Abstract
Multiple myeloma (MM) is a malignant disorder characterized by the neoplastic growth of plasma cells in the bone marrow. MicroRNAs (miRNAs/miRs) modulate key regulatory cell pathways via their influence on target genes, and may serve a crucial function in tumorigenesis. Previous studies have indicated that the downregulation of miR-15a and miR-16 contributes to MM pathogenesis. However, the functional mechanisms of miR-15a and miR-16 in MM remain unclear. In the present study, potential target sites for miR-15a and miR-16 were identified on the calcineurin-binding protein 1 (CABIN1) mRNA sequence from analyses of previously published crosslinking, ligation and sequencing of hybrids data. Again-of-function study was also performed, which determined that miR-15a/16 directly targeted CABIN1 mRNA and negatively regulated the expression of CABIN1 at the mRNA and protein level in MM cells. A cell proliferation assay demonstrated that the upregulation of miR-15a and miR-16 inhibited the proliferation of MM cells via targeting CABIN1. miR-15a and miR-16 were significantly decreased in MM specimens, compared with in normal specimens, whereas CABIN1 mRNA levels were significantly higher in MM samples compared with in normal samples. CABIN1 mRNA levels were negatively correlated with miR-15a and miR-16 expression levels in MM tissues, as determined using Pearson's correlation coefficient analysis. The results of the present study indicate that the downregulation of miR-15a and miR-16 promotes tumor proliferation in MM by increasing CABIN1 expression. The present study may aid elucidation of the functions of miR-15a and miR-16 and their function in MM carcinogenesis.
Keywords: multiple myeloma, microRNA-15a/16, calcineurin-binding protein-1, cell proliferation
Introduction
Multiple myeloma (MM) is a malignant disorder characterized by the neoplastic growth of plasma cells in the bone marrow (1). Plasma cells are a type of white blood cell normally responsible for producing antibodies; the accumulation of abnormal plasma cells interferes with the production of normal blood cells in the bone marrow (1). MM is also characterized by the production of a paraprotein, an abnormal antibody detected in the serum or urine in the majority patients, which can cause kidney problems (2). Patients with MM exhibit a range of clinical symptoms, including bone lesions, anemia and hypercalcemia (2).
Multiple myeloma is the second most common type of hematological malignancy (comprising 13% of these diseases), and constitutes ~1% of all neoplastic diseases (2). Although strategies for the prevention, diagnosis and treatment of MM have improved, the 5-year relative survival rate is only 39% for patients with MM in Europe (3). Therefore, elucidation of the mechanisms underlying the initiation and progression of MM is essential.
MicroRNAs (miRNAs/miRs) are highly conserved short non-coding sequences of ~20 nucleotides that are able to silence target genes by acting as inhibitors of protein translation or by degrading mRNA (4). miRNAs modulate regulatory cell pathways by influencing target genes and may serve crucial functions in oncogenesis (5). Evidence indicates that miRNAs regulate MM tumorigenesis and can be valuable markers for predicting diagnosis, risk stratification and clinical outcomes (6–8).
miR-15a and miR16 are clustered at chromosomal location 13q14 and possess similar sequences; they are considered to have similar tumor suppressor functions and to be involved in the regulation of cell differentiation, proliferation, apoptosis or angiogenesis in several types of human cancer, including MM (8–12). Previous studies have indicated that miR-15a/16-1 downregulation contributes to myeloma pathogenesis and mediates drug resistance in myeloma cells (13,14). However, the functional mechanism of miR-15a/16 in MM remains unclear.
In the present study, the expression of miR-15a/16 and their functional mechanism was investigated in MM cells and patients. To the best of our knowledge, the present study provides the first evidence to support miR-15a/16 directly targeting calcineurin-binding protein 1 (CABIN1) mRNA and negatively regulating them RNA and protein expression of CABIN1 in MM cells in a gain-of-function study. Using cell proliferation, the results demonstrated that the upregulation of miR-15a and miR-16 inhibited the proliferation of MM cells by targeting CABIN1. The present study also demonstrated that miR-15a and miR-16 are downregulated and negatively associated with CABIN1 mRNA levels in MM specimens. Data obtained in the present study may aid elucidation of the functions of miR-15a and miR-16 and their functions in multiple myeloma carcinogenesis.
Materials and methods
Specimens
Plasma specimens were obtained from 30 patients with MM and 10 healthy volunteers from Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine (Shanghai, China). Plasma cells from patients with MM and normal controls were purified from bone marrow aspirates using CD138 MicroBeads (cat. no. 130-051-301; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany), as described previously (15). All patients provided written informed consent to participate in the study. This study was approved by the Ethics Committee of Shanghai University of Traditional Chinese Medicine.
Cell culture
The U266 and RPMI-8226 MM plasma cell myeloma cell lines, and the 293 cell line, used in the present study were purchased from the American Type Culture Collection (Manassas, VA, USA). The U266 and RPMI-8226 cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Inc.). The 293 cells were cultured in Dulbecco's minimum essential medium (DMEM) (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS. Cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from MM tissues, adjacent non-tumor tissues and MM cell lines using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Purified mRNA and miRNA were detected using an RT-qPCR assay with the All-in-One miRNA RT-qPCR Detectionkit (GeneCopoeia, Inc., Rockville, MD, USA), according to the manufacturer's protocol. All primer sequences are listed in Table I. U6 small nuclear RNA was used as an internal control for the normalization and quantification of miRNA expression. β-actin was used as the internal control for the normalization and quantification of CABIN1 mRNA expression as described previously (16).
Table I.
Primers used in the present study.
| Name | Primer sequence |
|---|---|
| miR-15a F | 5′-TAGCAGCACATAATGGTTTGTG-3′ |
| miR-16 F | 5′-TAGCAGCACGTAAATATTGGCG-3′ |
| U6 F | 5′-CTCGCTTCGGCAGCACATATAC-3′ |
| U6 R | 5′-AATATGGAACGCTCACGAATTTG-3′ |
| β-actin F | 5′-CAGCAAGCAGGAGTATGACG-3′ |
| β-actin R | 5′-GAAAGGGTGTAACGCAACTAA-3′ |
| CABIN1 F | 5′-ATCCTCACTGTGAAGGTGCTCGAA-3′ |
| CABIN1 R | 5′-TTTGGTCTGCACTGTCTCCTGCAT-3′ |
| CABIN1 (reporter) F | 5′-AAACTAGTTTGCCTGGCACATGAA CCG-3′ |
| CABIN1 (reporter) R | 5′-GGAAGCTTGGTCCCGCAGCTGGGCCAGC-3′ |
miR-15a, microRNA-15a; F, forward primer; R, reverse primer; CABIN1, calcineurin-binding protein 1.
Dual-luciferase reporter assays
The luciferase reporter construct was created by cloningthehuman CABIN1 mRNA sequence into a pMIR-Report construct (Ambion; Thermo Fisher Scientific, Inc.). CABIN1 mRNA fragment (from 5178 to 5240) was amplified and cloned into the luciferase reporter via SpeI and HindIII sites using Ready-to-Use Seamless Cloning kit (Songon Biotech, Shanghai, China), according to the manufacturer's protocol. All primers are listed in Table I. 293 cells (1×104) were plated in a 96-well plate and co-transfected with 50 nM miRNA mimics or negative control oligonucleotides RNA oligonucleotides were chemically synthesized and purified by Shanghai GenePharma Co., Ltd. (Shanghai, China). A total of 30 ng firefly luciferase reporter and 10 ng pRL-TK (Promega Corporation, Madison, WI, USA) were transfected into the cells using the Interferin® reagent (Polyplus-transfection SA, Illkirch, France). Cells were collected 48 h after the last transfection and analyzed using the Dual-Luciferase Reporter Assay system (Promega Corporation), according to the manufacturer's protocol. The firefly luciferase activity of each sample was normalized to the Renilla luciferase activity.
