Abstract
Papillary thyroid carcinoma (PTC), the most common histological subtype of thyroid cancer, accounts for between 80 and 90% of all thyroid cancer cases. Previous studies have suggested that microRNAs (miRNAs/miRs) are involved in the development of PTC. The aim of the present study was to investigate whether miR-144 inhibits cellular proliferation in PTC. The expression of miR-144 was detected in PTC and corresponding adjacent non-cancerous tissues, and in the PTC cell line IHH4, using reverse transcription-quantitative polymerase chain reaction. Associations between miR-144 expression levels and the clinicopathological characteristics were analyzed. Receiver operating characteristic (ROC) curves were used to assess the diagnostic value of miR-144 expression, and the potential function of miR-144 was investigated in IHH4 cells using a Cell Counting Kit-8 and colony formation assays. Western blotting was applied to analyze the expression level of WW domain-containing transcription regulator 1 (WWTR1) in PTC tissues. miR-144 was significantly downregulated in PTC tissues and the PTC cell line. Low expression of miR-144 was associated with larger tumor sizes (P<0.001). The ROC curves demonstrated that miR-144 may be a potential biomarker for identifying PTC and non-cancerous diseases (sensitivity, 58.7%; specificity, 87.3%) as well as to differentiate PTC with tumor sizes ≥2 cm (sensitivity, 79.2%; specificity, 69.2%). Upregulation of miR-144 significantly suppressed proliferation in IHH4 cells. WWTR1 was overexpressed in PTC tissues compared with in adjacent non-cancerous tissues, and the ectopic expression of miR-144 downregulated WWTR1 in IHH4 cells. Co-transfection with pcDNA-WWTR1 and miR-144 ‘rescued’ the proliferation inhibition. The results of the present study collectively demonstrated that miR-144 is downregulated in PTC, that low expression levels of miR-144 are associated with larger tumor sizes and that miR-144 inhibits cellular proliferation in PTC by targeting WWTR1.
Keywords: papillary thyroid cancer, miR-144, cell proliferation, WW domain-containing transcription regulator protein 1, tumor sizes
Introduction
Thyroid cancer is the most common type of endocrine malignancy (1). The incidence of thyroid cancer has been increasing rapidly globally. For example, in the United States of America, thyroid cancer incidence rates have increased by 211% between 1975 and 2013 (2,3). This incidence of thyroid cancer is increasing, primarily due to a rise in the incidence of papillary thyroid carcinoma (PTC), which accounts for between 80 and 90% of all thyroid malignancies (4,5). Although the prognosis of patients with PTC is favorable, with 10- and 15-year survival rates of >91 and >87%, respectively (6,7), lymph node metastasis in the neck is observed in between 20 and 90% of all patients (8,9). Recurrence is observed in between 5 and 20% of patients who undergo a total thyroidectomy (10,11). Therefore, investigations into understanding the underlying molecular mechanisms of PTC are urgently required, in order to develop effective diagnosis and therapeutic strategies to improve patient prognosis.
MicroRNAs (miRNAs/miRs) are noncoding single stranded RNAs that regulate gene expression at the post-transcriptional level (12). A previous study identified that miRNAs serve important functions in tumorigenesis and may be applied as biomarkers in a variety of cancer types (13). miR-144 has been demonstrated to be downregulated and accompanied by suppressed proliferation and invasion in non-small cell lung cancer, breast cancer, hepatocellular carcinoma, prostate cancer, bladder cancer and laryngeal squamous cell carcinoma (14–18). The results of the present study further support these observations, as miR-144 was identified to be significantly downregulated in PTC tissues and cell lines. Low expression of miR-144 was associated with increased tumor sizes in PTC via reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and the ectopic expression of miR-144 significantly suppressed the proliferation of IHH4 cells. WW domain-containing transcription regulator 1 (WWTR1) was overexpressed in PTC tissues and associated with proliferation and invasion. Furthermore, WWTR1 was identified as a target of miR-144.
Materials and methods
Human tissue samples
PTC and adjacent non-cancerous tissues were collected from 63 patients (25 male, 38 female) who underwent thyroid cancer resection surgery at the First Hospital of China Medical University (Shenyang, China) between September 2009 and January 2015. Patient characteristics are presented in Table I. All specimens were frozen in liquid nitrogen immediately and stored at −80°C until use. A diagnosis of PTC was histologically confirmed at the First Hospital of China Medical University. The inclusion criteria were as follows: i) All patients had received primary surgery; ii) PTC was pathologically confirmed intraoperatively or postoperatively; and iii) none of the patients recruited in the present study had undergone prior oncological surgery or head and neck irradiation. The present study was approved by the Ethics Committee of the First Hospital of China Medical University and written informed consent was obtained from all study participants.
