Abstract
Dysregulation of microRNAs in various types of human cancer promote or suppress oncogenesis. MicroRNA (miR)-1 was previously revealed to function as a tumor suppressor in prostate cancer cells, and its expression was associated with reduced metastatic potential in lung cancer. The present study investigated the role of miR-1 and its association with phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α (PIK3CA) in the pathophysiology of esophageal squamous cell carcinoma (ESCC), and analyzed the effects of miR-1 inhibitor or mimics on sensitivity to epidermal growth factor receptor-tyrosine kinase inhibitors, the alterations of cell cycle distribution and apoptosis in ESCC cells. Compared with normal tissues, the level of miR-1 expression was significantly lower and PIK3CA expression was higher in ESCC tissues. The level of miR-1 expression was also inversely associated with the level of PIK3CA mRNA expression. Low miR-1 and high PIK3CA expression levels were strongly associated with lymph node metastasis, and the level of miR-1 expression was negatively associated with clinical Tumor-Node-Metastasis stage. Furthermore, exogenous expression of miR-1 inhibited growth, arrested cell cycle in the G1 phase and increased apoptosis in ESCC cells, whereas it decreased PIK3CA protein expression levels. Furthermore, overexpression of miR-1 increased the sensitivity of ESCC cells to the anticancer drug, gefitinib. A possible mechanism for this increased sensitivity to gefitinib may be inactivation of the PIK3CA signaling pathway. To the best of our knowledge, this is the first time that the results of the present study demonstrated that miR-1 upregulation may be a potential strategy for the treatment of human ESCC.
Keywords: esophageal squamous cell carcinoma; gefitinib; microRNA-1; phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α; tumor suppressor
Introduction
At present, esophageal cancer is the sixth most common cause of cancer-associated mortality worldwide (1). Based on the recent data collected from the National Central Cancer Registry in 2016, esophageal squamous cell carcinoma (ESCC) represents ~88% of esophageal cancer cases in China (2). Despite a myriad of improvement in therapeutic techniques, including chemotherapeutic, radiotherapeutic and surgical treatment during the previous 30 years, the prognosis of ESCC remains poor with a 5-year survival rate of 10–15% (3). Therefore, improved treatment strategies are urgently required. In recent years, molecular targeted therapy against key somatic alterations has become an important endeavor towards pathway-driven treatment. ESCC is frequently associated with the overexpression of the epidermal growth factor receptor (EGFR) gene (4). The dysregulation of EGFR has been implicated in the development of resistance to the conventional chemotherapy and poor clinical outcome (5). Therefore, inhibition of EGFR may be a promising approach for the management of ESCC. Previously, numerous novel compounds, including small molecule tyrosine kinase inhibitors, gefitinib and erlotinib, and monoclonal antibodies (cetuximab) targeting various proteins in the EGFR signaling cascade have been developed, and their therapeutic effects have been extensively evaluated in vitro and in clinical studies (6–10). Although EGFR inhibitors have higher efficacies and lower toxicities compared with conventional chemotherapeutic agents, patients have demonstrated highly variable responses to these inhibitors (11). Therefore, there is an urgent requirement to develop clinically useful agents with optimal therapeutic efficacies.
MicroRNAs (miRNAs) are a type of small non-coding RNAs that bind to the 3′ untranslated region of target mRNAs. miRNAs control gene expression by degrading the target mRNAs or inhibiting their translation into functional proteins (12). Emerging evidence has indicated that miRNAs may serve diverse roles in the regulation of cancer initiation and progression (13,14). miRNAs function as oncogenes or tumor suppressor genes depending on their specific target mRNAs (15–19). miR-1, which was first identified as a muscle-specific miRNA and is known to be abundantly expressed in cardiac and skeletal muscles, was previously identified as a tumor suppressor in various types of human cancer, including rhabdomyosarcoma, lung, thyroid, prostatic, bladder, colorectal and hepatocellular carcinomas (20–27). Additionally, a lower expression level of miR-1 in lung cancer is an indicator of poor prognosis (28). Previous studies have revealed that miR-1 is downregulated in human ESCC tissues and cell lines (29,30). However, the functional significance of miR-1 in ESCC has not yet been clarified. Insights into the association between miR-1 and its target oncogenes may further the understanding of the molecular mechanism underlying ESCC oncogenes and subsequently enable the development of improved therapies.