Oligonucleotide transfection
RNA oligonucleotides were chemically synthesized and purified by Shanghai GenePharma Co., Ltd. The sequence of human miR-15a was 5′-UAGCAGCACAUAAUGGUUUGUG-3′, and that of human miR-16 was 5′-UAGCAGCACGUAAAUAUUGGCG-3′. The negative-control oligonucleotide sequence was 5′-CAGUACUUUUGUGUAGUACAA-3′. The small interfering RNA (siRNA) sequences were as follows: CABIN1 sense, 5′-CUACAUUGAGGGAACUUCATT-3′ and antisense, 5′-UGAAGUUCCCUCAAUGUAGTT-3′; control sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGUAGAATT-3′. Transfections were performed using Interferin® reagent (Polyplus-transfection SA). The final concentration of miRNA was 50 nM; the final concentration of siRNAs was 20 nM.
Cell counting kit-8 (CCK-8) assay
Cell proliferation was measured using a CCK-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer's protocol. A total of 1.0×104 cells were plated into each well of a 96-well plate and transfected with miRNA or CABIN1 siRNA to a final concentration of 50 or 20 nM, respectively using Interferin® reagent (Polyplus-transfection SA), according to the manufacturer's protocol. Once the cells were harvested, 10 µl CCK-8 reagent was added to 90 µl RPMI-1640 medium. The cells were subsequently incubated for 2 h at 37°C and the optical density was measured at 450 nm. Three independent experiments were performed.
Western blot analysis
Total cellular extracts (20 µg) were separated by SDS-PAGE (4–20% Tris-glycine gel) then transferred onto a polyvinylidene fluoride membrane (Immobilon-P transfer membranes; EMD Millipore, Billerica, MA, USA). Following transfer, the blots were blocked with 5% non-fat dry milk in PBS and 0.1% Tween-20 for 2 h at 4°C and washed three times with PBS and 0.1% Tween-20 at 4°C. The blots were then probed with the primary anti-CABIN1 antibody (cat. no. ab3349; dilution, 1:200; Abcam, Cambridge, MA, USA), CDKN1A (cat. no. sc-6246; dilution, 1:200) and β-actin (cat. no. c-11;1: 200; Santa Cruz Biotechnology, Inc.). The blots were then probed with the horseradish peroxidase-conjugated goat anti-rabbit IgG H&L secondary antibody, (cat. no. ab6721, dilution, 1:750; Abcam) for 1 h at 4°C, followed by washes in PBS + 0.1% Tween-20, and then detected using an Odyssey Scanning system (Gene Company, Ltd., Hong Kong, China).
Statistical analysis
All statistical analyses were performed using the SPSS 16.0 statistical software package (SPSS, Inc., Chicago, IL, USA). Quantitative data is expressed as mean ± standard deviation. Data of mRNA expression levels in MM specimens compared to healthy volunteers were subjected to Student's t-test analysis. Data from dual-luciferase reporter assays and CCK-8 assay were analyzed by one-way analysis of variance followed by the Student-Newman-Keuls post-test. P<0.05 was considered to indicate a statistically significant difference. Pearson's correlation coefficient and Spearman's correlation coefficient was used forthe comparison of mRNA expression of miRNA and its target gene.
Results
miR-15a/16 have potential target sites on CABIN1 mRNA
miR-15a and miR-16, whose expression is decreased or absent in MM, have similar sequences and are considered to have the same target genes. To investigate the biological significance of these miRNAs and the underlying mechanisms of their downregulation in MM, the potential target genes of miR-15a/16 that have functions in MM pathogenesis were further analyzed.
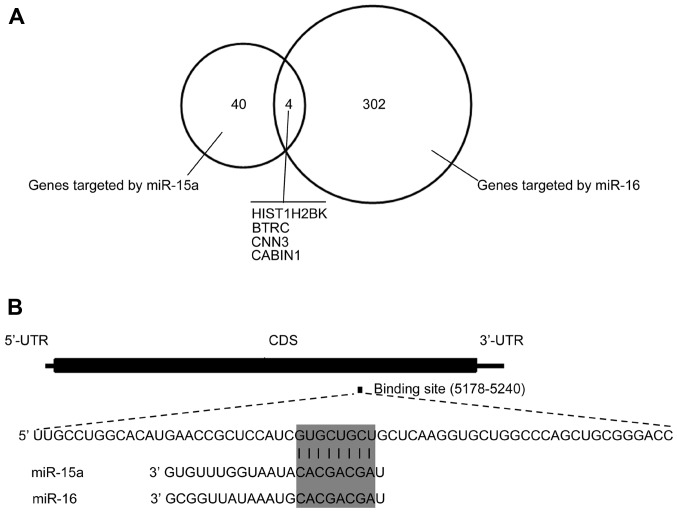
Previously published crosslinking, ligation, and sequencing of hybrids (CLASH) data in 293 cells provided direct evidence for the miRNA-mRNA pairing (17). The CLASH data revealed that four genes, HIST1H2K, BTRC, CNN3 and CABIN1, aretargeted by miR-15a and miR-16 (Fig. 1A). All genes targeted by miR-15aor miR-16 in the CLASH dataset (17) are listed in Table II. CABIN1 is involved in several cellular processes including cell proliferation and the restraint of p53 activity on chromatin (18); therefore, CABIN1 was selected for further investigation. Alignment results indicated that the potential targeting sequence for miR-15a/16 was within the protein-coding region of CABIN1 mRNA between nucleotides 5,178 and 5,240 (Fig. 1B).
Figure 1.
Bioinformatic analysis of the miR-15a/16 binding site in CABIN1 mRNA. (A) HIST1H2BK, BTRC, CNN3 and CABIN1 are targeted by miR-15a and miR-16, according to CLASH data (16). (B) Schematic diagrams of the CABIN1 mRNA and the potential binding site for the miR-15a and miR-16. The upper panel depicts a potential target site on the CDS of CABIN1; the lower panel illustrates the sequence alignment of miR-15a and miR-16 with the binding site on the CDS of CABIN1. CABIN1, calcineurin-binding protein 1, miR-15a, microRNA-15a; BTRC, β-transducin repeat containing e3 ubiquitin protein ligase; CNN3, calponin 3, UTR, untranslated region; CDS, coding sequence.
Table II.
