Abstract
Increasing evidence demonstrates that microRNAs (miRNAs/miRs), a type of non-coding small RNA, can regulate tumor cell migration, invasion and metastasis, and may therefore serve a major function in the occurrence and development of tumors. The present study investigated the effect of miR-383 on the proliferation, migration and invasion of colon cancer HT-29 and LoVo cell lines. The expression of miR-383 in colon cancer and adjacent non-tumor tissues was examined by reverse transcription-quantitative polymerase chain reaction. MiR-383 upregulation was stimulated by transfection with a miR-383 mimic. Cell proliferation was measured with MTT and colony formation assays, and cell migration and invasion potential were examined by Transwell chamber assays. A proliferating-inducing ligand (APRIL), myeloid cell leukemia-1 and cyclooxygenase-2 protein expression was analyzed by western blotting. The expression of miR-383 was decreased in colon cancer tissues compared with adjacent non-tumor tissues (P<0.05). Transfection with a miR-383 mimic suppressed proliferation and inhibited cell migration and invasion in HT-29 and LoVo colon cancer cell lines. Overexpression of miR-383 in HT-29 and LoVo cells resulted in the suppression of APRIL protein expression. In conclusion, miR-383 was downregulated in colon cancer. The upregulation of miR-383 inhibited proliferation, migration and invasion of colon cancer cells, potentially through the regulation of target gene APRIL.
Keywords: microRNA-383, colon cancer, cell proliferation, cell migration, cell invasion, A proliferating-inducing ligand
Introduction
Colon cancer is a common malignant tumor of the digestive tract, and the morbidity and mortality rates are high in China; recent environmental and lifestyle changes have increased the incidence rate of colorectal cancer in China (1). The 5-year survival rate for patients with primary colon cancer is 80–90%; however, the 5-year survival rate for patients with metastatic colon cancer is 5–15% (2). Therefore, it is important to investigate and clarify the molecular pathological mechanism of colon cancer metastasis and identify new therapeutic targets for the prevention and treatment of colon cancer (3).
MicroRNAs (miRNAs/miRs) are single stranded non-coding endogenous RNA molecules of ~22 nucleotides. miRNAs can bind with mRNA from a target gene at the 3′-untranslated region (3′-UTR), leading to post-transcriptional silencing, which regulates the expression of the target gene (4). It is established that the abnormal expression of miRNAs, including miR-511-3p (5) and miR-135b (6), serves an important function in the occurrence and development of colon cancer.
miR-383 is located in chromosome 8p22. Previous studies have demonstrated that miR-383 is associated with medulloblastoma (7), glioma (8) and hepatocellular carcinoma (9), and may serve important function(s) in these types of tumor. Previous studies have demonstrated that majority of miR-383's target genes in numerous tumors (7–9); however, target genes of miR-383 in colon cancer remain unclear. A proliferating-inducing ligand (APRIL) is a member of the tumor necrosis factor (TNF) super-family. Initially, it was discovered as a cytokine that was able to stimulate tumor cell growth, induce tumor cell proliferation and modulate tumor cell apoptosis (10). APRIL is predominantly expressed in tumor tissues in colon cancer, pancreatic cancer, gastric cancer and bladder cancer (11,12). The expression of APRIL promotes tumor growth and metastasis in colorectal cancer cells (13,14), and APRIL knockdown suppresses migration and invasion of colon caner cells (15). Cyclooxygenase-2 (COX-2) is markedly expressed in tumor tissues in colon cancer and breast cancer (16,17). Overexpression of COX-2 is associated with the aggressive and invasive potential of tumor cells (18) and is positively associated with the tumor-node-metastasis (TNM) staging and lymph node metastasis in colon cancer (16). Myeloid cell leukemia-1 (MCL-1) is an anti-apoptotic protein of the B cell lymphoma-2 (BCL-2) family (19). Over-expression of MCL-1 is associated with the inhibition of apoptosis in colon cancer cells and the promotion of drug resistance in colon cancer liver metastasis (20,21).
In the present study, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was used to determine expression levels of miR-383 in colon cancer and adjacent normal tissues. The associations between miR-383 expression and the clinicopathological characteristics of colon cancer were analyzed. A miR-383 mimic was used to overexpress miR-383 in colon cancer HT-29 and LoVo cell lines, and the effect of miR-383 on colorectal cancer cell proliferation, migration and invasion was analyzed. Western blot analysis assessed the effect of miR-383 upregulation on the protein expression levels of previously identified miR-383 target genes, including A proliferating-inducing ligand (APRIL), myeloid cell leukemia-1 (MCL-1) and cyclooxygenase-2 (COX-2) (9,22,23). The aim of the present study is to provide a theoretical basis for the future exploration of the molecular mechanism(s) of miR-383, and the identification of novel therapeutic targets in colon cancer.
Materials and methods
Patients
A total of 64 pairs of matched colon tumor and adjacent non-tumor mucosal tissues (>5 cm from the edge of the tumor) were obtained from patients with colon cancer at Zhejiang Provincial People's Hospital (Zhejiang, China) between March 2014 and January 2015.