The phosphatiditylinositide-3-kinase (PI3K) signaling pathway is central to growth and survival of numerous types of cancer, and PI3K signaling can be directly activated by genetic alterations. Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α (PIK3CA), the p110α subunit of PI3K, functions as an oncogene and serves an important role in numerous types of cancer, including ESCC (31,32). A previous study found that higher expression of PIK3CA is associated with a poor prognosis in non-small cell lung cancer (28). Previous studies also revealed that PIK3CA was amplified in ESCC (33) and that the expression level of PIK3CA mRNA and protein was associated with lymph node metastasis (32,34). A previous study by the present authors has demonstrated that miR-1 inhibited tumorigenic properties of lung cancer cells by targeting PIK3CA (35). The present study aimed to investigate the levels of miR-1 and PIK3CA expression in resected esophageal tumor tissue samples and determined the association between their expression levels and clinicopathological features of patients with ESCC.
Materials and methods
Clinical samples
A total of 74 patients (61 men and 13 women) with ESCC were included in this study, ranging in age between 38 and 76 years (mean, 62 years). All patients were clinically staged according to the seventh edition of the American Joint Committee on Cancer (AJCC) system for esophageal cancer (36). Human ESCC tissues and matched adjacent normal tissues (2 cm between tumor and normal tissue) were collected directly following surgical resection at the First Affiliated Hospital of Nanjing Medical University (Nanjing, China) between January 2011 and December 2012. None of the patients had received chemotherapy or radiotherapy prior to surgery. All samples were immediately frozen in liquid nitrogen and stored at −80°C. ESCC diagnosis was confirmed following histological evaluation, which was performed by a pathologist who was blinded to the aim of the study. The present study was approved by the Ethical Review Committee in the First Affiliated Hospital of Nanjing Medical University. Written informed consent was obtained from all patients prior to enrollment in the present study.
Cell culture
The TE-1 human ESCC cell line was obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were incubated in RPMI-1640 medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in a humidified incubator containing 5% CO2. The cells in the exponential phase were used for all subsequent assays.
Drug
Gefitinib was provided by AstraZeneca Pharmaceuticals (Macclesfield, UK). Gefitinib was dissolved in DMSO to obtain a stock concentration of 10 mM and stored at −20°C. The 10 mM stocks were diluted in fresh medium prior to each experiment. The control cells were treated with the medium supplemented with an equal concentration of DMSO (<0.1%).
Transfection
The cells were transiently transfected with 50 nM miR-1 mimics or the 50 nM negative control (Genepharma, Inc., Sunnyvale, CA, USA) using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) diluted in Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The negative control was a scrambled oligonucleotide not encoding any known miRNA. The sequences of miR-1 mimics and the negative control are presented in Table I. Transfection efficiency was confirmed by analyzing miR-1 expression level using the TaqMan real-time polymerase chain reaction (PCR) system (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Quantitative real-time PCR was run on ABI PRISM 7900HT (Applied Biosystems; Thermo Fisher Scientific, Inc.). Subsequent experimentation was performed 24–48 h after transfection.
Table I.
Sequences of miR-1 mimics and the negative control.
| Name of the primers | Sequences (5′-3′) |
|---|---|
| miR-1 mimics | |
| Forward | UGGAAUGUAAAGAAGUAUGUAU |
| Reverse | ACAUACUUCUUUACAUUCCAUU |
| Negative control | |
| Forward | UUCUCCGAACGUGUCACGUTT |
| Reverse | ACGUGACACGUUCGGAGAATT |
miR, microRNA.
Quantitative RT-PCR
Quantitative real time polymerase chain reaction (qRT-PCR) was performed to determine the expression of miR-1 and all related genes. Total RNA was extracted from tissues and cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total RNA (500 ng) was quantitated at 260 nm and reverse-transcribed into cDNA using the PrimeScript RT reagent kit (Takara Biotechnology, Co., Ltd., Dalian, China) at 37°C for 15 min and 85°C for 30 sec. qPCR was performed using the SYBR Premix Ex Taq™ kit (Takara Biotechnology, Co., Ltd.) in the ABI PRISM 7900HT (Applied Biosystems; Thermo Fisher Scientific, Inc.) system. The thermocycling conditions were: 50°C for 2 min, 95°C for 10 min followed by 40 cycles with each cycle consisting of 30s at 95°C, and 1 min at 60°C. Cycle threshold (Ct) values were determined using the SDS version 2.4 software (Applied Biosystems; Thermo Fisher Scientific, Inc.). PIK3CA expression levels were normalized to β-actin expression using the 2−ΔΔCt method (37). All primers (Invitrogen; Thermo Fisher Scientific, Inc.) are presented in Table II. These experiments were performed in triplicate.
