket side chains from the available X-ray structure (3PBL) was selected for visualization. The interactions show that all of the heavy atoms of 1 are in contact with 19 residues at a distance of 4 Å or less (Fig. 3). The primary amine in the morpholine ring forms the conserved salt bridge with D1103.32 while the rest of the ring is packed between residues T3697.39 and V1113.33. The 2-methylphenyl group packs between residues V1113.33, C1143.36, I183ECL2 and F3456.51 with the methyl substituent oriented toward T1153.37. The sulfonamide oxygen interacts with conserved S1925.42, S1935.43 and H3496.55, which together with I183ECL2 and Y3657.35, define the surface curvature of the upper side of the D3R ligand binding pocket. In contrast to the D3R X-ray crystal structure with 2, the 4-isopropylphenyl group displaces the aromatic residues F3466.52 and F1975.47 (Fig. 4a), opening a cryptic pocket delimited by residues F3386.44, W3426.48, L3436.49, F3456.51, and F3466.52.
Figure 3.
Binding mode of compound 1. Frontal (a) and back (b) views of the ligand pose and interacting residues within 4 Å. Polar interactions are depicted by blue dashed lines. Asp110 is shown in both representations as reference.
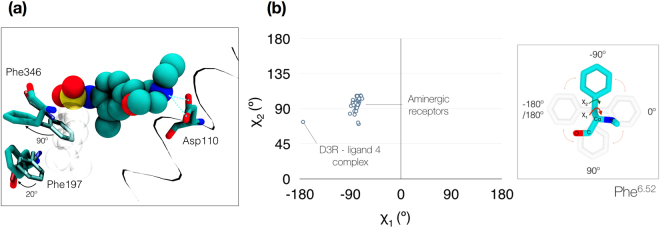
Figure 4.
Opening of a cryptic-pocket after displacement of F1975.47and F3466.52 by compound 1. (a) Displacement of the two phenylalanine residues in the D3R:1 complex versus the X-ray crystal structure by the 4-isopropylphenyl group in 1 (shown in transparent grey). (b) Structural analysis performed for the phenylalanine sidechain dihedrals (χ1 and χ2) as represented in the inset diagram. The conformation stabilized by compound 1 explores a χ1 angle close to −180°, previously unseen in the studied set of 39 crystallized aminergic receptors, which are close to −90°.
The predicted protein conformation of D3R in complex with 1 does not significantly differ from the experimentally determined co-crystal structure of D3R with 2, or with the recently crystallized D4R structure with the antagonist Nemonapride:34 overlays of the Cα backbone atoms with these two crystal structures result in RMSDs of 2.1 and 3.3 Å, respectively. The ionic lock salt bridge between R3.50 in the conserved D[E]RY motif and D/E6.30, a common feature in many GPCR structures and conserved in rhodopsin and D3R/D4R inactive structures, is also present in the predicted D3R structure with 1. The distance between the E6.30 oxygen and R3.50 nitrogen atoms is 2.7 Å, in line with the D3R and D4R X-ray structures (2.5 Å and 4.5 Å respectively). ICL2 is helical in the predicted structure of D3R with 1, similar to that seen in chain A of the D3R X-ray crystal structure with 2. The conserved NPxxY motif, recognized as a critical determinant of GPCR activation35 superimposes with both D3R and D4R structures. Y7.53 in the NPxxY motif plays a key role in GPCR activation, which takes part in the so-called tyrosine toggle switch along with Y5.58 36. During activation, the two sidechains rearrange and disrupt the ionic lock, whereas in the antagonist-induced structures of D3R and D4R, they are 10.7 Å and 9.7 Å apart, respectively. We observe that Y7.53 occupies a different rotamer state in our predicted D3R:1 structure, leading to a distance of 18.3 Å, in line with the recent MD study of D3R:3 complex36.