In total, 30 patients were <60 years, 34 patients were ≥60 years (range, 34–86 years), 36 patients were male and 28 patients were female. Overall, 26 patients had a left colon tumor and 38 patients had a right colon tumor. Furthermore, 45 patients presented with adenocarcinoma and 19 with mucinous carcinoma cases. A total of 55 patients were at tumor-node-metastasis stages I, II or III, and 9 were at TNM stage IV (24).
Written informed consent was obtained from all patients prior to surgery for the use of their tissues in the present study. The use of all specimens was approved by the Ethics Committee of Zhejiang Provincial People's Hospital (Zhejiang, China). No patients received radiotherapy or chemotherapy prior to surgery. Following resection, tissues were immediately frozen in liquid nitrogen and stored at −80°C until further use.
Cell lines
LoVo and HT-29 colon cancer cell lines were purchased from the Chinese Academy of Sciences, Shanghai Institute of Life Sciences Cell Bank (Shanghai, China). Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS, Dingguo Biotechnology Co., Ltd., Beijing, China) and incubated at 37°C in a 5% CO2 incubator with saturated humidity.
Reagents
TRIzol reagent was purchased from Invitrogen (Thermo Fisher Scientific, Inc.). The PrimeScript One Step miRNA cDNA Synthesis kit and the SYBR premix Ex Taq™ II qPCR reagents were purchased from Takara Bio, Inc. (Otsu, Japan). An rno-miR-383 mimic kit (cat. no., miR1003114-1-2) and a corresponding negative control (cat no., miR01201-1-2) were procured from Guangzhou Ruibo Biological Technology Co., Ltd. (Guangzhou, China). A Transwell chamber migration assay kit was obtained from BD Biosciences (Franklin Lakes, NJ, USA). A Boyden chamber invasion assay kit was purchased from EMD Millipore (Billerica, MA, USA). β-actin and primary antibodies for western blot against β-actin (ab8227), APRIL (ab64967), MCL-1 (ab28147) and COX-2 (ab102005) were obtained from Abcam (Cambridge, UK). Horseradish peroxidase-conjugated-Goat Anti-Rabbit immunoglobulin (Ig)G (cat. no. ab6721) used as a secondary antibody was obtained from Abcam.
Total RNA extraction and reverse transcription
Total RNA was extracted using RNA Extraction kit (Takara Bio. Inc., Otsu, Japan), according to the manufacturer's protocol. Total RNA (1 µg) was reverse transcribed to cDNA by a reverse transcription reaction, using PrimeScript One Step miRNA cDNA Synthesis kit (Takara Bio. Inc.), with the following reaction conditions: 37°C for 60 min, 85°C for 5 sec.
RT-qPCR assay for miR-383
Primer sequences were as follows: miR-383 forwards, 5′-CACGAAAGATCAGAAGGTGATTG-3′; U6B forwards, 5′-ACGCAAATTCGTGAAGCGTT-3′. The reverse primer was the universal primer provided by the reverse transcription kit. The qPCR reaction system contained 10 µl SYBR premix, 0.4 µl Rox, 0.5 µl reverse primer, 1 µl template and 7.6 µl distilled H2O. Reaction conditions were as follows: 94°C for 4 min; then 40 cycles of 94°C, 5 sec, 55°C, 20 sec and 72°C, 20 sec; melting curve program: 95°C, 1 min, 55°C, 30 sec and 95°C, 30 sec, 1 cycle. The expression of miR-383 was calculated relative to U6B with the 2−ΔΔCq method (25).
Transfection of the HT-29 and LoVo cell lines with an miR-383 mimic
HT-29 and LoVo cells in the logarithmic growth phase were seeded onto 6-well plates at a density of 2×105/well. The cells were transfected according to the protocol of the rno-miR-383 kit (Guangzhou Ruibo Biological Technology Co., Ltd.) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Cells transfected with the rno-miR-383 mimic were defined as the experimental group and the cells transfected with the mimic negative control were defined as the control group. At 24 h post-transfection, the efficiency was assessed using RT-qPCR.
MTT assay for cell proliferation
rno-miR-383-transfected HT-29 and LoVo cells were added to 96-well plates at a concentration of 2×103 cells/well in sextuplicate, and incubated at 37°C in a 5% CO2 incubator with saturated humidity. Following 24, 48 or 72 h incubation, 20 µl MTT reagent (5 mg/ml) was added to each well and incubated for a further 4 h. Dimethyl sulfoxide (150 µl) was added to dissolve the reaction products, and the absorbance (A) was measured at 570 and 630 nm using a microplate reader. The relative cell proliferation rate was calculated as follows: Relative proliferation rate (%)=[(A570-A630) of study group/(A570-A630) of control group] × 100.
Colony formation assay
A total of 200 cells from the experimental and control groups were cultured on a 6-well plate, and media was replenished every 5 days. At 2 weeks, the colonies were fixed with 4% poly methanol for 15 min at room temperature, and stained with 10% Giemsa stain for 20 min at room temperature.