Table II.
Sequences of primers for reverse transcription-quantitative polymerase chain reaction.
| Name of gene | Sequences (5′-3′) |
|---|---|
| PIK3CA | |
| Forward | CCACGACCATCATCAGGTGAA |
| Reverse | CCTCACGGAGGCATTCTAAAGT |
| Akt | |
| Forward | GCGGCATCCACGAAACTAC |
| Reverse | TGATCTCCTTCTGCATCCTGTC |
| Survivin | |
| Forward | GGCTCTTTCTCTGTCCAGTT |
| Reverse | ACCACCGCATCTCTACATTC |
| β-actin | |
| Forward | CCAACCGCGAGAAGATGA |
| Reverse | CCAGAGGCGTACAGGGATAG |
PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α.
For miR-1 detection, 1 µg total RNA extracted from clinical samples was converted to cDNA using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The resulting cDNA was diluted in the ratio 1:40 and mixed with 1 µl miR-1 or U6 TaqMan primers in triplicate wells using TaqMan Universal Master Mix II without Uracil DNA glycosylase (Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction condition was as follows: Denaturation at 95°C for 30 sec, and followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec, and extension at 95°C for 15 sec. The plates were read using the ABI PRISM 7900HT system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Ct values were calculated using the SDS version 2.4 software (Applied Biosystems; Thermo Fisher Scientific, Inc.). miR-1 expression level was normalized to that of U6 using the 2−ΔΔCt method. The TaqMan probes for miR-1 (assay ID, 002222) and U6 (assay ID, 001973) were purchased from Applied Biosystems (Thermo Fisher Scientific, Inc.). The assay was performed in triplicate.
Western blotting
A total of 48 h after transfection, the cells were lysed in lysis buffer [50 mM Tris (pH 7.4), 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 1.0 mM EDTA, 1.0 mM Na3VO4 and 1 µg/ml freshly added leupeptin]. The protein concentration of the lysates was determined using a BCA protein assay (Bio-Rad, Laboratories Inc., Hercules, CA, USA). Equivalent amounts of protein lysates (30 µg per lane) and loading buffer were loaded onto 7.5 or 12.5% polyacrylamide gels, separated by SDS-PAGE and electrophoretically transferred to polyvinylidene membranes. The membranes were blocked for 1 h with 5% non-fat milk in TBST buffer [20 mM Tris (pH 8.0), 150 nM NaCl and 0.05% Tween-20] at room temperature and incubated overnight at 4°C with one of the following primary antibodies: Anti-PIK3CA (cat. no. AP80166; Abgent, Inc., San Diego, CA, USA) at 1:200, anti-Akt (cat. no. 9272; Cell Signaling Technology, Inc., Danvers, MA, USA) at 1:1,000, rabbit monoclonal anti-phosphorylated (p)-Akt (cat. no. 4056; Thr308; 244F9; Cell Signaling Technology, Inc.) at 1:1,000, biotinylated anti-human survivin (cat. no., BAF6471, R&D Systems, Inc., Minneapolis, MN, USA) at 1:2,000 or anti-GAPDH (cat. no., 5014; Cell Signaling Technology Inc.). The membranes were incubated with the corresponding horseradish peroxidase-conjugated rabbit anti-goat or goat anti-rat secondary antibodies (cat. nos. MR-G100 and 2B-2305; Beyotime Institute of Biotechnology) at 1:5,000 for 1 h at room temperature, and proteins were visualized using an ECL Chemiluminescence kit (EMD Millipore, Billerica, MA, USA), detected and analyzed using the Bio-Rad Gel Doc XR system (Bio-Rad, Laboratories Inc.) and ImageJ 2.1.4.7 software (National Institutes of Health, Bethesda, MD, USA).
Cell proliferation assay
Cell proliferation was assessed using Cell Counting kit-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Briefly, the transfected cells (5,000 cells/well) were seeded in 96-well plates and incubated overnight at 37°C. The cells were then treated with various concentrations of gefitinib (0, 0.01, 0.1, 1 or 10 µM) for 48 h at 37°C. The water-soluble tetrazolium salt WST-8 (10 µl) was added to each well and incubated for 1 h at 37°C. The optical density was evaluated at 450 nm using a microplate reader to determine the number of viable cells. Each experiment was performed in triplicate.