Table I.
Association between miR-144 expression and clinicopathological features in PTC.
| Characteristics | n | High expression, n (%) | Low expression, n (%) | P-value |
|---|---|---|---|---|
| Sex | 0.027a | |||
| Male | 25 | 8 (32.0) | 17 (68.0) | |
| Female | 38 | 23 (60.5) | 15 (39.5) | |
| Age, years | 0.884 | |||
| <45 | 36 | 18 (50.0) | 18 (50.0) | |
| ≥45 | 27 | 13 (48.1) | 14 (51.9) | |
| Extrathyroidal extension | 0.701 | |||
| Yes | 30 | 14 (46.7) | 16 (53.3) | |
| No | 33 | 17 (51.5) | 16 (48.5) | |
| TNM staging | 0.503 | |||
| I–II | 38 | 20 (52.6) | 18 (47.4) | |
| III–IV | 25 | 11 (44.0) | 14 (56.0) | |
| Lymph node metastasis | 0.214 | |||
| Yes | 47 | 25 (53.2) | 22 (46.8) | |
| No | 16 | 6 (37.5) | 10 (62.5) | |
| Multicentricity | 0.383 | |||
| Yes | 27 | 15 (55.6) | 12 (44.4) | |
| No | 36 | 16 (44.4) | 20 (55.6) | |
| Tumor size, cm | <0.001a | |||
| <2 | 24 | 19 (79.2) | 5 (20.8) | |
| ≥2 | 39 | 12 (30.8) | 27 (69.2) |
Data that is statistically significant, P<0.05 using χ2.
Cells culture and transfection
Nthy-ori 3–1 normal human thyroid follicular epithelial cells were obtained from The European Collection of Authenticated Cell Cultures (Salisbury, UK); IHH4 cells were obtained from the Health Science Research Resources Bank (Osaka, Japan). The Nthy-ori 3–1 cells were maintained in RPMI-1640 (Hyclone, GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The IHH4 cells were maintained in a 1:1 mixture of RPMI-1640 and Dulbecco's modified Eagle's medium supplemented with 10% FBS at 37°C, in a humidified atmosphere with 5% CO2. The miR-144 mimic and negative control (NC) were purchased from GenePharma Co., Ltd. (Suzhou, China). The sequences of the miR-144 mimics were as follows: Sense (S); 5′-UACAGUAUAGAUGAUGUACU-3′ and antisense (AS); 5′-UACAUCAUCUAUACUGUAUU-3′. The sequences of the negative control (NC) were: S; 5′-UUCUCCGAACGUGUCACGUTT-3′ and AS; 5′-ACGUGACACGUUCGGAGAATT-3′. The pcDNA-WWTR1 and the pcDNA empty vector were purchased from Shanghai GeneChem, Inc. (Shanghai, China). IHH4 cells were transfected using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Proteins were extracted 48 h post-transfection and used for western blot analysis.
Western blotting
Proteins were extracted from IHH4 cells, as well as PTC and adjacent normal tissue samples from 15 patients (6 male, 9 female), using a Total Protein Extraction kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). Ten patients were <45 years old, 5 patients were >45 years old. These 15 patients were selected based on identical inclusion criteria to the aforementioned 63 patients. A separate study population was used as the processing performed on the other tissue samples may have made protein expression difficult to detect. Protein extracts were quantified with a BCA protein assay (Beyotime Institute of Biotechnology, Haimen, China). Proteins (20–30 µg/lane) were separated by 10% SDS-PAGE and then electrophoretically transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% non-fat milk in Tris-buffered saline for 2 h at room temperature, and then incubated with primary antibodies against rabbit anti-WWTR1 (1:1,000; cat. no; 23306-1-AP; ProteinTech Group, Inc., Chicago, IL, USA) and rabbit anti-GAPDH (1:1,000; cat. no; 10494-1-AP; ProteinTech Group, Inc.) overnight at 4°C. Following incubation with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:10,000 dilution; cat. no; 7074; Cell Signaling Technology, Inc., Danvers, MA, USA) for 2 h at room temperature, the protein bands were visualized by chemiluminescence (SuperSignal@ West Pico Chemiluminescent Substrate, Thermo Fisher Scientific, Inc.) and quantified by Image J (version no. 1.43, National Institutes of Health, Bethesda, MD, USA).