List of genes that paired with miR-15a and miR-16 in CLASH data.
| miRNA | Target gene |
|---|---|
| miR-15a | ENSG00000099991_ENST00000398319_CABIN1 |
| miR-16 | ENSG00000099991_ENST00000398319_CABIN1 |
| miR-15a | ENSG00000110429_ENST00000265651_FBXO3 |
| miR-16 | ENSG00000110429_ENST00000265651_FBXO3 |
| miR-15a | ENSG00000117519_ENST00000370206_CNN3 |
| miR-16 | ENSG00000117519_ENST00000370206_CNN3 |
| miR-15a | ENSG00000197903_ENST00000396891_HIST1H2BK |
| miR-16 | ENSG00000197903_ENST00000396891_HIST1H2BK |
| miR-15a | ENSG00000013275_ENST00000157812_PSMC4 |
| miR-15a | ENSG00000015568_ENST00000016946_RGPD5 |
| miR-15a | ENSG00000023909_ENST00000370238_GCLM |
| miR-15a | ENSG00000025796_ENST00000369002_SEC63 |
| miR-15a | ENSG00000057608_ENST00000380191_GDI2 |
| miR-15a | ENSG00000069956_ENST00000261845_MAPK6 |
| miR-15a | ENSG00000071553_ENST00000369762_ATP6AP1 |
| miR-15a | ENSG00000073009_ENST00000369606_IKBKG |
| miR-15a | ENSG00000110092_ENST00000227507_CCND1 |
| miR-15a | ENSG00000110435_ENST00000227868_PDHX |
| miR-15a | ENSG00000111641_ENST00000399466_NOP2 |
| miR-15a | ENSG00000111642_ENST00000357008_CHD4 |
| miR-15a | ENSG00000113312_ENST00000231238_TTC1 |
| miR-15a | ENSG00000115758_ENST00000234111_ODC1 |
| miR-15a | ENSG00000115966_ENST00000264110_ATF2 |
| miR-15a | ENSG00000117215_ENST00000375105_PLA2G2D |
| miR-15a | ENSG00000118971_ENST00000261254_CCND2 |
| miR-15a | ENSG00000119777_ENST00000238788_TMEM214 |
| miR-15a | ENSG00000121940_ENST00000415331_CLCC1 |
| miR-15a | ENSG00000122786_ENST00000361901_CALD1 |
| miR-15a | ENSG00000129515_ENST00000396526_SNX6 |
| miR-15a | ENSG00000130294_ENST00000320389_KIF1A |
| miR-15a | ENSG00000130726_ENST00000253024_TRIM28 |
| miR-15a | ENSG00000131023_ENST00000253339_BX276089-1 |
| miR-15a | ENSG00000132141_ENST00000314144_CCT6B |
| miR-15a | ENSG00000133059_ENST00000367162_DSTYK |
| miR-15a | ENSG00000135503_ENST00000257963_ACVR1B |
| miR-15a | ENSG00000138073_ENST00000260643_PREB |
| miR-15a | ENSG00000138381_ENST00000260952_ASNSD1 |
| miR-15a | ENSG00000138386_ENST00000337386_NAB1 |
| miR-15a | ENSG00000148688_ENST00000413330_RPP30 |
| miR-15a | ENSG00000150907_ENST00000379561_FOXO1 |
| miR-15a | ENSG00000163249_ENST00000295414_CCNYL1 |
| miR-15a | ENSG00000165494_ENST00000298281_PCF11 |
| miR-15a | ENSG00000165672_ENST00000298510_PRDX3 |
| miR-15a | ENSG00000166167_ENST00000370187_BTRC |
| miR-15a | ENSG00000169710_ENST00000306749_FASN |
| miR-15a | ENSG00000185658_ENST00000341322_BRWD1 |
| miR-15a | ENSG00000185697_ENST00000402282_MYBL1 |
| miR-15a | ENSG00000188647_ENST00000415701_PTAR1 |
| miR-16 | ENSG00000005302_ENST00000337339_MSL3 |
| miR-16 | ENSG00000005700_ENST00000306270_IBTK |
| miR-16 | ENSG00000008438_ENST00000008938_PGLYRP1 |
| miR-16 | ENSG00000008516_ENST00000336577_MMP25 |
| miR-16 | ENSG00000008838_ENST00000394126_MED24 |
| miR-16 | ENSG00000009954_ENST00000339594_BAZ1B |
| miR-16 | ENSG00000010322_ENST00000345716_NISCH |
| miR-16 | ENSG00000010404_ENST00000340855_IDS |
| miR-16 | ENSG00000011028_ENST00000303375_MRC2 |
| miR-16 | ENSG00000035403_ENST00000372755_VCL |
| miR-16 | ENSG00000037474_ENST00000264670_NSUN2 |
| miR-16 | ENSG00000050405_ENST00000341247_LIMA1 |
| miR-16 | ENSG00000056277_ENST00000370978_ZNF280C |
| miR-16 | ENSG00000063046_ENST00000262056_EIF4B |
| miR-16 | ENSG00000064419_ENST00000265388_TNPO3 |
| miR-16 | ENSG00000065150_ENST00000261574_IPO5 |
| miR-16 | ENSG00000065526_ENST00000375759_SPEN |
| miR-16 | ENSG00000065665_ENST00000379033_SEC61A2 |
| miR-16 | ENSG00000066926_ENST00000262093_FECH |
| miR-16 | ENSG00000067836_ENST00000322048_ROGDI |
| miR-16 | ENSG00000069998_ENST00000336737_CECR5 |
| miR-16 | ENSG00000070501_ENST00000265421_POLB |
| miR-16 | ENSG00000070882_ENST00000313367_OSBPL3 |
| miR-16 | ENSG00000072110_ENST00000193403_ACTN1 |
| miR-16 | ENSG00000072135_ENST00000175756_PTPN18 |
| miR-16 | ENSG00000072274_ENST00000360110_TFRC |
| miR-16 | ENSG00000072803_ENST00000296933_FBXW11 |
| miR-16 | ENSG00000074755_ENST00000381638_ZZEF1 |
| miR-16 | ENSG00000075292_ENST00000409544_ZNF638 |
| miR-16 | ENSG00000075624_ENST00000331789_ACTB |
| miR-16 | ENSG00000076604_ENST00000262395_TRAF4 |
| miR-16 | ENSG00000076826_ENST00000446248_ KIAA1543 |
| miR-16 | ENSG00000083544_ENST00000411610_TDRD3 |
| miR-16 | ENSG00000085872_ENST00000198939_CHERP |
| miR-16 | ENSG00000086570_ENST00000261800_FAT2 |
| miR-16 | ENSG00000086712_ENST00000380122_CXorf15 |
| miR-16 | ENSG00000088387_ENST00000357329_DOCK9 |
| miR-16 | ENSG00000088899_ENST00000329152_RP5-1187M17-10 |
| miR-16 | ENSG00000089157_ENST00000228306_RPLP0 |
| miR-16 | ENSG00000089693_ENST00000203630_MLF2 |
| miR-16 | ENSG00000090376_ENST00000457197_IRAK3 |
| miR-16 | ENSG00000090621_ENST00000372857_PABPC4 |
| miR-16 | ENSG00000091164_ENST00000217515_TXNL1 |
| miR-16 | ENSG00000093000_ENST00000347635_NUP50 |
| miR-16 | ENSG00000093010_ENST00000361682_COMT |
| miR-16 | ENSG00000095564_ENST00000265990_BTAF1 |
| miR-16 | ENSG00000095739_ENST00000375533_BAMBI |
| miR-16 | ENSG00000096968_ENST00000381652_JAK2 |
| miR-16 | ENSG00000099203_ENST00000214869_TMED1 |
| miR-16 | ENSG00000099991_ENST00000398319_CABIN1 |