Migration assay
Migration assays were performed using a 24-well Transwell chamber kit. In the upper chamber, 200 µl of cell suspension containing 4×104 experimental or control cells was added to serum-free Dulbecco's modified Eagle's medium (DMEM) culture medium; the lower chamber contained DMEM with 10% FBS. Following 24 h of incubation, cells remaining on the upper surface of the membrane were removed with a cotton swab, whereas adhered cells on the lower membrane surface were fixed in 4% poly methanol for 15 min at room temperature and stained with 10% Giemsa stain for 20 min at room temperature. Subsequently, five fields of view were selected at random and observed under a microscope (brightfield; magnification, ×200). The number of cells was counted and the mean was calculated.
Invasion assay
A Matrigel invasion chamber kit was used. A total of 300 µl serum-free DMEM culture medium was added to the upper chamber, including 5×105 experimental or control cells. DMEM with 10% FBS was added to the lower chamber. Following incubation for 48 h, the cells were fixed in 4% poly methanol for 15 min at room temperature and stained with 10% Giemsa stain for 20 min at room temperature. Subsequently, five fields of view were selected at random and observed under a microscope (brightfield, magnification, ×200). The number of cells was counted and the median was calculated.
Western blotting
Western blotting was employed to examine the protein expression of APRIL, MCL-1 and COX-2, target genes of miR-383. Total protein was extracted from cells with TRIzol, according to the manufacturer's protocol. Use BCA protein assay kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA) to quantify protein concentration. Total protein (30 µg per lane) was resolved by 15% SDS-PAGE gel electrophoresis for 2 h and transferred to a polyvinylidene fluoride membrane. The membrane was washed once in TBST (20 mM Tris, 150 mM NaCl, containing 0.05% Tween-20, pH 7.4), for 5 min. The membrane was blocked by incubation for 1 h in 1% BSA (bovine serum albumin) at room temperature. Following 3 washes in TBST for 10 min each, the membrane was probed overnight at 4°C with APRIL, MCL-1 or COX-2 antibodies (dilution, 1:2,000), and β-actin (dilution, 1:5,000) as a loading control. The membrane was washed 3 times in TBST for 10 min each. Membranes were then incubated with the enzyme-labeled secondary antibody (Goat Anti-Rabbit IgG (HRP), dilution, 1:5,000) at room temperature for 1 h prior to detection with enhanced chemiluminscence (Beyotime Institute of Biotechnology, Shanghai, China) and X-ray film.
Statistical analysis
Statistical analysis was performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). Measurement data are presented as the mean ± standard deviation. Paired measurement data were analyzed with a paired samples t-test; categorical data were compared using a χ2 or Fisher's exact test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-383 expression is decreased in colon cancer tissue
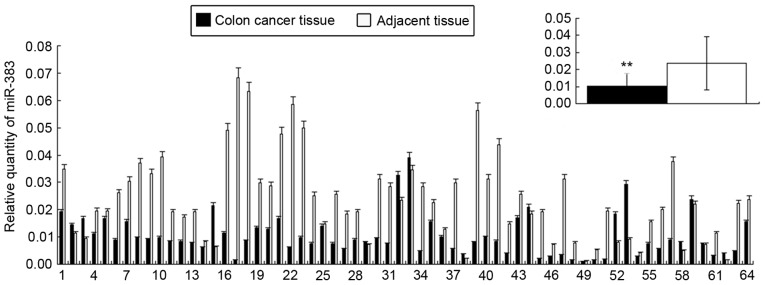
The relative expression of miR-383 in colon cancer tissues was 1.04x10−2±7.54x10−3, significantly lower than in adjacent tissues (2.57x10−2±1.42x10−3; Fig. 1; P<0.01). In addition, the expression of miR-383 was negatively associated with the pathological type and TNM staging of patients with colon cancer (P<0.05), whereas it was not associated with patient sex or age, or tumor location and differentiation (Table I).
Figure 1.
Expression of miR-383 in corresponding colon cancer and adjacent tissue samples, including for each patient and overall, relative to U6B. **P<0.01 vs. adjacent tissue. miR-383, microRNA-383.
Table I.
Association of clinical features with the relative expression of miR-383, as calculated with the χ2 test.
| Clinical feature | n | Relative expression of miR-383, mean ± standard deviation | P-value |
|---|---|---|---|
| Sex | 0.374 | ||
| Male | 36 | 1.07×10−2±8.03×10−3 | |
| Female | 28 | 1.01×10−2±6.98×10−3 | |
| Age, years | 0.215 | ||
| <60 | 30 | 9.58×10−2±8.75×10−3 | |
| ≥60 | 34 | 1.11×10−2±6.32×10−3 | |
| Tumor location | 0.458 | ||
| Left hemicolon | 26 | 1.03×10−2±8.17×10−3 | |
| Right hemicolon | 38 | 1.05×10−3±4.41×10−3 | |
| Pathological type | <0.001 | ||
| Adenocarcinoma | 45 | 1.24×10−2±7.68×10−3 | |
| Mucinous carcinoma | 19 | 5.56×10−3±4.41×10−3 | |
| Differentiation degree | 0.052 | ||
| Low | 16 | 8.46×10−3±4.36×10−3 | |
| High/medium | 48 | 1.11×10−2±8.26×10−3 | |
| Tumor, node, metastasis stage | 0.005 | ||
| I, II or III | 55 | 1.42×10−2±7.26×10−3 | |
| IV | 9 | 4.16×10−3±4.09×10−4 |
miR-383, microRNA-383.