Total RNA isolation and RT-qPCR
Total RNA, including miRNA, was extracted from the frozen PTC and adjacent non-cancerous tissue specimens collected from 63 patients, as well as from Nthy-ori 3-1 and IHH4 cell lines, using RNAiso (Takara Biotechnology Co., Ltd., Dalian, China). The RR716 Reverse Transcription kit (Takara Biotechnology Co., Ltd.) was used to reverse transcribe RNA into cDNA and the temperature protocol for reverse transcription was 37°C for 60 min, and then 85°C for 5 sec. RT-qPCR was performed using SYBR® Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.) on a LightCycler 480 system (Roche Diagnostics, Indianapolis, IN, USA). The thermocycling conditions included an initial denaturation step at 95°C for 30 sec, denaturation at 95°C for 5 sec and annealing at 60°C for 30 sec for 40 cycles, dissociation stage at 95°C for 60 sec, 55°C for 1 min, 95°C for 30 sec. All primers were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). The miR-144 primer sequences were as follows: forward, 5′-CGGCGGTACAGTATAGATGATG-3′. The reverse primer of miR-144 was supplied as part of the RR716 Reverse Transcription kit. The U6 primer sequences were as follows: Forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The endogenous control for normalization of the input RNA was U6. The double-standard curves method was used to analyze the relative expression of miR-144 (19).
Cellular proliferation assay
Cellular proliferation was assessed using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan). IHH4 cells (3×103 cells/well) were seeded into a 96-well plate at a final volume of 100 µl and then transfected with miR-144 mimics, negative control (NC) and pcDNA-WWTR1, as aforementioned. The samples were detected at 0, 24, 48 and 72 h following gene transfection. CCK-8 solution (10 µl) was added into each well and incubated for 3 h at 37°C. Absorbance was measured at 450 nm to calculate the number of viable cells.
Colony formation assays
For the colony formation assay, IHH4 cells transfected with miR-144 mimic and NC were seeded into 6-well plates at 5×102/well and cultured for 2 weeks at 37°C, in a humidified atmosphere with 5% CO2. Colonies were washed with PBS, fixed in 4% methanol for 20 min at room temperature and stained with 0.5% crystal violet for 20 min at room temperature. The number of colonies (>50 cells/colony) was counted (original magnification, ×40; Leica Microsystems, Wetzlar, Germany). The images were captured using a digital camera (Nikon D5300; Nikon, Toyko, Japan).
Bioinformatics analysis
The following software programs were used for the prediction of putative miR-144 targets: TargetScan (www.targetscan.org, version no. 6.0–6.2) and miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html). The date of access was June 2014.
Statistical analysis
SPSS software (version 13.0; SPSS, Inc., Chicago, IL, USA) was applied to perform statistical analyses. Data are presented as the mean ± standard deviation, obtained from at least three independent experiments. The χ2 test was applied to assess the association between miR-144 expression and the clinicopathological characteristics. A paired Student's t-test was used to assess inter-group comparisons. Two receiver operating characteristic curves (ROCs) were established to evaluate the diagnostic value of miR-144 for benign or malignant status, and larger tumors (≥2 cm). Youden's index was used to calculate the threshold value to predict benign or malignant status, and larger tumors (≥2 cm). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-144 was downregulated in human PTC tissues and cell line
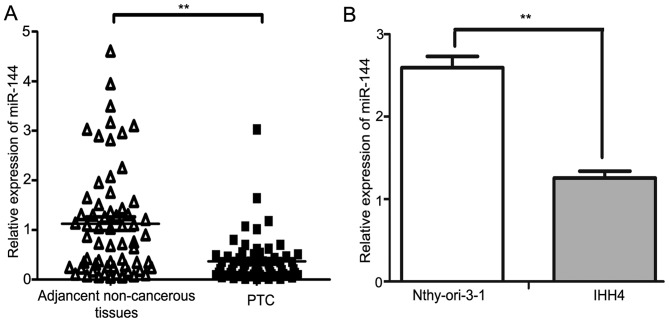
The expression of miR-144 in PTC tissues and matched non-cancerous tissues from 63 patients were investigated using RT-qPCR. The results demonstrated that miR-144 was significantly downregulated in PTC tissues compared with in the adjacent non-cancerous tissues (P<0.01; Fig. 1A). The expression of miR-144 in the IHH4 PTC cell line was decreased compared with in the normal human Nthy-ori 3-1 thyroid follicular epithelial cell line (P<0.01; Fig. 1B). The median fold-change value of miR-144 between PTC tissues and the adjacent non-cancerous tissues was 0.28, which was then used as a threshold value to separate the results into two groups: High miR-144 expression group (≥0.28; n=31); low miR-144 expression group (<0.28; n=32). The downregulation of miR-144 was associated with larger tumor sizes when the threshold value for tumor sizes was 2 cm (P<0.001), whereas no significant difference was observed between miR-144 expression level and patient age, extrathyroidal extension, TNM stage, lymph node metastasis or multicentricity (Table I). Patients were staged according to the TNM staging system (7th edition) of the American Joint Committee on Cancer (20).