| miR-16 | ENSG00000100234_ENST00000266085_TIMP3 |
| miR-16 | ENSG00000100241_ENST00000380817_SBF1 |
| miR-16 | ENSG00000100304_ENST00000216129_TTLL12 |
| miR-16 | ENSG00000100304_ENST00000216129_TTLL12 |
| miR-16 | ENSG00000100697_ENST00000343455_DICER1 |
| miR-16 | ENSG00000100764_ENST00000261303_PSMC1 |
| miR-16 | ENSG00000101166_ENST00000355937_SLMO2 |
| miR-16 | ENSG00000101190_ENST00000335351_TCFL5 |
| miR-16 | ENSG00000101294_ENST00000340852_HM13 |
| miR-16 | ENSG00000101452_ENST00000252011_DHX35 |
| miR-16 | ENSG00000101782_ENST00000339486_RIOK3 |
| miR-16 | ENSG00000102174_ENST00000379374_PHEX |
| miR-16 | ENSG00000102931_ENST00000219204_ARL2BP |
| miR-16 | ENSG00000103148_ENST00000399953_C16orf35 |
| miR-16 | ENSG00000103222_ENST00000399410_ABCC1 |
| miR-16 | ENSG00000103319_ENST00000263026_EEF2K |
| miR-16 | ENSG00000103489_ENST00000261381_XYLT1 |
| miR-16 | ENSG00000104331_ENST00000262644_IMPAD1 |
| miR-16 | ENSG00000104341_ENST00000445593_LAPTM4B |
| miR-16 | ENSG00000104824_ENST00000221419_HNRNPL |
| miR-16 | ENSG00000104824_ENST00000221419_HNRNPL |
| miR-16 | ENSG00000105173_ENST00000262643_CCNE1 |
| miR-16 | ENSG00000105223_ENST00000409587_PLD3 |
| miR-16 | ENSG00000105281_ENST00000306894_SLC1A5 |
| miR-16 | ENSG00000105429_ENST00000334370_MEGF8 |
| miR-16 | ENSG00000105784_ENST00000338056_RUNDC3B |
| miR-16 | ENSG00000106009_ENST00000340611_C7orf27 |
| miR-16 | ENSG00000106355_ENST00000450169_LSM5 |
| miR-16 | ENSG00000106665_ENST00000361545_CLIP2 |
| miR-16 | ENSG00000107937_ENST00000360803_GTPBP4 |
| miR-16 | ENSG00000108510_ENST00000397786_MED13 |
| miR-16 | ENSG00000109775_ENST00000264689_UFSP2 |
| miR-16 | ENSG00000109971_ENST00000227378_HSPA8 |
| miR-16 | ENSG00000109971_ENST00000227378_HSPA8 |
| miR-16 | ENSG00000110367_ENST00000264018_DDX6 |
| miR-16 | ENSG00000110713_ENST00000324932_NUP98 |
| miR-16 | ENSG00000111012_ENST00000228606_CYP27B1 |
| miR-16 | ENSG00000112242_ENST00000346618_E2F3 |
| miR-16 | ENSG00000112306_ENST00000230050_RPS12 |
| miR-16 | ENSG00000112592_ENST00000392092_TBP |
| miR-16 | ENSG00000112664_ENST00000358797_NUDT3 |
| miR-16 | ENSG00000112701_ENST00000447266_SENP6 |
| miR-16 | ENSG00000113569_ENST00000231498_NUP155 |
| miR-16 | ENSG00000113732_ENST00000519374_ATP6V0E1 |
| miR-16 | ENSG00000114030_ENST00000344337_KPNA1 |
| miR-16 | ENSG00000114933_ENST00000403263_INO80D |
| miR-16 | ENSG00000114982_ENST00000354204_KIAA1310 |
| miR-16 | ENSG00000115307_ENST00000377526_AUP1 |
| miR-16 | ENSG00000115616_ENST00000233969_SLC9A2 |
| miR-16 | ENSG00000116396_ENST00000369787_KCNC4 |
| miR-16 | ENSG00000116685_ENST00000376572_KIAA2013 |
| miR-16 | ENSG00000116748_ENST00000369538_AMPD1 |
| miR-16 | ENSG00000116786_ENST00000420314_PLEKHM2 |
| miR-16 | ENSG00000117984_ENST00000236671_CTSD |
| miR-16 | ENSG00000120158_ENST00000381750_RCL1 |
| miR-16 | ENSG00000120438_ENST00000321394_TCP1 |
| miR-16 | ENSG00000120833_ENST00000340600_SOCS2 |
| miR-16 | ENSG00000122482_ENST00000370440_ZNF644 |
| miR-16 | ENSG00000122566_ENST00000354667_HNRNPA2B1 |
| miR-16 | ENSG00000123384_ENST00000243077_AC137834-1 |
| miR-16 | ENSG00000123411_ENST00000262032_IKZF4 |
| miR-16 | ENSG00000124529_ENST00000377364_HIST1H4B |
| miR-16 | ENSG00000125166_ENST00000245206_GOT2 |
| miR-16 | ENSG00000125166_ENST00000245206_GOT2 |
| miR-16 | ENSG00000126016_ENST00000304758_AMOT |
| miR-16 | ENSG00000126464_ENST00000418929_PRR12 |
| miR-16 | ENSG00000126602_ENST00000246957_TRAP1 |
| miR-16 | ENSG00000127152_ENST00000345514_BCL11B |
| miR-16 | ENSG00000127616_ENST00000429416_SMARCA4 |
| miR-16 | ENSG00000128714_ENST00000392539_HOXD13 |
| miR-16 | ENSG00000129566_ENST00000262715_TEP1 |
| miR-16 | ENSG00000130054_ENST00000252338_FAM155B |
| miR-16 | ENSG00000130227_ENST00000252512_XPO7 |
| miR-16 | ENSG00000130402_ENST00000252699_ACTN4 |
| miR-16 | ENSG00000130640_ENST00000368563_TUBGCP2 |
| miR-16 | ENSG00000130733_ENST00000393508_YIPF2 |
| miR-16 | ENSG00000131269_ENST00000253577_ABCB7 |
| miR-16 | ENSG00000131467_ENST00000441946_PSME3 |
| miR-16 | ENSG00000132153_ENST00000443212_DHX30 |
| miR-16 | ENSG00000132341_ENST00000448750_RAN |
| miR-16 | ENSG00000132423_ENST00000254759_COQ3 |
| miR-16 | ENSG00000132676_ENST00000368336_DAP3 |
| miR-16 | ENSG00000132718_ENST00000368324_SYT11 |
| miR-16 | ENSG00000132953_ENST00000255305_XPO4 |
| miR-16 | ENSG00000133124_ENST00000372129_IRS4 |
| miR-16 | ENSG00000133243_ENST00000255608_BTBD2 |
| miR-16 | ENSG00000133433_ENST00000290765_GSTT2B |
| miR-16 | ENSG00000133773_ENST00000256151_CCDC59 |
| miR-16 | ENSG00000134049_ENST00000256433_IER3IP1 |
| miR-16 | ENSG00000134817_ENST00000257254_APLNR |
| miR-16 | ENSG00000134871_ENST00000360467_COL4A2 |
| miR-16 | ENSG00000134970_ENST00000456936_TMED7 |
| miR-16 | ENSG00000135269_ENST00000358204_TES |
| miR-16 | ENSG00000135341_ENST00000369329_MAP3K7 |
| miR-16 | ENSG00000135898_ENST00000392040_GPR55 |
| miR-16 | ENSG00000136695_ENST00000393200_IL1F5 |
| miR-16 | ENSG00000136827_ENST00000437532_TOR1A |
| miR-16 | ENSG00000137713_ENST00000427203_PPP2R1B |
| miR-16 | ENSG00000138041_ENST00000272313_SMEK2 |
| miR-16 | ENSG00000138326_ENST00000435275_RPS24 |
| miR-16 | ENSG00000138326_ENST00000435275_RPS24 |