Confirmation of transfection with a miR-383 mimic
The RT-qPCR results demonstrated that following the transfection with an rno-miR-383 mimic, the relative expression miR-383 level was 7.43×10−2±3.31×10−3 in the HT-29 experimental group, and 3.46×10−2±5.42×10−3 in the negative control group (P<0.01); in LoVo cells, the relative expression level was 5.68x10−2±4.25x10−3 in the experimental group, and 2.03x10−2±6.57x10−3 in the control group (Fig. 2; P<0.01). These results suggest that transfection with an rno-miR-383 mimic effectively upregulated miR-383 expression in colorectal cancer cell lines.
Figure 2.
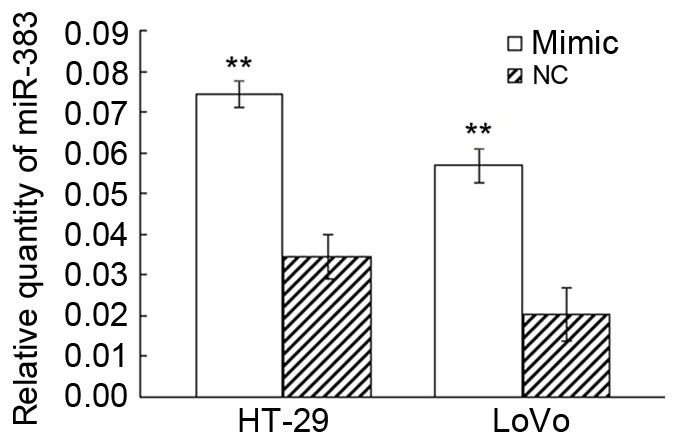
Detectable miR-383 expression was upregulated by transfection with an rno-miR-383 mimic, relative to U6B. **P<0.01 vs. NC. miR-383, microRNA-383; NC, negative control.
Effect of miR-383 on the proliferation of HT-29 and LoVo cells
A colony formation assay was used to determine the cell proliferation ability at 2 weeks. HT-29 and LoVo cell lines with overexpressed miR-383 exhibited a relative colony formation ability of 71.2±12.7, 26.4±7.8, respectively (P<0.05) compared with the negative control groups; the colony formation ability of the negative control groups were 125.3±18.9, 59.6±11.3; Compared with the negative group, the colony formation ability of the experimental group was significantly decreased (HT-29, P=0.0328; LoVo, P=0.0162), as demonstrated in Fig. 3A.
Figure 3.
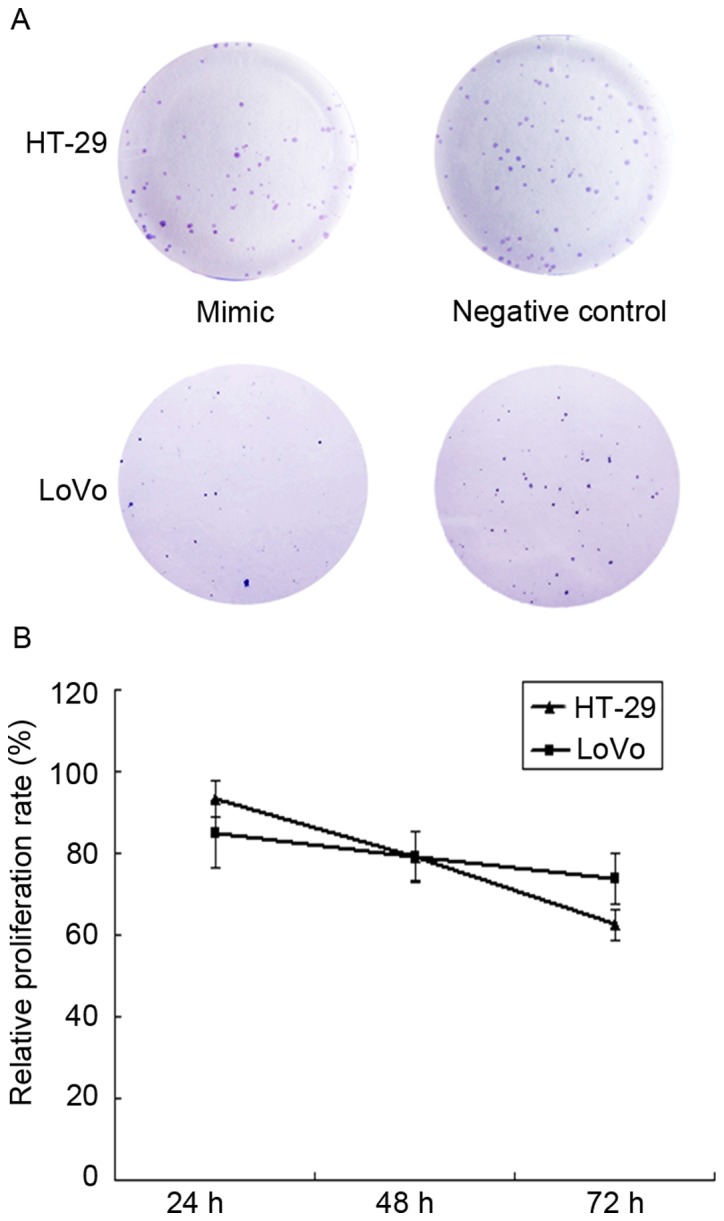
Upregulated microRNA-383 inhibits the proliferation of HT-29 and LoVo cells. (A) Representative images from the colony formation assay, (B) quantification of the MTT assay.