Cell cycle arrest and apoptosis assays
Transfected cells were treated with 3.0 µM gefitinib for 24 h at 37°C. The cells were harvested and washed with PBS and fixed with 70% ice-cold ethanol at −20°C for 10 min. Fixed cells were rehydrated in PBS, stained with propidium iodide (PI)/RNase (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 30 min at 37°C and analyzed by fluorescence-activated cell sorter (BD Biosciences, San Jose, CA, USA). The percentage of cells in the G0/G1, S and G2/M phases were counted and compared. The experiments were performed in triplicate.
Flow cytometry was used to determine the percentage of apoptotic cells using the Annexin V/fluorescein isothiocyanate and PI Apoptosis Detection kit (BD Biosciences), according to the manufacturer's instructions. The cells were classified as viable, dead, early apoptotic or late apoptotic. Flow cytometry was performed using FACSCalibur flow cytometer and analyzed using the automated CellQuest Pro 5.1 software (BD Biosciences). The experiments were performed in triplicate.
Statistical analysis
Statistical analysis was performed using the GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Values are presented as the mean ± standard deviation. Differences between two groups were calculated using the Student's t-test or the Mann-Whitney U test. The correlation between the expression levels of miR-1 and PIK3CA were analyzed using Pearson's correlation analysis. ESCC tissues expressing miR-1 and PIK3CA at levels less than the median expression level were assigned to the low expression group, and those with expression above the median value were assigned to the high expression group. Associations between clinicopathological features and the levels of miR-1 and PIK3CA expression were analyzed using the χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
Levels of miR-1 and PIK3CA expression in human ESCC samples
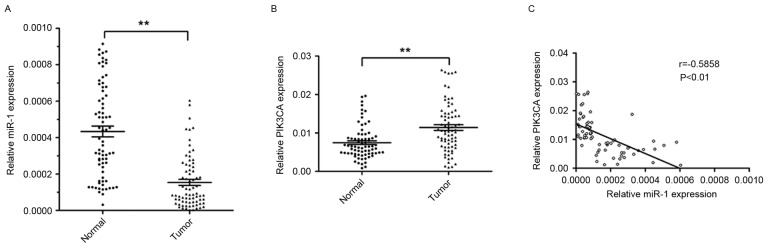
RT-qPCR was performed to analyze the levels of miR-l and PIK3CA expression in 74 ESCC tissues and corresponding non-tumor tissues. The relative expression level of miR-1 in ESCC tissues was significantly lower (~64.5%) compared with the corresponding non-tumor tissues (P<0.01; Fig. 1A). However, the level of PIK3CA expression was significantly higher in ESCC tissues (1.5 fold) compared with corresponding non-tumor tissues (P<0.01; Fig. 1B).
Figure 1.
Inverse correlation between the levels of miR-1 expression and PIK3CA expression in ESCC tissues. (A) The relative level of miR-1 expression was analyzed by TaqMan RT-qPCR in 74 ESCC tissues and corresponding non-tumor tissues. (B) The level of PIK3CA mRNA expression was analyzed by RT-qPCR in 74 ESCC tissues and corresponding non-tumor tissues. (C) Correlation between the levels of miR-1 and PIK3CA mRNA expression in ESCC tissues. Data are presented as the mean ± standard error from three independent experiments. **P<0.01. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR, microRNA; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α; ESCC, esophageal squamous cell carcinoma.
The present study further investigated the association between the level of miR-1 and PIK3CA expression. The expression of miR-1 was inversely correlated with PIK3CA mRNA expression (r=−0.5858; P<0.01; Fig. 1C). These results suggested that miR-1 may serve an important role in suppressing the expression of PIK3CA in patients with ESCC.
Association between the expression of miR-1 and PIK3CA and clinicopathological characteristics of ESCC
All patients were divided into four groups (miR-1 high, miR-1 low, PIK3CA high and PIK3CA low) on the basis of the mean levels of miR-1 and PIK3CA expression in 74 ESCC samples. The clinicopathological characteristics of these groups are summarized in Table III. Low levels of miR-1 (P<0.001) and high levels of PIK3CA (P=0.006) expression were strongly correlated with lymph node metastasis. Of the 41 tissue samples without lymph node metastasis, 25 samples (60.98%) exhibited high miR-1 expression levels and 13 samples (31.71%) exhibited high PIK3CA expression. Of the 33 tissue samples with lymph node metastasis, 5 samples (15.15%) exhibited high miR-1 expression and 21 samples (63.64%) exhibited high PIK3CA expression. Furthermore, low miR-1 expression was associated with high TNM stage (P<0.001). However, miR-1 and PIK3CA expression levels were not associated with other patient characteristics, including sex, age, pathological grading and invasion depth. These results suggested that low miR-1 expression and high PIK3CA expression may be associated with the pathophysiology of ESCC.