| miR-16 | ENSG00000138399_ENST00000453153_FASTKD1 |
| miR-16 | ENSG00000138592_ENST00000433963_USP8 |
| miR-16 | ENSG00000138802_ENST00000265175_SEC24B |
| miR-16 | ENSG00000139117_ENST00000331366_CPNE8 |
| miR-16 | ENSG00000140391_ENST00000267970_TSPAN3 |
| miR-16 | ENSG00000140526_ENST00000355100_ABHD2 |
| miR-16 | ENSG00000140829_ENST00000268482_DHX38 |
| miR-16 | ENSG00000140955_ENST00000315906_ADAD2 |
| miR-16 | ENSG00000141101_ENST00000268802_NOB1 |
| miR-16 | ENSG00000141140_ENST00000378326_MYO19 |
| miR-16 | ENSG00000141367_ENST00000269122_CLTC |
| miR-16 | ENSG00000141367_ENST00000269122_CLTC |
| miR-16 | ENSG00000141404_ENST00000334049_GNAL |
| miR-16 | ENSG00000141720_ENST00000269554_PIP5K2B |
| miR-16 | ENSG00000142235_ENST00000270238_LMTK3 |
| miR-16 | ENSG00000142961_ENST00000371940_MOBKL2C |
| miR-16 | ENSG00000144227_ENST00000272641_NXPH2 |
| miR-16 | ENSG00000144659_ENST00000273158_SLC25A38 |
| miR-16 | ENSG00000144852_ENST00000393716_NR1I2 |
| miR-16 | ENSG00000145495_ENST00000274140_MARCH6 |
| miR-16 | ENSG00000145495_ENST00000274140_MARCH6 |
| miR-16 | ENSG00000146833_ENST00000349062_TRIM4 |
| miR-16 | ENSG00000147255_ENST00000370904_IGSF1 |
| miR-16 | ENSG00000147403_ENST00000424325_RPL10 |
| miR-16 | ENSG00000147408_ENST00000454498_CSGALNACT1 |
| miR-16 | ENSG00000147533_ENST00000520817_GOLGA7 |
| miR-16 | ENSG00000149273_ENST00000278572_RPS3 |
| miR-16 | ENSG00000150764_ENST00000440460_DIXDC1 |
| miR-16 | ENSG00000151612_ENST00000379448_ZNF827 |
| miR-16 | ENSG00000152234_ENST00000282050_ATP5A1 |
| miR-16 | ENSG00000152270_ENST00000282096_PDE3B |
| miR-16 | ENSG00000152291_ENST00000377386_TGOLN2 |
| miR-16 | ENSG00000154124_ENST00000284274_FAM105B |
| miR-16 | ENSG00000154760_ENST00000285013_SLFN13 |
| miR-16 | ENSG00000155393_ENST00000299192_HEATR3 |
| miR-16 | ENSG00000156110_ENST00000372734_ADK |
| miR-16 | ENSG00000156261_ENST00000286788_CCT8 |
| miR-16 | ENSG00000156304_ENST00000286835_SFRS15 |
| miR-16 | ENSG00000156486_ENST00000521839_KCNS2 |
| miR-16 | ENSG00000156508_ENST00000316292_EEF1A1 |
| miR-16 | ENSG00000159202_ENST00000360943_UBE2Z |
| miR-16 | ENSG00000159348_ENST00000367249_CYB5R1 |
| miR-16 | ENSG00000160209_ENST00000291565_PDXK |
| miR-16 | ENSG00000160767_ENST00000361361_FAM189B |
| miR-16 | ENSG00000160991_ENST00000356387_ORAI2 |
| miR-16 | ENSG00000161542_ENST00000446526_PRPSAP1 |
| miR-16 | ENSG00000162300_ENST00000294258_ZFPL1 |
| miR-16 | ENSG00000163013_ENST00000521871_FBXO41 |
| miR-16 | ENSG00000163113_ENST00000369135_OTUD7B |
| miR-16 | ENSG00000163527_ENST00000295770_STT3B |
| miR-16 | ENSG00000163946_ENST00000431842_C3orf63 |
| miR-16 | ENSG00000164076_ENST00000477224_CAMKV |
| miR-16 | ENSG00000164300_ENST00000507668_SERINC5 |
| miR-16 | ENSG00000164818_ENST00000297440_HEATR2 |
| miR-16 | ENSG00000164889_ENST00000485713_SLC4A2 |
| miR-16 | ENSG00000165629_ENST00000356708_ATP5C1 |
| miR-16 | ENSG00000165995_ENST00000377315_CACNB2 |
| miR-16 | ENSG00000165995_ENST00000396576_CACNB2 |
| miR-16 | ENSG00000166105_ENST00000431683_GLB1L3 |
| miR-16 | ENSG00000166167_ENST00000370187_BTRC |
| miR-16 | ENSG00000166681_ENST00000372645_NGFRAP1 |
| miR-16 | ENSG00000166833_ENST00000396085_NAV2 |
| miR-16 | ENSG00000167397_ENST00000394975_VKORC1 |
| miR-16 | ENSG00000167552_ENST00000301071_TUBA1A |
| miR-16 | ENSG00000167721_ENST00000301364_TSR1 |
| miR-16 | ENSG00000167842_ENST00000381165_MIS12 |
| miR-16 | ENSG00000168137_ENST00000402198_SETD5 |
| miR-16 | ENSG00000168268_ENST00000422318_NT5DC2 |
| miR-16 | ENSG00000168488_ENST00000336783_ATXN2L |
| miR-16 | ENSG00000168758_ENST00000305476_SEMA4C |
| miR-16 | ENSG00000169180_ENST00000304658_XPO6 |
| miR-16 | ENSG00000169251_ENST00000351193_NMD3 |
| miR-16 | ENSG00000169813_ENST00000443950_HNRNPF |
| miR-16 | ENSG00000170004_ENST00000380358_CHD3 |
| miR-16 | ENSG00000170006_ENST00000304385_TMEM154 |
| miR-16 | ENSG00000170500_ENST00000393437_LONRF2 |
| miR-16 | ENSG00000170759_ENST00000302418_KIF5B |
| miR-16 | ENSG00000170776_ENST00000394518_AKAP13 |
| miR-16 | ENSG00000171150_ENST00000306503_SOCS5 |
| miR-16 | ENSG00000171385_ENST00000369697_KCND3 |
| miR-16 | ENSG00000171490_ENST00000396503_RSL1D1 |
| miR-16 | ENSG00000171570_ENST00000406058_EGLN2 |
| miR-16 | ENSG00000172354_ENST00000393926_GNB2 |
| miR-16 | ENSG00000173068_ENST00000380672_BNC2 |
| miR-16 | ENSG00000173638_ENST00000311124_SLC19A1 |
| miR-16 | ENSG00000173786_ENST00000393892_CNP |
| miR-16 | ENSG00000173825_ENST00000309880_TIGD3 |
| miR-16 | ENSG00000173875_ENST00000343325_ZNF791 |
| miR-16 | ENSG00000173915_ENST00000337003_USMG5 |
| miR-16 | ENSG00000174231_ENST00000304992_PRPF8 |
| miR-16 | ENSG00000174292_ENST00000311668_TNK1 |
| miR-16 | ENSG00000174405_ENST00000356922_LIG4 |
| miR-16 | ENSG00000174595_ENST00000310992_KLF14 |
| miR-16 | ENSG00000175414_ENST00000310389_ARL10 |
| miR-16 | ENSG00000175467_ENST00000312397_SART1 |
| miR-16 | ENSG00000177169_ENST00000321867_ULK1 |
| miR-16 | ENSG00000179915_ENST00000406316_NRXN1 |
| miR-16 | ENSG00000180596_ENST00000396984_HIST1H2BC |
| miR-16 | ENSG00000180758_ENST00000377411_GPR157 |
| miR-16 | ENSG00000181031_ENST00000323434_RPH3AL |
| miR-16 | ENSG00000181192_ENST00000263035_DHTKD1 |