Based on the MTT assay, subsequent to being cultured for 24, 48 and 72 h, the relative proliferation rates of HT-29 cells in the experimental group were 93.3±4.27%, 78.9±6.23% and 62.5±3.62%, respectively (P<0.05) compared with the negative control group; the relative proliferation rates in the LoVo cells experimental group were 84.7±8.23%, 79.2±5.97% and 73.6±6.24% (Fig. 3B; P<0.05).
These results indicated that the upregulation of miR-383 expression inhibited the proliferation of HT-29 and LoVo colon cancer cells.
Effect of miR-383 on the migration and invasion of HT-29 and LoVo cells
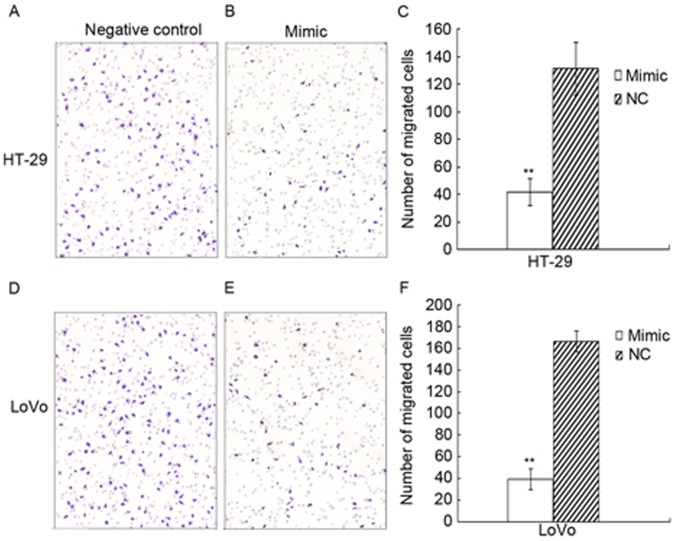
In the migration assay, the number of cells on the lower membrane surface in the HT-29 experimental group was 41.5±9.6, whereas it was 131.3±19.2 for the control group; therefore, the migration capacity decreased by 68.4% (P=0.031). The number of lower surface cells in the LoVo experimental group was 39.6±9.7, whereas it was 166.2±23.9 in the control group; therefore, the migtarion capacity decreased by 76.2% (P=0.016; Fig. 4).
Figure 4.
Upregulated microRNA-383 suppresses the migratory ability of HT-29 and LoVo cells, including representative images and quantified results. (A) Negative control group of HT-29, (B) experimental group of HT-29, (C) quantified results for HT-29, (D) negative control group of LoVo, (E) experimental group of LoVo, all at ×200 magnification (F) quantified results for LoVo. **P<0.05 vs. NC. NC, negative control.
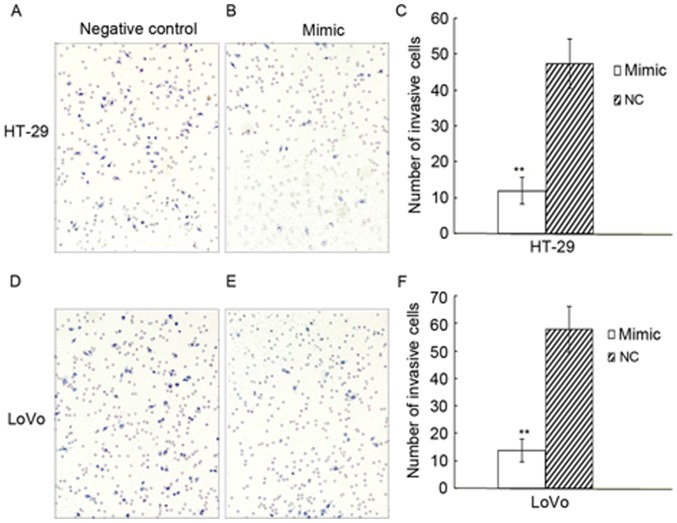
In the invasion assay, the number of cells invading across the membrane in the HT-29 experimental group of was 11.9±3.7, whereas it was 47.3±6.9 in the negative control group; the invasion capacity decreased by 74.8% (P=0.004). The number of invading cells in the LoVo experimental group was 13.7±4.1, whereas it was 57.9±8.2 in the negative control group; the invasion ability decreased by 76.4% (P=0.006; Fig. 5). In conclusion, the migratory and invasive capacity of HT-29 and LoVo cells transfected with rno-miR-383 mimic was reduced compared with the control group.