Aside from the different rotamer states in the tyrosine toggle switch, the formation of the aromatic cryptic pocket between helices V and VI comprises the greatest conformational difference between the predicted D3R:1 structure and the D3R:2 X-ray crystal structure. Residues F1975.47 and F3466.52 are pushed away from their original X-ray positions by 20 and 90 degrees, respectively, allowing the 4-isopropylphenyl group to fit deeper in the OBS. To our knowledge, such a conformation has not been observed in published crystal structures of other aminergic GPCRs. In order to validate this assumption, we performed a structural superimposition of publicly available aminergic receptors. From the six existing aminergic subfamilies, four of them present a phenylalanine at position 6.52, totaling six representative receptors with X-ray data: serotonin (5HT1B, 5HT2B), adrenergic (β1 A, β2 A), dopamine (D3R), and histamine (H1R). We used 39 available crystal structures for our analysis (Table S2). After superimposing all crystal structures, we measured the phenylalanine sidechain dihedral angles χ1 and χ2, which provided distributions of (−90.8°, −68.9°) and (70.2°, 106.8°), respectively. The value for χ2 in our binding mode is within this array (72.9°), but the χ1 value lies outside of the previously observed conformations (−174.4°) (Fig. 4b). While this new conformation has not been detected in any solved X-ray co-crystal structure to date, we found a similar conformation of this aromatic pocket in a recently disclosed large-scale MD study by Thomas et al.24. Although the authors did not discuss this explicitly in the manuscript, we were able to find a pose of clozapine bound to D3R within the Supplementary Information that is consistent with the F3466.52 rotamer identified in this study.
The pose identified for 1 based on this extensive simulation data provides a rationale for the potency loss observed with the I183ECL2F and V1895.39A mutations in the OBS. The superposition of the poses obtained by docking and the simulation data along with the performed mutations is shown in Fig. 5a. Although both poses interact with D1103.32 via the morpholine salt bridge, they adopt very different orientations. The docking pose (blue) extends towards the extracellular vestibule, while the simulation data pose (red) fits tightly into the OBS and opens a cryptic-pocket among helices V and VI. The sidechain of V1895.39 is in contact with the sulfonamide group in 1, and the heavy-atom reduction to Ala decreases favorable hydrophobic interactions. Mutation of a small, neutral amino acid such as I183ECL2 to a larger one such as Phe would have detrimental effects by clashing with compound 1 as shown in Fig. 5b. Indeed a phenylalanine at this position would either clash with the 2-methylphenyl group or with the nearby sidechain of V1895.39.
Figure 5.
Comparison of docking and MSM poses and effect of mutations. (a) Overlay of the pose obtained by high-throughput molecular dynamics (red), the docking prediction (blue) and the residues implicated in binding in the 3PBL coordinates. The new pose obtained by MSM fills the pocket formed by V1895.39A, and I183ECL2F and would clash with F1975.47and F3466.52 in their crystal structure coordinates. (b) The pose obtained by molecular dynamics provides a rationale for the effect of the two mutations (V1895.39A, I183ECL2F). The mutated residues are shown with an asterisk. Rotation of the engineered F183ECL2 would clash with either the ligand or residue 189, affecting protein stability and ligand affinity. Mutant A1895.39 would lose favorable packing (van der Waals) interactions by reduction of its sidechain.
Discussion
In this work, the binding poses for compounds 1–4, which are known to have affinity for D3R, were studied. Docking poses generated for compound 1 were not consistent with results from mutagenesis experiments, suggesting it may adopt a different pose compared to 2 in the D3R X-ray co-crystal structure. By performing high-throughput molecular dynamics simulations, a new binding mode was identified that was consistent with mutation results. This pose stabilized an inactive D3R conformation by displacing F3466.52 to a conformation that has not yet been observed in aminergic GPCRs. Compound 1 binds by opening a cryptic pocket, which can be exploited in the design of D3R antagonists and, by extension, other aminergic GPCR ligands.