| miR-16 | ENSG00000181222_ENST00000322644_POLR2A |
| miR-16 | ENSG00000181418_ENST00000421952_DDN |
| miR-16 | ENSG00000181472_ENST00000325144_ZBTB2 |
| miR-16 | ENSG00000182149_ENST00000329908_KIAA0174 |
| miR-16 | ENSG00000182158_ENST00000330387_CREB3L2 |
| miR-16 | ENSG00000182447_ENST00000327928_OTOL1 |
| miR-16 | ENSG00000182611_ENST00000333151_HIST1H2AJ |
| miR-16 | ENSG00000183638_ENST00000382483_RP1L1 |
| miR-16 | ENSG00000184009_ENST00000331925_ACTG1 |
| miR-16 | ENSG00000184436_ENST00000215742_THAP7 |
| miR-16 | ENSG00000184634_ENST00000374080_MED12 |
| miR-16 | ENSG00000184675_ENST00000330258_FAM123B |
| miR-16 | ENSG00000184779_ENST00000330339_RPS17 |
| miR-16 | ENSG00000184867_ENST00000328766_ARMCX2 |
| miR-16 | ENSG00000185176_ENST00000407834_AQP12B |
| miR-16 | ENSG00000185818_ENST00000331662_NAT8L |
| miR-16 | ENSG00000185915_ENST00000379499_KLHL34 |
| miR-16 | ENSG00000186765_ENST00000334850_FSCN2 |
| miR-16 | ENSG00000187239_ENST00000446176_FNBP1 |
| miR-16 | ENSG00000187498_ENST00000375820_COL4A1 |
| miR-16 | ENSG00000187583_ENST00000379410_PLEKHN1 |
| miR-16 | ENSG00000187837_ENST00000343677_HIST1H1C |
| miR-16 | ENSG00000187837_ENST00000343677_HIST1H1C |
| miR-16 | ENSG00000188486_ENST00000375167_H2AFX |
| miR-16 | ENSG00000188493_ENST00000378313_C19orf54 |
| miR-16 | ENSG00000189091_ENST00000302516_SF3B3 |
| miR-16 | ENSG00000189292_ENST00000403610_FAM150B |
| miR-16 | ENSG00000189334_ENST00000368702_S100A14 |
| miR-16 | ENSG00000196090_ENST00000373198_PTPRT |
| miR-16 | ENSG00000196235_ENST00000378524_SUPT5H |
| miR-16 | ENSG00000196365_ENST00000360614_LONP1 |
| miR-16 | ENSG00000196604_ENST00000357462_A26C1B |
| miR-16 | ENSG00000196924_ENST00000369863_FLNA |
| miR-16 | ENSG00000196981_ENST00000330689_WDR5B |
| miR-16 | ENSG00000197157_ENST00000354725_SND1 |
| miR-16 | ENSG00000197353_ENST00000359228_LYPD2 |
| miR-16 | ENSG00000197386_ENST00000355072_HTT |
| miR-16 | ENSG00000197858_ENST00000355091_GPAA1 |
| miR-16 | ENSG00000198586_ENST00000431350_TLK1 |
| miR-16 | ENSG00000198804_ENST00000361624_MT-CO1 |
| miR-16 | ENSG00000205581_ENST00000380749_HMGN1 |
| miR-16 | ENSG00000205707_ENST00000381356_LYRM5 |
| miR-16 | ENSG00000215041_ENST00000399464_NEURL4 |
| miR-16 | ENSG00000215114_ENST00000399598_UBXN2B |
| miR-16 | ENSG00000231555_ENST00000445736_HSPA1B |
| miR-16 | ENSG00000232804_ENST00000450744_HSPA1B |
| miR-16 | ENSG00000235067_ENST00000419792_TUBB |
| miR-16 | ENSG00000237405_ENST00000447921_AGER |
| miR-16 | ENSG00000244476_ENST00000472091_RP4-761I2-4 |
| miR-16 | ENSG00000253953_ENST00000519479_PCDHGB4 |
miR-15a, microRNA-15a.
miR-15a/16 directly target and downregulate CABIN1 expression in MM cells
Previous studies have reported that the expression of miR-15 and miR-16 was downregulated in MM tissues (8,19). To determine whether miR-15a and miR-16 are capable of targeting and regulating CABIN1 expression in MM, a gain-of-function study was performed.
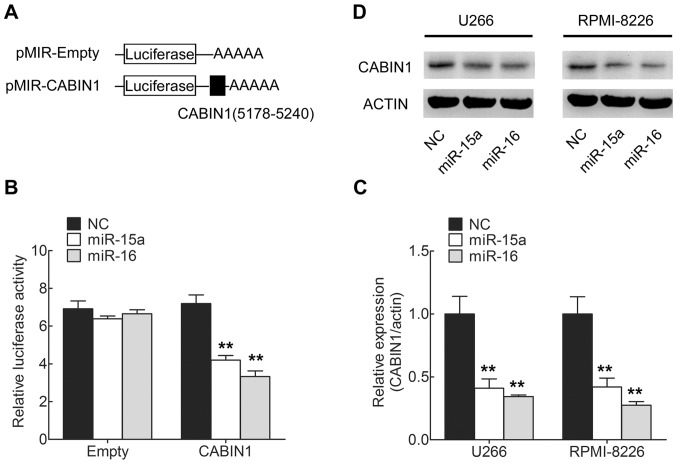
Luciferase reporter plasmids were created with targeting sequences of CABIN1 mRNA (Fig. 2A). miR-15a and miR-16 were transfected into 293 cells and a dual-luciferase reporter assay was used to assess the miRNA regulation of CABIN1. The results revealed that the upregulation of miR-15a or miR-16 decreased luciferase activity, whereas a reporter plasmid without the presence of CABIN1 sequence produced no change in luciferase activity (Fig. 2B). These results indicated miR-15a and miR-16 negatively regulated CABIN1 expression.
Figure 2.
miR-15a and miR-16 target CABIN1 in MM cells. (A) Schematic representation of pMIR firefly luciferase reporter construction. The potential binding site of miR-15a and miR-16 is indicated in Fig. 1B. (B) Analysis of luciferase activity in 293 cells. Cells were co-transfected with a firefly luciferase reporter plasmid containing putative miR-15a- and miR-16-targeting sequences; 48 h after transfection, cell lysates were assayed for luciferase activity and normalized to Renilla luciferase activity. (C and D) Effects of miR-15a and miR-16 on the endogenous expression level of CABIN1. U266 and RPMI-8226 MM cells were co-transfected with miR-15a or miR-16 and negative control oligonucleotides and 48 h after transfection, cells were isolated. The (C) mRNA levels and (D) protein levels of CABIN1 were analyzed by RT-qPCR and western blot analysis, respectively. **P<0.01 by one-way analysis of variance followed by the Student Newman-Keuls test. miR-15a, microRNA-15a; CANIB1, calcineurin-binding protein 3; MM, multiple myeloma; NC, negative control.
Whether the endogenous CABIN1 in MM cells was similarly regulated was subsequently investigated. The MMU266 and RPMI-8226 cells lines were transfected with miR-15a and miR-16, and CABIN1 mRNA and protein levels were examined by RT-qPCR and western blotting, respectively. The levels of CABIN1 mRNA (Fig. 2C) and protein (Fig. 2D) were consistently and substantially downregulated by miR-15a and miR-16 in MM cells. The results of the present study indicated that miR-15a and miR-16 directly targeted CABIN1 mRNA and negatively regulated expression of CABIN1 at the mRNA and protein level.