Figure 5.
Upregulated miR-383 suppresses the invasion ability of HT-29 and LoVo cells. (A) Negative control group of HT-29, (B) experimental group of HT-29, (C) quantified results for HT-29, (D) negative control group of LoVo, (E) experimental group of LoVo, all at ×200 magnification (F) quantified results for LoVo. **P<0.01 vs. NC. NC, negative control.
miR-383 inhibits the expression of APRIL
Western blot analysis (Fig. 6) demonstrated that APRIL bands were narrower and less intense in the HT-29 and LoVo experimental groups, compared with the negative control groups and the β-actin reference. By contrast, MCL-1 and COX-2 bands exhibited no clear change. This suggested that the expression of APRIL decreased, whereas the expression of MCL-1 and COX-2 remained unaltered, indicating that miR-383 may regulate the expression of APRIL in HT-29 and LoVo colon cancer cells.
Figure 6.
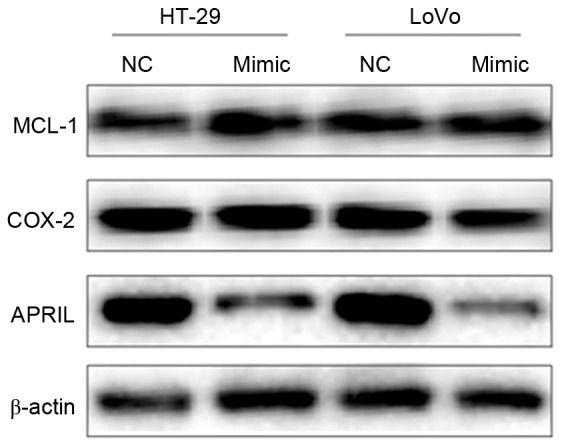
Expression of APRIL, MCL-1 and COX-2 proteins in HT-29 and LoVo as detected by western blot analysis. APRIL, A proliferating-inducing ligand; MCL-1, myeloid cell leukemia-1; COX-2, cyclooxygenase-2; NC, negative control.
Discussion
MiRNAs are small non-coding RNAs composed of 18–25 nucleotides. MiRNAs can bind the 3′UTR of their target mRNAs to induce the degradation or inhibition of transcription, resulting in the regulation of the target gene function (4). A number of miRNAs may regulate cell migration, invasion and metastasis, including miR-1246 and miR-145 (26). MiRNA can also regulate the differentiation, proliferation and apoptosis of cells; therefore, alterations in expression can lead to disease, including gastric, bladder and colon cancer (27). MiRNAs serve a pivotal role in the occurrence and development of colon cancer (5,6). For example, miR-34a can inhibit the proliferation and metastasis of colon cancer cells by inhibiting platelet growth factor receptor (28). Another study revealed that miR-381 was significantly downregulated in human colon cancer tissues (29). A nuclear receptor, liver receptor homolog 1, is a target gene of miR-381, and the downregulation of miR-381 is associated with colon cancer progression (29).
MiR-383, with the sequence for has-miR-383 is CACGAAAGATCAGAAGGTGATTG, has been the subject of previous studies, although there is little evidence to demonstrate an association between miR-383 expression and tumor development. miR-383 is considered to be a tumor suppressor gene. For example, a previous study demonstrated that miR-383 expression was suppressed in hepatocellular carcinoma, and was significantly associated with tumor size, lymph node metastasis and the overall survival of patients with liver cancer (9). Further experiments indicated that miR-383 may have inhibited the growth of liver cancer cells by downregulating the expression of APRIL (9). This result is similar to the results of the present study; the present study confirmed that miR-383 expression was relatively low in colon cancer tissue, and that miR-383 expression was associated with the pathological type and TNM stage of colon cancer.
In order to confirm that miR-383 can inhibit the development of colon cancer, an rno-miR-383 mimic was transfected into HT-29 and LoVo cells and the proliferation, migration and invasion rates were determined. It was identified that the upregulation of miR-383 suppressed proliferation and inhibited cell migration and invasion in the cell lines. This is consistent with the effect of miR-383 in medulloblastoma (7), glioma (8) and hepatocellular carcinoma (9), confirming the role of miR-383 as a tumor suppressor gene. The further exploration of the biological significance of miR-383 in colon cancer and the mechanism for interaction between miR-383 and target genes may provide novel ideas for clinical treatment.