The recent advances in GPCR structural biology have led to unprecedented high-resolution pictures of GPCR-ligand complexes. These structures have been of tremendous importance in drug design34, with some of these efforts leading to drugs currently in clinical trials. This work suggests that characterizing the intrinsic flexibility of GPCRs is of considerable importance in structure-based drug design (SBDD), even for sampling different inactive conformational states. Docking of compound 1 or analogous compounds into the previously crystallized D3R structure would have missed this promising compound in a virtual screening protocol, as single-structure GPCR SBDD only provides conformational accessibility to a subset of ligands37. In the near future, we expect that approaches similar to the one presented here will become common in the preclinical stage of the drug discovery pipeline and will be particularly enabling for GPCR preclinical programs where obtaining X-ray structures is challenging.
Methods
D3R construct
The wild type template D3R construct was a kind gift from Ellen Y. T. Chien16, subcloned into a pFastbac vector containing a polyhedran promoter. Full sequence is presented in Text S3.
Mutagenesis
Point mutants were made by site-directed mutagenesis using 0.5 μl Takara’s PrimeStar Hot start polymerase with 20ng D3R wild type template plasmid DNA, 5 × PrimeStar buffer, 4 μl dNTP mix (2.5 mM each dNTP), plus 0.5 μg forward and reverse primers. 18 cycles were performed at 98 °C for 30 seconds, 55 °C for 30 seconds and 72 °C for 6.5 minutes. The template was degraded with DpnI for 1 hr and transformed into DH5α competent cells.
Expression
Mutants were expressed in SF9 insect cells for 48 hrs with an approximate MOI of 5. A list of mutagenesis primers used can be found in Table S3.
In Vitro Receptor Binding Assay
Binding assays were performed on membranes prepared from transiently transfected human D3 receptor mutants in SF9 insect cells. Frozen cell paste was homogenized by Polytron in 50 mM Tris HCl buffer (pH 7.4 at 4 degrees C) containing 2.0 mM MgCl2. The cell membranes were spun in a high-speed centrifuge at 40,000 × g for ten minutes. The final pellet was re-suspended in assay buffer (50 mM Tris buffer, 120 mM NaCl, 5 mM MgCl2, 5 mM KCl, 2 mM CaCl2, pH 7.4 at 37 °C). Incubation was initiated by the addition of cell membrane solution to 96-well plates containing test compounds and [3H] 7-OH-DPAT (1 nM final concentration). The final concentration of protein in the assay was between 2 and 30 μg/well, depending on receptor expression of the transient mutant. Non-specific binding was determined by the presence of a saturating concentration (10 μM) of 3, a potent D3 antagonist. After a 20 minutes incubation period at 37 °C, assay samples were rapidly filtered through PEI coated, GF/B fired Unifilter plates (PerkinElmer) and rinsed with ice-cold 50 mM Tris buffer (pH 7.4). Filterplates were allowed to dry overnight. Membrane bound [3H] 7-OH-DPAT levels were determined by liquid scintillation counting of the filterplates in Ecolume scintillation fluid. The IC50 value (concentration at which 50% inhibition of specific binding occurs) was calculated by curve fitting of the concentration response, percent inhibition data. Ki values were calculated according to the Cheng- Prusoff equation, Ki = IC50/(1 + (L/Kd)), where L is the concentration of the radioligand used in the experiment and the Kd value is the dissociation constant for the radioligand at the mutant receptor (determined previously by saturation analysis).
Docking
Molecular docking experiments were performed with GLIDE (version 6.9 Schrödinger package)38–40. The protein 3D model was prepared using the Schrödinger Protein Preparation Wizard workflow41, starting from the D3R X-ray structure 3PBL and adding hydrogens at pH = 7.0. Taking the prepared receptor, Glide docking experiments were performed following receptor grid generation. The center of the grid box was defined by the center of the bound ligand as described in the original PDB entry. The enclosing box dimensions, automatically deduced from the ligand size, fit the entire active site. Concrete dimensions were 10 × 10 × 10 and 40 × 40 × 40 Å, for the inner and outer boxes, respectively. The standard precision mode (SP) of Glide was chosen, sampling ligands as flexible, saving 10 maximum structures per ligand and setting D110 as H-bond donor constraint. A post-docking optimization of the docking poses was performed choosing 0.5 kcal/mol as the cutoff for rejecting the minimized poses. For each processed compound, we considered the best scoring docking model for comparison with the mutagenesis and MD data. Flexible docking was also performed using Schrödinger´s Induced Fit Docking (IFD) Workflow42,43. IFD initial docking was done by Glide set to standard precision. The docking site was also defined by the original ligand and the receptor optimization by Prime was set to the residues within 5 Å from 2, excluding those forming hydrogen bonds with the ligand.