Figure 1.
The expression of miR-144 in PTC and adjacent non-cancerous tissues from 63 patients, as well as the PTC cell line IHH4, were analyzed using reverse transcription-quantitative polymerase chain reaction. (A) The expression levels of miR-144 were demonstrated to be significantly downregulated in PTC compared with in paired adjacent non-cancerous tissue (P<0.01). (B) The expression levels of miR-144 were significantly downregulated in the IHH4 cells compared with in the normal human thyroid follicular epithelial cell line Nthy-ori 3-1. **P<0.01. PTC, papillary thyroid carcinoma.
Diagnostic value of miR-144
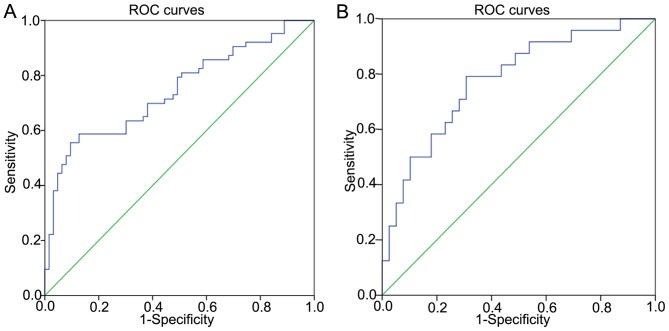
The diagnostic value of miR-144 was also investigated. ROCs curves were applied to evaluate whether miR-144 may serve as a biomarker to predict benign or malignant status. The area under the curve (AUC) was 0.743 [95% confidence interval (CI), 0.657–0.829; P<0.001], suggesting that miR-144 may be applied as a potential biomarker for PTC (Fig. 2A). The sensitivity was 58.7%, and the specificity was 87.3% when the threshold value was 0.63. The threshold values for predicting benign or malignant status refer to the relative expression in PTC tissues and the adjacent non-cancerous tissues, investigated by RT-qPCR. In addition, miR-144 was revealed to be a potential diagnostic biomarker for whether the tumor size is ≥2 cm (Fig. 2B). The AUC was 0.779 (95% CI, 0.661–0.896; P<0.001). The sensitivity was 79.2%, and the specificity was 69.2% when the threshold value was 0.28. The threshold values for predicting whether the tumor sizes ≥2 cm refer to the fold-change between the PTC tissues and the adjacent non-cancerous tissues, determined by RT-qPCR.
Figure 2.
ROC curves demonstrating the diagnostic value of miR-144. (A) Diagnostic value for differentiating between PTC and non-cancerous tissues (AUC, 0.743; 95% CI, 0.657–0.829; P<0.001). The sensitivity was 58.7%, and the specificity was 87.3%. (B) Diagnostic value of miR-144 for whether the tumor sizes ≥2 cm (AUC; 0.779; 95% CI, 0.661–0.896; P<0.001). The sensitivity was 79.2%, and specificity was 69.2%. ROC, receiver operating characteristic; PTC, papillary thyroid carcinoma; AUC, area under the curve.
miR-144 inhibited proliferation in a PTC cell line
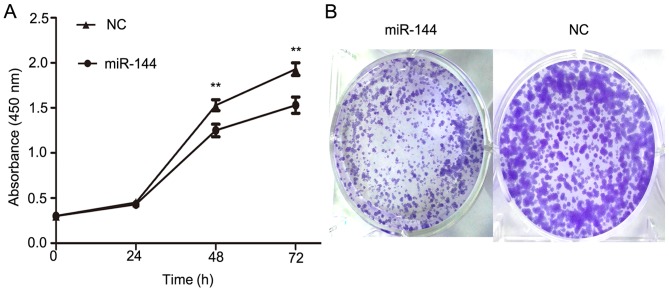
To further evaluate the function of miR-144 on PTC development, miR-144 was overexpressed in IHH4 cells using miR-144 mimics and a CCK-8 assay was used to determine its effects on cellular proliferation. The overexpression of miR-144 in IHH4 cells resulted in a significant decrease in cellular proliferation when compared with the NC group (P<0.01; Fig. 3A). Furthermore, a colony formation assay was conducted to provide further evidence of the effect on the proliferation of IHH4 cells (Fig. 3B). The colony formation assay indicated that miR-144 overexpression significantly inhibited the ability of proliferation in IHH4. These results suggested that miR-144 exhibits a suppressive effect on tumor proliferation in PTC.