APRIL is a member of the tumor necrosis factor family, and consists of 250 amino acids encoded by a gene located at chromosome 17p13.1 (30). Previous studies have confirmed that APRIL is highly expressed in malignant tumors, and that its expression is associated with invasion and metastasis (12,30). APRIL promotes the invasion and metastasis of colon cancer by upregulating the expression of matrix metalloproteinases (31). Chen et al (9) revealed that miR-383 may downregulate the expression of APRIL to inhibit the proliferation of hepatocellular carcinoma cells. Previous studies have also indicated that miR-383 may regulate COX-2 (22) and MCL-1 (23). In the present study, western blot analysis revealed that the expression of APRIL was decreased in cells with upregulated miR-383 compared with the control group, whereas COX-2 and MCL-1 were not altered. At present, the association between miR-383, and APRIL, MCL-1 and COX-2 expression in colon cancer has not been confirmed. The results of the present study suggest that miR-383 may be associated with the downregulation of APRIL expression, which may inhibit the invasion, migration and proliferation of colon cancer cells.
In conclusion, the expression of miR-383 in colon cancer tissue was decreased, and its expression was associated with the pathological type and TNM stage of colon cancer. The upregulated expression of miR-383 in colon cancer cells inhibited proliferation, migration and invasion. The mechanism for the effect of miR-383 may be mediated by the downregulation of APRIL. In-depth study of the interaction between miR-383 and APRIL may provide insight into the mechanism of tumor suppression by miR-383.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Fritzmann J, Morkel M, Besser D, Budczies J, Kosel F, Brembeck FH, Stein U, Fichtner I, Schlag PM, Birchmeier W. A colorectal cancer expression profile that includes transforming growth factor inhibitor BAMBI predicts metastatic potential. Gastroenterology. 2009;137:165–175. doi: 10.1053/j.gastro.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 3.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Heinsbroek SE, Squadrito ML, Schilderink R, Hilbers FW, Verseijden C, Hofmann M, Helmke A, Boon L, Wildenberg ME, Roelofs JJ, et al. MiR-511-3p, embedded in the macrophage mannose receptor gene, contributes to intestinal inflammation. Mucosal Immunol. 2016;9:960–973. doi: 10.1038/mi.2015.113. [DOI] [PubMed] [Google Scholar]
- 6.Akbari A, Ghahremani MH, Mobini GR, Abastabar M, Akhtari J, Bolhassani M, Heidari M. Down-regulation of miR-135b in colon adenocarcinoma induced by a TGF-β receptor I kinase inhibitor (SD-208) Ran J Basic Med Sci. 2015;18:856–861. [PMC free article] [PubMed] [Google Scholar]
- 7.Wang XM, Zhang SF, Cheng ZQ, Peng QZ, Hu JT, Gao LK, Xu J, Jin HT, Liu HY. MicroRNA-383 regulates expression of PRDX3 in human medulloblastomas. Zhonghua Bing Li Xue Za Zhi. 2012;41:547–552. doi: 10.3760/cma.j.issn.0529-5807.2012.08.009. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 8.Zhao L, Gu H, Chang J, Wu J, Wang D, Chen S, Yang X, Qian B. MicroRNA-383 regulates the apoptosis of tumor cells through targeting Gadd45g. PLoS One. 2014;9:e110472. doi: 10.1371/journal.pone.0110472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Guan H, Gu C, Cao Y, Shao J, Wang F. miR-383 inhibits hepatocellular carcinoma cell proliferation via targeting APRIL. Tumour Boil. 2016;37:2497–2507. doi: 10.1007/s13277-015-4071-1. [DOI] [PubMed] [Google Scholar]
- 10.Hahne M, Kataoka T, Schröter M, Hofmann K, Irmler M, Bodmer JL, Schneider P, Bornand T, Holler N, French LE, et al. APRIL, a new ligand of the tumor necrosis factor family, stimulates tumor cell growth, stimulates tumor cell growth. J Exp Med. 1998;188:1185–1190. doi: 10.1084/jem.188.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mhawech-Fauceglia P, Kaya G, Sauter G, McKee T, Donze O, Schwaller J, Huard B. The source of APRIL up-regulation in human solid tumor lesions. J Leukoc Biol. 2006;80:697–704. doi: 10.1189/jlb.1105655. [DOI] [PubMed] [Google Scholar]