System setup and MD production
Systems were built, equilibrated and run with the software HTMD31. The protein coordinates were taken from the PDB code 3PBL16. L119 was mutated to Trp according to the wild type sequence. The lysozyme T4 protein was removed for the simulation and the resulting gap in the third cytoplasmic loop (ICL3) capped, by addition of neutral caps at residues 220 and 320. Protonation states were determined with the propKa tool at pH = 7.244. The membrane was formed by a 80 × 80 A2 POPC lipid bilayer and its location was set using the OPM database45. The ligand was randomly placed around the protein in the extracellular part, with random x and y coordinates while keeping the z coordinate at 43 Å. The system was parameterized with the Charmm36 forcefield for the protein atoms and lipids46. Parameterization of the ligand was performed using a derivative of the Gaamp tool47 implemented in HTMD (Fig. S4). The system was solvated using TIP3P water molecules48 and neutralized with NaCl at a 0.150 M concentration, giving a total of 48650 atoms. Four different systems were built and used as input for four independent minimizations and equilibrations.
Energy minimization and equilibration were conducted under NPT conditions at 1 atm, 298 K, using a time step of 4 fs for thermalization and a cutoff of 9 Å, with rigid bonds and PME for long-range electrostatics. Energy minimization was run for 250 fs and equilibration for volume relaxation was run for 40 ns. In these two phases, the heavy protein atoms were restrained by a 1 kcal·mol−1·Å−2 spring constant that linearly decreased until 20 ns, when it was set to zero. During the production runs, a flat-bottom potential was set using a cuboid box with dimensions [−20, 20, −20, 20, −20, 35] for the x1, x2, y1, y2, z1, and z2 coordinates, respectively. Whenever the ligand explored an area outside the box, it was pushed back with a force proportional to its distance from V1113.33’s alpha carbon and a constant of 5 kcal/mol·Å2. The flat-bottom potential kept the ligand from crossing to other periodic boxes, and from inefficiently exploring the receptor’s intracellular region or the lipid bilayer for large times. The effective concentration of the simulation box computes as 18.9 mM, taking into account the volume of the flat-bottom box (88 nm3). Minimizations and equilibrations were performed on local workstations whereas the productions were run with ACEMD30 on GPUGRID.net29. By using an adaptive sampling protocol32, a total of 680 μs were run from which 220 μs were used for the analysis in the main text (Text S2).
Simulation data and binding pose may be distributed upon request.
Markov State Models
Markov modeling proceeds from the discretization of the conformational space of the system of interest and the description of the dynamics of the system as a sequence of transitions between all discrete states. An optimally discretized MSM shows converging timescales with high probability of transition among kinetically similar states, and lower probability between kinetically separated states. From this model, the pathways and kinetic rates between distinct conformations may be derived49. In this case the conformations were represented by means of two separate protein-ligand contact representations. The first is a contact map, which accounts for interactions of all residues in the protein with the ligand. The distances between all heavy atoms of the protein to all oxygens, nitrogens and carbons not attached to hydrogens in the ligand are calculated. If any residue has a distance of less than 5 Å from the considered ligand atoms, the residue is considered to be in contact with the ligand. The second contact representation is a single flag, which is set to true if any of the above-mentioned atoms in the ligand is closer than 10 Å to any CA atom in the protein, thus creating an extra dimension which can be used to separate the bulk from the interacting states.