Figure 3.
Ectopic overexpression of miR-144 inhibits cellular proliferation in the IHH4 PTC cell line. (A) A CCK-8 cell counting assay was applied to analyze proliferative function in IHH4 cells. Cellular proliferation was significantly inhibited in the miR-144-overexpressing group compared with in the NC group. (B) Colony formation assays indicated that the ectopic overexpression of miR-144 markedly inhibited colony formation. **P<0.01 vs. NC. PTC, papillary thyroid carcinoma; NC, negative control.
WWTR1 is a target of miR-144
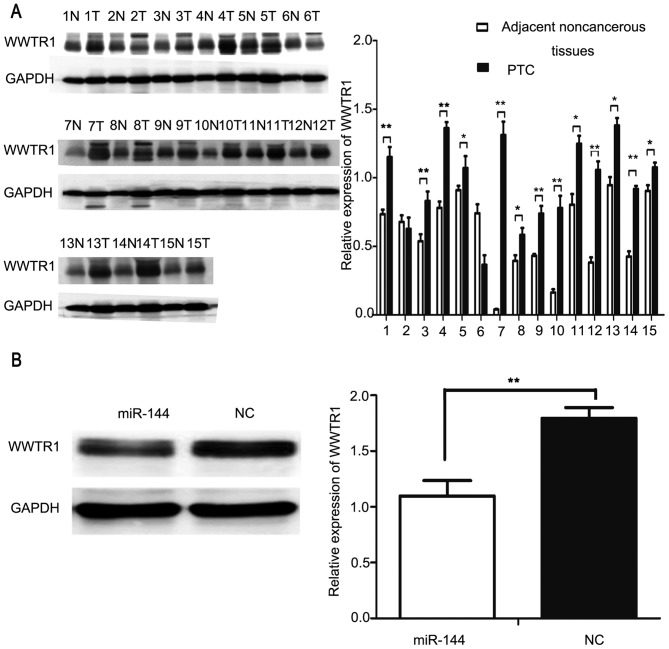
TargetScan (www.targetscan.org) and StarBase (starbase.sysu.edu.cn) were used to identify the potential targets of miR-144, and revealed that WWTR1 may be a target of miR-144. Previous studies have identified that the expression of WWTR1 is upregulated in PTC tissues as compared with in the adjacent non-cancerous tissues (21,22). The upregulation of WWTR1 was demonstrated to be positively associated with tumor size and lymph node metastasis in PTC (22). Western blot analysis was used to investigate the expression of WWTR1 in PTC and adjacent normal tissues from 15 patients. The results of the present study indicated that WWTR1 was significantly overexpressed in PTC tissues compared with in the adjacent normal tissues in 13/15 patients (P<0.05; Fig. 4A). Subsequently, miR-144 and NC were transfected into IHH4 cells and western blot analysis was applied to detect the protein expression of WWTR1. The expression of WWTR1 was significantly decreased in the miR-144-overexpressing groups compared with the NC group (P<0.01; Fig. 4B).
Figure 4.
WWTR1 is a target of miR-144. (A) WWTR1 was detected using western blot analysis in PTC, denoted by T, and adjacent non-cancerous tissues, denoted by N, from 15 (1–15) patients. WWTR1 was significantly overexpressed in PTC tissues compared with the adjacent non-cancerous tissue. (B) Ectopic overexpression of miR-144 resulted in the downregulation of WWTR1 in the PTC cell line IHH4. The protein expression of WWTR1 was significantly downregulated in the overexpressed miR-144 group compared with in the NC group. *P<0.05, **P<0.01. WWTR1, WW domain containing transcription regulator 1; PTC, papillary thyroid carcinoma; NC, negative control.