Upregulation of miR-15a and miR-16 inhibits the proliferation of MM cells by targeting CABIN1
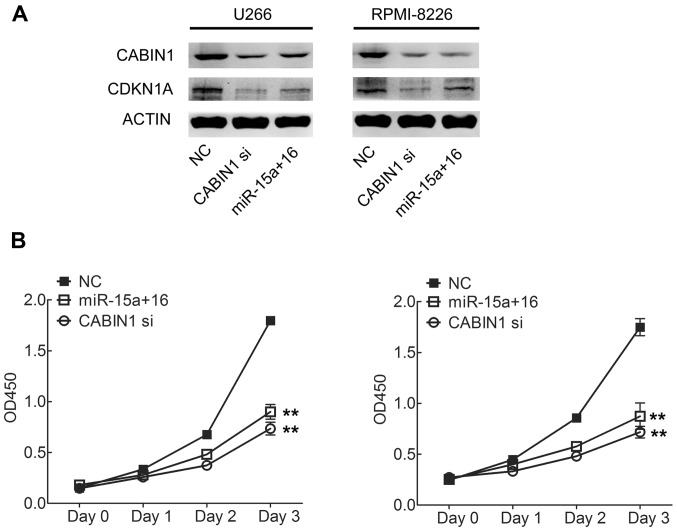
Overexpression of CABIN1 increased the proliferation of cancer cells by physically interacting with p53 and repressing p53 transcriptional activity on target genes, including CDKN1A (18). Given that CABIN1 is a direct target of miR-15a/16, it was hypothesized that miR-15a and miR-16 may affect MM cell viability through CABIN1. miR-15aand miR-16a or CABIN1 siRNA was transiently transfected into U266 and RPMI-8226 cells. The results of western blotting revealed that transfection with miR-15a and miR-16or CABIN1-targeted siRNA decreased the expression of CABIN1 and p53 downstream gene CDKN1A in U266 and RPMI-8226 cells (Fig. 3A).
Figure 3.
Upregulation of miR-15a and miR-16 inhibits proliferation of MM cells through targeting of CABIN1. (A) Western blot analysis was performed to determine the expression level of CABIN1 and CDKN1A following transfection of CABIN1 siRNA or miR-15a and miR-16 in U266 and RPMI-8226 cells. (B) The effects of CABIN1 siRNA or miR-15a and miR-16 transfection on U266 and RPMI-8226 cell proliferation were determined using a Cell Counting kit-8 assay at the indicated times following transfection. **P<0.01 by one-way analysis of variance followed by the Student Newman-Keuls test. miR-15a, microRNA-15a; CANIB1, calcineurin-binding protein 1; CDKN1A, cyclin-dependent kinase inhibitor 1α; siRNA, small interfering RNA.
Using a cell proliferation assay, the overexpression of miR-15a and miR-16 in U266 and RPMI-8226 cells was determined to result in the significant suppression of cell proliferation, similar to CABIN1 siRNA (Fig. 3B). The results of the present study demonstrated that the downregulation of CABIN1 expression by miR-15a and miR-16 contributes, partially, to the suppression of MM cell growth.
miR-15a and miR-16 are downregulated and negatively associated with CABIN1 mRNA levels in MM specimens
Previous studies have reported that miR-15a and miR-16 downregulated in several types of cancer, including MM (8,19). Overexpression of CABIN1 is associated with poorer patient outcomes in a number of cancer types (18,20,21). The results of the present study demonstrated that CABIN1 is negatively regulated by miR-15a and miR-16 in MM cells. Next, the correlation between miR-15a and miR-16 expression levels and CABIN1 mRNA levels was assessed in MM specimens.
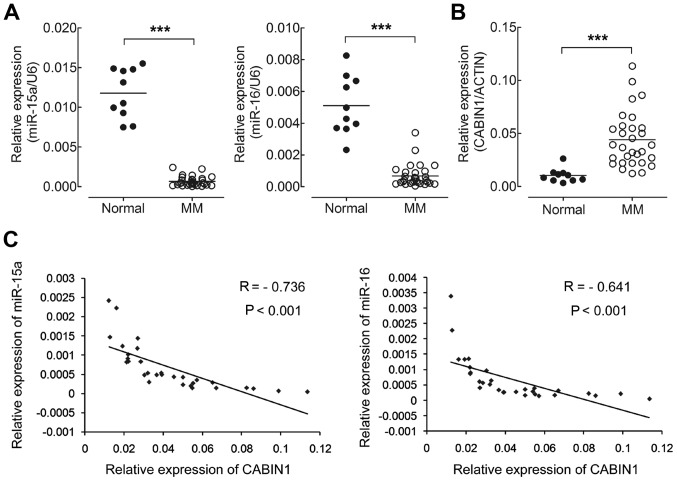
To determine the levels of miR-15a and miR-16 in MM samples, total RNA was extracted from MM specimens (n=30), following which the expression levels of miR-15a and miR-16 were analyzed using RT-qPCR and normalized to an endogenous control (U6 RNA). miR-15a and miR-16levels were significantly decreased in MM specimens compared with in healthy volunteers (n=10; Fig. 4A; P<0.001), which was concordant with the results of certain previous studies (8,19). RT-qPCR analysis was performed to detect the mRNA expression of CABIN1 in the same MM specimens and normalized to β-actin. As presented in Fig. 4B, CABIN1 mRNA levels were significantly increased in MM specimens compared with healthy volunteers. RNA levels of miR-15a and miR-16, and mRNA levels of CABIN1 in MM tissues were analyzed by Pearson's correlation coefficient analysis. CABIN1 mRNA expression levels were negatively correlated with those of miR-15a (R=−0.736; P<0.001) and miR-16 (R=−0.641; P<0.001) in MM tissues (Fig. 4C). Collectively, these results suggested that high endogenous miR-15a and miR-16levels may target and downregulate the expression of CABIN1 in MM.
Figure 4.
miR-15a and miR-16 downregulate CABIN1 expression and expression of miR-15a and miR-16 are negatively associated with CABIN1 mRNA expression in MM samples. (A) RT-qPCR analysis of miR-15a and miR-16 expression in 30 MM specimens and 10 normal specimens. The expression of miR-15a and miR-16 was normalized to U6 small nuclear RNA. (B) RT-qPCR analysis of CABIN1 expression in the same MM and healthy volunteers as in (A). The expression of CABIN1 was normalized to β-actin. (C) A negative Spearman's correlation between miR-15a/miR-16 and CABIN1 mRNA levels was observed in 30 MM specimens. ***P<0.01 by one-way analysis of variance followed by the Student Newman-Keuls test. T, tumor specimens; N, normal specimens; MM, multiple myeloma; miR-15a, microRNA-15a; CANIB1, calcineurin-binding protein 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
MM is characterized by multi-stage cellular transformation and complex genomic and epigenetic abnormalities (22). The deregulation of miRNAs in cancer often arises following changes to the genome and epigenome (23). Numerous miRNA genes are located at cancer-associated fragile genomic loci that frequently undergo mutations and could represent novel biomarkers (5,24). Several studies suggest that miRNAs regulate MM oncogenesis, providing functional information on the function of specific miRNAs inpatients with MM, characterized by increased oncogenic miRNA expression (19,25,26). Previous studies have reported that miR-15a and miR-16 are downregulated in numerous types of cancer, including MM (8,19). However, the biological relevance and underlying mechanisms of the downregulation of miR-15a and miR-16 in MM are not yet clear.