One of the most important requirements for constructing Markov models is to be able to finely discretize the slowest order parameters. TICA50 (time-structure independent component analysis) is a method that allows projecting the data on the slow order parameters, yielding very good discretization. After projecting the first protein-ligand contact representation onto the five slowest processes found by TICA, the 5-dimensional projected data was clustered into 800 clusters using the mini batch k-means algorithm51. Although this fine discretization provides very accurate Markov models, it is necessary to reduce the amount of states to obtain a humanly interpretable model of the system in question. Hence, the produced clusters were then lumped together into 4 macrostates using the PCCA algorithm33, each consisting of a set of kinetically similar clusters. Next, the protein-ligand contact-flag used to indicate bulk states was used to split a 5th bulk state out of the other 4 macrostates to obtain cleaner kinetics of binding. Finally, core sets were used to improve the model quality. This was done by identifying clusters that belonged to the cores of the 5 macrostates and recalculating the Markov model accounting only for transitions between the core clusters. This provided improved estimates of kinetics between states, as spurious transitions between states caused by inaccuracies in cluster borders are discounted. The converged timescales of the final model can be seen in Fig. 2c.
Electronic supplementary material
Acknowledgements
We thank Ellen Y.T. Chien for the D3R template construct. N.F. acknowledges support from Generalitat de Catalunya (FI-Agaur). GDF acknowledges support from MINECO (BIO2014-53095-P) and FEDER. We also thank all the volunteers of GPUGRID who donated GPU computing time to the project.
Author Contributions
N.F., S.S. and G.D.F. designed the research and wrote the paper. N.F. generated the simulation data. N.F., S.D. and G.D.F. analysed the simulation data. R.T.N. made the D3R mutants by site-directed mutagenesis. M.A.V. generated the in vitro receptor binding assay data. Y.Z., X.C., E.S.M., B.L.K. T.T.W. X.H. and A.V. contributed to the discussion of the results. All the authors contributed to the final edition of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
4/10/2019
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has not been fixed in the paper.
Contributor Information
Simone Sciabola, Email: simone.sciabola@pfizer.com.
Gianni De Fabritiis, Email: gianni.defabritiis@upf.edu.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19345-7.
References
- 1.Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov. 2017;16:829. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dror RO, et al. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc. Natl. Acad. Sci. 2011;108:13118–13123. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bockenhauer S, Fürstenberg A, Yao XJ, Kobilka BK, Moerner WE. Conformational dynamics of single G protein-coupled receptors in solution. J. Phys. Chem. B. 2011;115:13328–13338. doi: 10.1021/jp204843r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dror RO, et al. Activation mechanism of the β2-adrenergic receptor. Proc. Natl. Acad. Sci. USA. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michino M, et al. Toward Understanding the Structural Basis of Partial Agonism at the Dopamine D3 Receptor. J. Med. Chem. 2017;60:580–593. doi: 10.1021/acs.jmedchem.6b01148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 7.Munk C, et al. GPCRdb: the G protein-coupled receptor database – an introduction. Br. J. Pharmacol. 2016;173:2195–2207. doi: 10.1111/bph.13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michino M, et al. What Can Crystal Structures of Aminergic Receptors Tell Us about Designing Subtype-Selective Ligands? Pharmacol. Rev. 2015;67:198–213. doi: 10.1124/pr.114.009944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokoloff P, Giros B, Martres M-P, Bouthenet M-L, Schwartz J-C. Molecular cloning and characterization of a novel dopamine receptor (D3) as a target for neuroleptics. Nature. 1990;347:146–151. doi: 10.1038/347146a0. [DOI] [PubMed] [Google Scholar]