- 12.Mhawech-Fauceglia P, Allal A, Odunsi K, Andrews C, Herrmann FR, Huard B. Role of the tumour necrosis family ligand APRIL in solid tumour development: Retrospective studies in bladder, ovarian and head and neck carcinomas. Eur J Cancer. 2008;44:2097–2100. doi: 10.1016/j.ejca.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Lascano V, Zabalegui LF, Cameron K, Guadagnoli M, Jansen M, Burggraaf M, Versloot M, Rodermond H, van der Loos C, Carvalho-Pinto CE, et al. The TNF family member APRIL promotes colorectal tumorigenesis. Cell Death Differ. 2012;19:1826–1835. doi: 10.1038/cdd.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang GH, Lu MH, Wang JC, Wang F, Ding WF, Wang YG, Ju SQ, Wang HM. Abnormal expression of APRIL in colorectal cancer cells promotes tumor growth and metastasis. Zhonghua Zhong Liu Za Zhi. 2013;35:249–255. doi: 10.3760/cma.j.issn.0253-3766.2013.04.003. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 15.Ding W, Wang J, Sun B, Ju S, Yuan H, Wang X, Wang Y, Wang H. APRIL knockdown suppress migration and invasion of human colon caner cells. Clin Biochem. 2009;42:1694–1698. doi: 10.1016/j.clinbiochem.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 16.Zhan J, Liu JP, Zhu ZH, Yao HR, Chen CY. Relationship between COX-2 expression and clinicopathological features of colorectal cancers. Chin Med J. 2004;117:1151–1154. [PubMed] [Google Scholar]
- 17.Basu GD, Liang WS, Stephan DA, Wegener LT, Conley CR, Pockaj BA, Mukherjee P. A novel role for cyclooxygenase-2 in regulating vascular channel formation by human breast cancer cells. Breast Cancer Res. 2006;8:R69. doi: 10.1186/bcr1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsujli M, Kawano S, Tsuji S, Swaoka H, Hori M, DuBois RN. Cyclooxygenase regulates angiogenesis induced by colon cancer cells. Cell. 1998;93:705–716. doi: 10.1016/S0092-8674(00)81433-6. [DOI] [PubMed] [Google Scholar]
- 19.Akgul C. Mcl-1is a potential therapeutic target in multiple types of cancer. Cell Mol Life Sci. 2008;66:1326–1336. doi: 10.1007/s00018-008-8637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma XT, Yu LW, Wang S, Du RY, Cui ZR. Molecular mechanism of Stat5b signaling pathway in regulating the expression of bcl-2 family members and promoting survival of human colon cancer cells. Zhonghua Shi Yan Wai Ke Za Zhi. 2005;22:1157–1169. (In Chinese) [Google Scholar]
- 21.Yoon JH, Werneburg NW, Higuchi H, Canbay AE, Kaufmann SH, Akgul C, Edwards SW, Gores GJ. Bile acids inhibit Mcl-1 protein turnover via an epidermal growth factor receptor/Raf-1-dependent mechanism. Cancer Res. 2002;62:6500–6505. [PubMed] [Google Scholar]
- 22.Fan G, Jiang X, Wu X, Fordjour PA, Miao L, Zhang H, Zhu Y, Gao X. Anti-Inflammatory Activity of Tanshinone IIA in LPS-Stimulated RAW264.7 Macrophages via miRNAs and TLR4-NF-κB pathway. Inflammation. 2016;39:375–384. doi: 10.1007/s10753-015-0259-1. [DOI] [PubMed] [Google Scholar]
- 23.Liao XH, Zheng L, He HP, Zheng DL, Wei ZQ, Wang N, Dong J, Ma WJ, Zhang TC. STAT3 regulated ATR via microRNA-383 to control DNA damage to affect apoptosis in A431 cells. Cell Signal. 2015;27:2285–2295. doi: 10.1016/j.cellsig.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Sobin LH, Fleming ID. TNM classification of malignant tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 1997;80:1803–1804. doi: 10.1002/(SICI)1097-0142(19971101)80:9<1803::AID-CNCR16>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancer. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 27.Orellana EA, Kasinski AL. MicroRNAs in cancer: A historical perspective on the path from discovery to therapy. Cancers (Basel) 2015;7:1388–1405. doi: 10.3390/cancers7030842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Wang Y, Lu S, Zhang Z, Meng H, Liang L, Zhang Y, Song B. MiR-34a inhibits colon cancer proliferation and metastasis by inhibiting platelet-derived growth factor receptor α. Mol Med Rep. 2015;12:7072–7078. doi: 10.3892/mmr.2015.4263. [DOI] [PubMed] [Google Scholar]
- 29.Liang Y, Zhao Q, Fan L, Zhang Z, Tan B, Liu Y, Li Y. Down-regulation of microRNA-381 promotes cell proliferation and invasion in colon cancer through up-regulation of LRH-1. Biomed Pharmacother. 2015;75:137–141. doi: 10.1016/j.biopha.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 30.Roosnek E, Burjanadze M, Dietrich PY, Matthes T, Passweg J, Huard B. Tumors that look for their springtime in APRIL. Crit Rev Oncol Hematol. 2009;72:91–97. doi: 10.1016/j.critrevonc.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Wang G, Wang F, Ding W, Wang X, Xu J, Li H, Wang H. APRIL mediates migration and invasion of colorectal cancer cell via regulation of matrix metalloproteinases. Chin J Microbiol Immunol. 2012;32:128–133. (In Chinese) [Google Scholar]