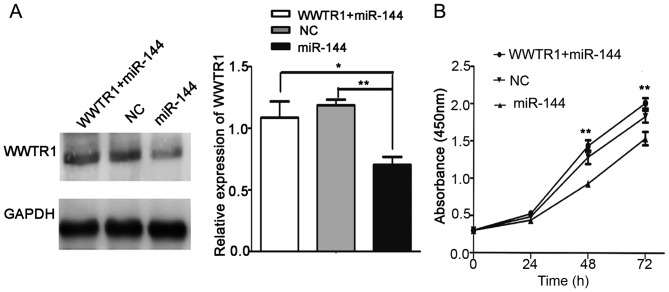
Overexpression of WWTR1 impairs the miR-144-induced inhibition of proliferation
A ‘rescue’ strategy was adopted in order to investigate the functional relevance of WWTR1 targeting by miR-144 in IHH4 cells. pcDNA-WWTR1 was first transfected into IHH4 cells. Then, miR-144 mimics were co-transfected into the cells with pcDNA-WWTR1. CCK-8 was used to evaluate the proliferation of IHH4 cells. As presented in Fig. 5A, western blot analysis was used to analyze the expression of WWTR1 in the WWTR1+miR-144, NC and miR-144 groups. The CCK-8 assay results indicated that pcDNA-WWTR1 significantly increased WWTR1 expression and rescued the inhibition of proliferation in the presence of miR-144 mimics (P<0.01; Fig. 5B). These results suggested that WWTR1 may be a target of miR-144 in PTC.
Figure 5.
Overexpression of WWTR1 impaired miR-144-induced inhibition of cellular proliferation. (A) Western blot analysis of WWTR1 expression in the miR-144 mimics+pcDNA-WWTR1 group, miR-144 mimics group and NC group in IHH4 cells. (B) A CCK-8 assay demonstrated that cellular proliferation in the miR-144 mimics+pcDNA-WWTR1 group was significantly increased compared with the miR-144 mimic group. *P<0.05, **P<0.01. WWTR1, WW domain containing transcription regulator 1; NC, negative control.
Discussion
In the USA, the overall incidence of thyroid cancer increased by 6.6% annually between 2000 and 2009; the highest increase among all types of cancer (23,24). Although PTC is a relatively indolent disease compared with hepatocellular carcinoma and gastric cancer, with low mortality, lymph node metastasis and recurrence frequently occur, leading to a poor prognosis. Therefore, the development of novel strategies for the diagnosis and treatment of PTC is urgently required (7).
miRNAs regulate the expression efficiency and stability of mRNAs at the post-transcriptional level by primarily binding to the 3′-untranslated region of target mRNAs, leading to mRNA degradation or translation inhibition (25,26). Increasing evidence has demonstrated the function of miRNAs in the development of PTC. An miRNA-chromatin immunoprecipitation microarray assay revealed a set of miRNAs that were upregulated in PTC tissues compared with those in normal thyroid tissues, including miR-21, miR-146, miR-221, miR-222, miR-155, miR-181a and miR-181b, whereas miRNAs such as miR-26a-1, miR-219-5p and miR-345 were identified to be downregulated (27,28). Unique miRNA profiles have demonstrated potential for the development of cancer biomarkers and novel therapeutics. Decreased miR-144 expression is demonstrated in numerous other types of cancer, acting as a tumor suppressor; for example, by inhibiting TP53-induced glycolysis regulatory phosphatase in lung cancer (16), inhibiting hepatocellular carcinoma by targeting E2F transcription factor 3 (17) and inhibiting enhancer of zeste homolog 2 in bladder cancer (18).
In the present study, miR-144 was revealed to inhibit cellular proliferation by targeting WWTR1 in PTC. First, the expression levels of miR-144 in PTC and corresponding adjacent normal tissues were analyzed, and the results indicated that miR-144 was significantly downregulated in PTC tissues compared with the adjacent non-cancerous tissues and was negatively associated with tumor size. miR-144 was also downregulated in PTC cell lines. Secondly, miR-144 demonstrated potential as a biomarker for diagnosing thyroid cancer (sensitivity, 58.7%; specificity, 87.3%). The expression fold-change of miR-144 between PTC tissues and adjacent non-cancerous tissues was a predictive marker for tumor sizes ≥2 cm (sensitivity, 79.2%; specificity, 69.2%). miR-144 may be applied as a biomarker in thyroid cancer, particularly for tumor size. Thirdly, the function of miR-144 was explored in the IHH4 PTC cell line. Cellular proliferation was investigated using a CCK-8 and colony formation assays. The results indicated that miR-144 significantly inhibited the proliferation ability of IHH4 cells. Subsequently, TargetScan and miRWalk were used to analyze miR-144 target genes in PTC.