- 10.Newman AH, Grundt P, Nader MA. Dopamine D3 Receptor Partial Agonists and Antagonists as Potential Drug Abuse Therapeutic Agents. J. Med. Chem. 2005;48:3663–3679. doi: 10.1021/jm040190e. [DOI] [PubMed] [Google Scholar]
- 11.Heidbreder CA, Newman AH. Current perspectives on selective dopamine D(3) receptor antagonists as pharmacotherapeutics for addictions and related disorders. Ann. N. Y. Acad. Sci. 2010;1187:4–34. doi: 10.1111/j.1749-6632.2009.05149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho DI, Zheng M, Kim K-M. Current perspectives on the selective regulation of dopamine D2 and D3 receptors. Arch. Pharm. Res. 2010;33:1521–1538. doi: 10.1007/s12272-010-1005-8. [DOI] [PubMed] [Google Scholar]
- 13.Keck TM, John WS, Czoty PW, Nader MA, Newman AH. Identifying Medication Targets for Psychostimulant Addiction: Unraveling the Dopamine D3 Receptor Hypothesis. J. Med. Chem. 2015;58:5361–5380. doi: 10.1021/jm501512b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman AH, et al. Molecular Determinants of Selectivity and Efficacy at the Dopamine D3 Receptor. J. Med. Chem. 2012;55:6689–6699. doi: 10.1021/jm300482h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wager TT, et al. Dopamine D3/D2 Receptor Antagonist PF-4363467 Attenuates Opioid Drug-Seeking Behavior without Concomitant D2 Side Effects. ACS Chem. Neurosci. 2017;8:165–177. doi: 10.1021/acschemneuro.6b00297. [DOI] [PubMed] [Google Scholar]
- 16.Chien EYT, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buch I, Giorgino T, De Fabritiis G. Complete reconstruction of an enzyme-inhibitor binding process by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2011;108:10184–10189. doi: 10.1073/pnas.1103547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferruz N, Tresadern G, Pineda-Lucena A, De Fabritiis G. Multibody cofactor and substrate molecular recognition in the myo-inositol monophosphatase enzyme. Sci. Rep. 2016;6:30275. doi: 10.1038/srep30275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferruz N, Harvey MJ, Mestres J, De Fabritiis G. Insights from Fragment Hit Binding Assays by Molecular Simulations. J. Chem. Inf. Model. 2015;55:2200–2205. doi: 10.1021/acs.jcim.5b00453. [DOI] [PubMed] [Google Scholar]
- 20.Plattner N, Noé F. Protein conformational plasticity and complex ligand-binding kinetics explored by atomistic simulations and Markov models. Nat. Commun. 2015;6:7653. doi: 10.1038/ncomms8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohlhoff KJ, et al. Cloud-based simulations on Google Exacycle reveal ligand modulation of GPCR activation pathways. Nat. Chem. 2014;6:15–21. doi: 10.1038/nchem.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plattner N, Doerr S, De Fabritiis G, Noé F. Complete protein-protein association kinetics in atomic detail revealed by molecular dynamics simulations and Markov modelling. Nat. Chem. 2017;9:1005–1011. doi: 10.1038/nchem.2785. [DOI] [PubMed] [Google Scholar]
- 23.Braden MR, Nichols DE. Assessment of the Roles of Serines 5.43(239) and 5.46(242) for Binding and Potency of Agonist Ligands at the Human Serotonin 5-HT2A Receptor. Mol. Pharmacol. 2007;72:1200–1209. doi: 10.1124/mol.107.039255. [DOI] [PubMed] [Google Scholar]
- 24.Thomas T, Fang Y, Yuriev E, Chalmers DK. Ligand Binding Pathways of Clozapine and Haloperidol in the Dopamine D2 and D3 Receptors. J. Chem. Inf. Model. 2016;56:308–321. doi: 10.1021/acs.jcim.5b00457. [DOI] [PubMed] [Google Scholar]
- 25.Zanatta G, et al. ACS Chem. Neurosci. 2014. Antipsychotic haloperidol binding to the human dopamine D3receptor: beyond docking through QM/MM refinement toward the design of improved schizophrenia medicines; pp. 1041–1054. [DOI] [PubMed] [Google Scholar]