Expression of miR-15a and miR-16 are decreased or totally absent in MM and are hypothesized to have same target genes owing to their sequence similarity. In order to investigate the biological significance and the underlying mechanisms of the downregulation of miR-15a and miR-16 in MM, previously published CLASH data was screened for potential miR-15a/miR-16 target sites in CABIN1 mRNA.
As the overexpression of CABIN1 increases the proliferation of cancer cells by physically interacting with p53 and repressing the transcriptional activity of p53 (18), and considering that CABIN1 is a direct target of miR-15a and miR-16, it was hypothesized that miR-15a and miR-16 may affect MM cell viability through CABIN1.
To assess this hypothesis, again-of-function study was performed, the results of which indicated that miR-15a and miR-16 directly target CABIN1 mRNA and negatively regulate the expression of CABIN1 at the mRNA and protein level in MM cells. Using a cell proliferation assay, upregulation of miR-15a and miR-16 was revealed to inhibit the proliferation of MM cells by targeting CABIN1. Furthermore, the correlation between miR-15a and miR-16 expression levels and the level of CABIN1 mRNA in MM specimens was analyzed. miR-15a and miR-16 expression was significantly lower in MM specimens than in normal specimens, whereas the levels of CABIN1 mRNA were significantly higher in MM specimens than in normal specimens. CABIN1 mRNA levels were negatively correlated with miR-15a (R=−0.736; P<0.001) and miR-16 (R=−0.641; P<0.001) expression levels in MM tissues. Collectively, the results of the present study indicate that miR-15a and miR-16 are downregulated and negatively associated with CABIN1 mRNA levels in MM specimens.
Future studies must investigate gene expression changes in the p53-p21 signaling pathway, in order to verify that miR-15a miR-16 affects MM cell viability through CABIN1, and that CABIN1 increases the proliferation of cancer cells via suppressing p53 transcriptional activity towards its target genes, including CDKN1A. Other studies have indicated that miR-15 could inhibit protein kinase B, nuclear factor κ-light-chain-enhancer of activated B cells and vascular endothelial growth factor activity, thus exerting antitumor effects (8,18), which should be considered in future studies.
In summary, the results of the present study suggest that miR-15a and miR-16 may serve as tumor suppressor genes in MM pathogenesis, and that miR-15a and miR-16 may be a promising therapeutic target in the treatment of MM.
References
- 1.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Raab MS, Podar K, Breitkreutz I, Richardson PG, Anderson KC. Multiple myeloma. Lancet. 2009;374:324–339. doi: 10.1016/S0140-6736(09)60221-X. [DOI] [PubMed] [Google Scholar]
- 3.De Angelis R, Minicozzi P, Sant M, Dal Maso L, Brewster DH, Osca-Gelis G, Visser O, Maynadié M, Marcos-Gragera R, Troussard X, et al. Survival variations by country and age for lymphoid and myeloid malignancies in Europe 2000–2007: Results of EUROCARE-5 population-based study. Eur J Cancer. 2015;51:2254–2268. doi: 10.1016/j.ejca.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Lionetti M, Agnelli L, Mosca L, Fabris S, Andronache A, Todoerti K, Ronchetti D, Deliliers GL, Neri A. Integrative high-resolution microarray analysis of human myeloma cell lines reveals deregulated miRNA expression associated with allelic imbalances and gene expression profiles. Genes Chromosomes Cancer. 2009;48:521–531. doi: 10.1002/gcc.20660. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers; Proc Natl Acad Sci USA; 2004; pp. 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Li C, Zhang R, Gao X, Qu X, Zhao M, Qiao C, Xu J, Li J. MiR-17-92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011;309:62–70. doi: 10.1016/j.canlet.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 7.Gao X, Zhang R, Qu X, Zhao M, Zhang S, Wu H, Jianyong L, Chen L. MiR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myeloma. Leuk Res. 2012;36:1505–1509. doi: 10.1016/j.leukres.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Sun CY, She XM, Qin Y, Chu ZB, Chen L, Ai LS, Zhang L, Hu Y. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis. 2013;34:426–435. doi: 10.1093/carcin/bgs333. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia; Proc Natl Acad Sci USA; 2002; pp. 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tung YT, Huang PW, Chou YC, Lai CW, Wang HP, Ho HC, Yen CC, Tu CY, Tsai TC, Yeh DC, et al. Lung tumorigenesis induced by human vascular endothelial growth factor (hVEGF)-A165 overexpression in transgenic mice and amelioration of tumor formation by miR-16. Oncotarget. 2015;6:10222–10238. doi: 10.18632/oncotarget.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang W, Tong JH, Lung RW, Dong Y, Zhao J, Liang Q, Zhang L, Pan Y, Yang W, Pang JC, et al. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol Cancer. 2015;14:52. doi: 10.1186/s12943-015-0323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang E, Liu R, Chu Y. miRNA-15a/16: As tumor suppressors and more. Future Oncol. 2015;11:2351–2363. doi: 10.2217/fon.15.101. [DOI] [PubMed] [Google Scholar]
- 13.Hao M, Zhang L, An G, Sui W, Yu Z, Zou D, Xu Y, Chang H, Qiu L. Suppressing miRNA-15a/−16 expression by interleukin-6 enhances drug-resistance in myeloma cells. J Hematol Oncol. 2011;4:37. doi: 10.1186/1756-8722-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao M, Zhang L, An G, Meng H, Han Y, Xie Z, Xu Y, Li C, Yu Z, Chang H, Qiu L. Bone marrow stromal cells protect myeloma cells from bortezomib induced apoptosis by suppressing microRNA-15a expression. Leuk Lymphoma. 2011;52:787–794. doi: 10.3109/10428194.2011.576791. [DOI] [PubMed] [Google Scholar]
- 15.Hideshima T, Mitsiades C, Akiyama M, Hayashi T, Chauhan D, Richardson P, Schlossman R, Podar K, Munshi NC, Mitsiades N, Anderson KC. Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood. 2003;101:1530–1534. doi: 10.1182/blood-2002-08-2543. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang H, Choi SY, Cho EJ, Youn HD. Cabin1 restrains p53 activity on chromatin. Nat StructMolBio. 2009;16:910–915. doi: 10.1038/nsmb.1657. [DOI] [PubMed] [Google Scholar]
- 19.Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, Runnels J, Jia X, Ngo HT, Melhem MR, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–6680. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W, Youn HD, Liu JO. Thapsigargin-induced apoptosis involves cabin1-MEF2-mediated induction of Nur77. Eur J Immunol. 2001;31:1757–1764. doi: 10.1002/1521-4141(200106)31:6<1757::AID-IMMU1757>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 21.Choi SY, Jang H, Roe JS, Kim ST, Cho EJ, Youn HD. Phosphorylation and ubiquitination-dependent degradation of CABIN1 releases p53 for transactivation upon genotoxic stress. Nucleic Acids Res. 2013;41:2180–2190. doi: 10.1093/nar/gks1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chng WJ, Glebov O, Bergsagel PL, Kuehl WM. Genetic events in the pathogenesis of multiple myeloma. Best Pract Res Clin Haematol. 2007;20:571–596. doi: 10.1016/j.beha.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP, et al. MicroRNAs regulate critical genes associated with multiplemyeloma pathogenesis; Proc Natl Acad Sci USA; 2008; pp. 12885–12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misiewicz-Krzeminska I, Sarasquete ME, Quwaider D, Krzeminski P, Ticona FV, Paíno T, Delgado M, Aires A, Ocio EM, García-Sanz R, et al. Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica. 2013;98:640–648. doi: 10.3324/haematol.2012.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]