The transcriptional coactivator WWTR1 was initially identified through its ability to interact with 14-3-3 proteins and is a downstream member of the Hippo pathway (29). The Hippo pathway regulates cellular proliferation, survival and differentiation in normal tissues and cells. Aberrant activation of WWTR1 is also implicated in a variety of cancer types, including breast cancer, hepatocellular carcinoma and lung cancer (30,31). In addition, the overexpression of WWTR1 has been demonstrated in PTC tissues, and is able to confer a proliferation advantage to thyroid cells and to induce the epithelial-to-mesenchymal transition (21,22). In the present study, western blot analysis was applied to analyze the expression of WWTR1. WWTR1 was significantly overexpressed in PTC tissues compared with in the adjacent non-cancerous tissues. Subsequently, miR-144 was overexpressed in IHH4 cells, and the WWTR1 protein level was revealed to be significantly decreased in the miR-144 overexpressed group compared with in the NC group. Furthermore, IHH4 cells were co-transfected with pcDNA-WWTR1 and miR-144, and it was demonstrated that the overexpression of WWTR1 was able to ‘rescue’ the inhibition of proliferation caused by miR-144.
In conclusion, the results of the present study demonstrated that miR-144 was frequently downregulated in PTC and the expression of miR-144 was associated with larger tumor sizes. miR-144 exhibited tumor suppressor activity by inhibiting IHH4 cell proliferation via targeting WWTR1. These results assist in improving understanding of the underlying molecular mechanisms in PTC, as well as providing further avenues to investigate whether miR-144 may be a potential biomarker and therapeutic target for PTC.
Acknowledgements
The present study was supported by the Liaoning BaiQianWan Talents Program (grant no. 2014921033), the Science and Technology Project of Shenyang City (grant no. F16-205-1-41), the Natural Science Foundation of Liaoning Province (grant no. 2015020536), the Liaoning Province PhD Start-up Fund (grant nos. 20141042 and 201501008) and the National Natural Science Foundation of China (grant nos. 81402208 and 81502319).
References
- 1.Sun W, Lan X, Zhang H, Dong W, Wang Z, He L, Zhang T, Liu S. Risk factors for central lymph node metastasis in CN0 papillary thyroid carcinoma: A systematic review and meta-analysis. PLoS One. 2015;10:e0139021. doi: 10.1371/journal.pone.0139021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lundgren CI, Hall P, Dickman PW, Zedenius J. Clinically significant prognostic factors for differentiated thyroid carcinoma: A population-based, nested case-control study. Cancer. 2006;106:524–531. doi: 10.1002/cncr.21653. [DOI] [PubMed] [Google Scholar]
- 3.Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. Trends in thyroid cancer incidence and mortality in the United States, 1974–2013. JAMA. 2017;317:1338–1348. doi: 10.1001/jama.2017.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hundahl SA, Fleming ID, Fremgen AM, Menck HR. A National Cancer Data Base report on 53,856 cases of thyroid carcinoma treated in the U.S., 1985–1995 [see comments] Cancer. 1998;83:2638–2648. doi: 10.1002/(SICI)1097-0142(19981215)83:12<2638::AID-CNCR31>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 5.Mao Y, Xing M. Recent incidences and differential trends of thyroid cancer in the USA. Endocr Relat Cancer. 2016;23:313–322. doi: 10.1530/ERC-15-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sciuto R, Romano L, Rea S, Marandino F, Sperduti I, Maini CL. Natural history and clinical outcome of differentiated thyroid carcinoma: A retrospective analysis of 1503 patients treated at a single institution. Ann Oncol. 2009;20:1728–1735. doi: 10.1093/annonc/mdp050. [DOI] [PubMed] [Google Scholar]
- 7.Toniato A, Boschin I, Casara D, Mazzarotto R, Rubello D, Pelizzo M. Papillary thyroid carcinoma: Factors influencing recurrence and survival. Ann Surg Oncol. 2008;15:1518–1522. doi: 10.1245/s10434-008-9859-4. [DOI] [PubMed] [Google Scholar]
- 8.Grebe SK, Hay ID. Thyroid cancer nodal metastases: Biologic significance and therapeutic considerations. Surg Oncol Clin N Am. 1996;5:43–63. [PubMed] [Google Scholar]