- 26.Vangveravong S, et al. Synthesis and characterization of selective dopamine D2 receptor ligands using aripiprazole as the lead compound. Bioorg. Med. Chem. 2011;19:3502–3511. doi: 10.1016/j.bmc.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alberts GL, Pregenzer JF, Bin Im W. Contributions of cysteine 114 of the human D3 dopamine receptor to ligand binding and sensitivity to external oxidizing agents. Br. J. Pharmacol. 1998;125:705–710. doi: 10.1038/sj.bjp.0702136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Micheli F, et al. 1,2,4-Triazolyl octahydropyrrolo[2,3-b]pyrroles: A new series of potent and selective dopamine D3 receptor antagonists. Bioorg. Med. Chem. 2016;24:1619–1636. doi: 10.1016/j.bmc.2016.02.031. [DOI] [PubMed] [Google Scholar]
- 29.Buch I, Harvey MJ, Giorgino T, Anderson DP, De Fabritiis G. High-Throughput All-Atom Molecular Dynamics Simulations Using Distributed Computing. J. Chem. Inf. Model. 2010;50:397–403. doi: 10.1021/ci900455r. [DOI] [PubMed] [Google Scholar]
- 30.Harvey MJ, Giupponi G, De Fabritiis G. ACEMD: Accelerating Biomolecular Dynamics in the Microsecond Time Scale. J. Chem. Theory Comput. 2009;5:1632–1639. doi: 10.1021/ct9000685. [DOI] [PubMed] [Google Scholar]
- 31.Doerr S, Harvey MJ, Noé F, De Fabritiis G. HTMD: High-Throughput Molecular Dynamics for Molecular Discovery. J. Chem. Theory Comput. 2016;12:1845–1852. doi: 10.1021/acs.jctc.6b00049. [DOI] [PubMed] [Google Scholar]
- 32.Doerr S, De Fabritiis G. On-the-Fly Learning and Sampling of Ligand Binding by High-Throughput Molecular Simulations. J. Chem. Theory Comput. 2014;10:2064–2069. doi: 10.1021/ct400919u. [DOI] [PubMed] [Google Scholar]
- 33.Deuflhard P, Weber M. Robust Perron cluster analysis in conformation dynamics. Linear Algebra Its Appl. 2005;398:161–184. doi: 10.1016/j.laa.2004.10.026. [DOI] [Google Scholar]
- 34.Wang S, et al. D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science. 2017;358:381–386. doi: 10.1126/science.aan5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wescott MP, et al. Signal transmission through the CXC chemokine receptor 4 (CXCR4) transmembrane helices. Proc. Natl. Acad. Sci. USA. 2016;113:9928–9933. doi: 10.1073/pnas.1601278113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weng W-H, Li Y-T, Hsu H-J. Activation-Induced Conformational Changes of Dopamine D3 Receptor Promote the Formation of the Internal Water Channel. Sci. Rep. 2017;7:12792. doi: 10.1038/s41598-017-13155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR Dynamics: Structures in Motion. Chem. Rev. 2017;117:139–155. doi: 10.1021/acs.chemrev.6b00177. [DOI] [PubMed] [Google Scholar]
- 38.Friesner RA, et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 39.Halgren TA, et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 40.Friesner RA, et al. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 41.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 42.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J. Med. Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 43.Farid R, Day T, Friesner RA, Pearlstein RA. New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorg. Med. Chem. 2006;14:3160–3173. doi: 10.1016/j.bmc.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 44.Dolinsky TJ, et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of Proteins in Membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 46.Klauda JB, Monje V, Kim T, Im W. Improving the CHARMM force field for polyunsaturated fatty acid chains. J. Phys. Chem. B. 2012;116:9424–9431. doi: 10.1021/jp304056p. [DOI] [PubMed] [Google Scholar]
- 47.Huang L, Roux B. Automated Force Field Parameterization for Nonpolarizable and Polarizable Atomic Models Based on Ab Initio Target Data. J. Chem. Theory Comput. 2013;9:3543–3556. doi: 10.1021/ct4003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mark P, Nilsson L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J Phys Chem A. 2001;105:9954–9960. doi: 10.1021/jp003020w. [DOI] [Google Scholar]
- 49.Bowman, G. R., Beauchamp, K. A., Boxer, G. & Pande, V. S. Progress and challenges in the automated construction of Markov state models for full protein systems. J. Chem. Phys. 131, (2009). [DOI] [PMC free article] [PubMed]
- 50.Pérez-Hernández G, Paul F, Giorgino T, De Fabritiis G, Noé F. Identification of slow molecular order parameters for Markov model construction. J. Chem. Phys. 2013;139:15102. doi: 10.1063/1.4811489. [DOI] [PubMed] [Google Scholar]
- 51.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.