- 9.Kouvaraki MA, Shapiro SE, Fornage BD, Edeiken-Monro BS, Sherman SI, Vassilopoulou-Sellin R, Lee JE, Evans DB. Role of preoperative ultrasonography in the surgical management of patients with thyroid cancer. Surgery. 2003;134:946–955. doi: 10.1016/S0039-6060(03)00424-0. [DOI] [PubMed] [Google Scholar]
- 10.Mazzaferri EL, Kloos RT. Clinical review 128: Current approaches to primary therapy for papillary and follicular thyroid cancer. J Clin Endocrinol Metab. 2001;86:1447–1463. doi: 10.1210/jcem.86.4.7407. [DOI] [PubMed] [Google Scholar]
- 11.Hay ID, Thompson GB, Grant CS, Bergstralh EJ, Dvorak CE, Gorman CA, Maurer MS, McIver B, Mullan BP, Oberg AL, et al. Papillary thyroid carcinoma managed at the Mayo Clinic during six decades (1940–1999): Temporal trends in initial therapy and long-term outcome in 2444 consecutively treated patients. World J Surg. 2002;26:879–885. doi: 10.1007/s00268-002-6612-1. [DOI] [PubMed] [Google Scholar]
- 12.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 13.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 14.Yu L, Yang Y, Hou J, Zhai C, Song Y, Zhang Z, Qiu L, Jia X. MicroRNA-144 affects radiotherapy sensitivity by promoting proliferation, migration and invasion of breast cancer cells. Oncol Rep. 2015;34:1845–1852. doi: 10.3892/or.2015.4173. [DOI] [PubMed] [Google Scholar]
- 15.Zhang SY, Lu ZM, Lin YF, Chen LS, Luo XN, Song XH, Chen SH, Wu YL. miR-144-3p, a tumor suppressive microRNA targeting ETS-1 in laryngeal squamous cell carcinoma. Oncotarget. 2016;7:11637–11650. doi: 10.18632/oncotarget.7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Li P, Li J, Wang Y, Du Y, Chen X, Zang W, Wang H, Chu H, Zhao G, Zhang G. MiR-144 inhibits proliferation and induces apoptosis and autophagy in lung cancer cells by targeting TIGAR. Cell Physiol Biochem. 2015;35:997–1007. doi: 10.1159/000369755. [DOI] [PubMed] [Google Scholar]
- 17.Cao T, Li H, Hu Y, Ma D, Cai X. miR-144 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting E2F3. Tumour Biol. 2014;35:10759–10764. doi: 10.1007/s13277-014-2017-7. [DOI] [PubMed] [Google Scholar]
- 18.Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang Z, Qiu F, Lin J. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 2013;280:4531–4538. doi: 10.1111/febs.12417. [DOI] [PubMed] [Google Scholar]
- 19.Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual. 7th. Springer; New York, NY: 2010. [Google Scholar]
- 21.Liu C, Huang W, Lei Q. Regulation and function of the TAZ transcription co-activator. Int J Biochem Mol Biol. 2011;2:247–256. [PMC free article] [PubMed] [Google Scholar]
- 22.de Cristofaro T, Di Palma T, Ferraro A, Corrado A, Lucci V, Franco R, Fusco A, Zannini M. TAZ/WWTR1 is overexpressed in papillary thyroid carcinoma. Eur J Cancer. 2011;47:926–933. doi: 10.1016/j.ejca.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 24.Chen AY, Jemal A, Ward EM. Increasing incidence of differentiated thyroid cancer in the United States, 1988–2005. Cancer. 2009;115:3801–3807. doi: 10.1002/cncr.24416. [DOI] [PubMed] [Google Scholar]
- 25.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 26.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets; Proc Natl Acad Sci USA; 2006; pp. 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma; Proc Natl Acad Sci USA; 2005; pp. 19075–19080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 29.Santinon G, Pocaterra A, Dupont S. Control of YAP/TAZ activity by metabolic and nutrient-sensing pathways. Trends Cell Biol. 2016;26:289–299. doi: 10.1016/j.tcb.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Hansen CG, Moroishi T, Guan KL. YAP and TAZ: A nexus for Hippo signaling and beyond. Trends Cell Biol. 2015;25:499–513. doi: 10.1016/j.tcb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santucci M, Vignudelli T, Ferrari S, Mor M, Scalvini L, Bolognesi ML, Uliassi E, Costi MP. The Hippo pathway and YAP/TAZ-TEAD protein-protein interaction as targets for regenerative medicine and cancer treatment. J Med Chem. 2015;58:4857–4873. doi: 10.1021/jm501615v. [DOI] [PubMed] [Google Scholar]