Table III.
Association between the levels of miR-1 and PIK3CA expression, and clinicopathological characteristics in patients with esophageal squamous cell carcinoma.
| miR-1 expression | PIK3CA expression | |||||
|---|---|---|---|---|---|---|
| Factors | High | Low | P-value | High | Low | P-value |
| Sex | 0.650 | 0.551 | ||||
| Male | 24 | 37 | 29 | 32 | ||
| Female | 6 | 7 | 5 | 8 | ||
| Age, years | 0.255 | 0.801 | ||||
| <60 | 17 | 19 | 16 | 20 | ||
| ≥60 | 13 | 25 | 18 | 20 | ||
| Pathological grading | 0.450 | 0.830 | ||||
| Well-moderately | 17 | 21 | 17 | 21 | ||
| Poorly | 13 | 23 | 17 | 19 | ||
| Invasion depth | 0.080 | 0.653 | ||||
| T1/T2 | 10 | 7 | 7 | 10 | ||
| T3/T4 | 20 | 37 | 27 | 30 | ||
| Lymph node metastasis | <0.001 | 0.006 | ||||
| Positive | 5 | 28 | 21 | 12 | ||
| Negative | 25 | 16 | 13 | 28 | ||
| TNM stage | 0.001 | 0.641 | ||||
| I/II | 22 | 15 | 16 | 21 | ||
| III/IV | 8 | 29 | 18 | 19 | ||
miR, microRNA; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α.
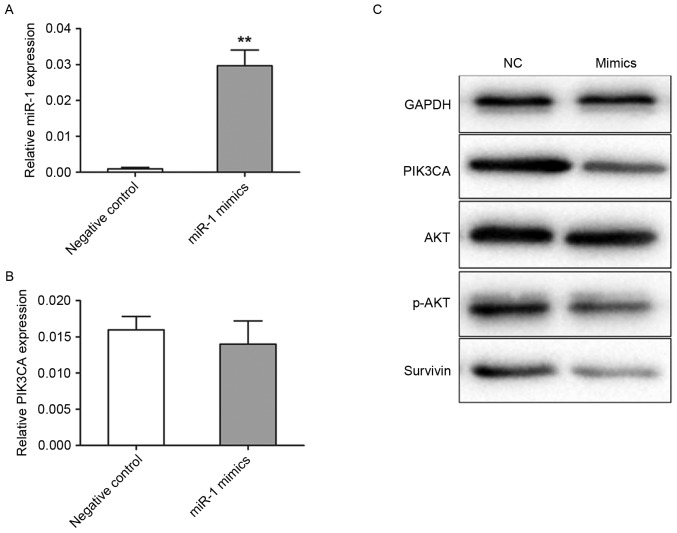
Levels of miR-1 and PIK3CA expression in transfected cells
Transfection efficiency was detected using TaqMan RT-qPCR. In the miR-1 mimics group, miR-1 expression level was 20.8-fold (P<0.001) higher compared with the negative control group (Fig. 2A). The present study further analyzed the levels of PIK3CA protein and mRNA expression by western blotting and RT-qPCR in transfected TE-1 cells, respectively. The levels of PIK3CA mRNA expression were not significantly different between the miR-1 mimic and negative control groups (Fig. 2B). However, the level of PIK3CA protein expression was revealed to be markedly decreased in the cells transfected with miR-1 mimics compared with the cells transfected with control miRNA (Fig. 2C). Akt and survivin are important downstream targets of PIK3CA, therefore in the present study the activation of Akt and survivin following PIK3CA regulation were analyzed. PIK3CA downregulation by miR-1 mimics induced a marked reduction in the levels of p-Akt and survivin expression, but not in the levels of total Akt expression, when compared with the negative control (Fig. 2C).
Figure 2.
Expression of miR-1 and PIK3CA in transfected cells. TE-1 cells were transiently transfected with miR-1 mimics (50 nM) or the negative control. The cells were obtained after 48 h for analysis. (A) TaqMan RT-qPCR detection of miR-1 expression levels in TE-1 cells. (B) The level of PIK3CA mRNA expression was detected by RT-qPCR in TE-1 cells. (C) Cell lysates were prepared and used for western blotting with antibodies specific for PIK3CA, total Akt, p-Akt and survivin. Data are presented as the mean ± standard error from three independent experiments. **P<0.01 vs. the negative control. NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR, microRNA; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α; Akt, protein kinase B; p, phosphorylated.
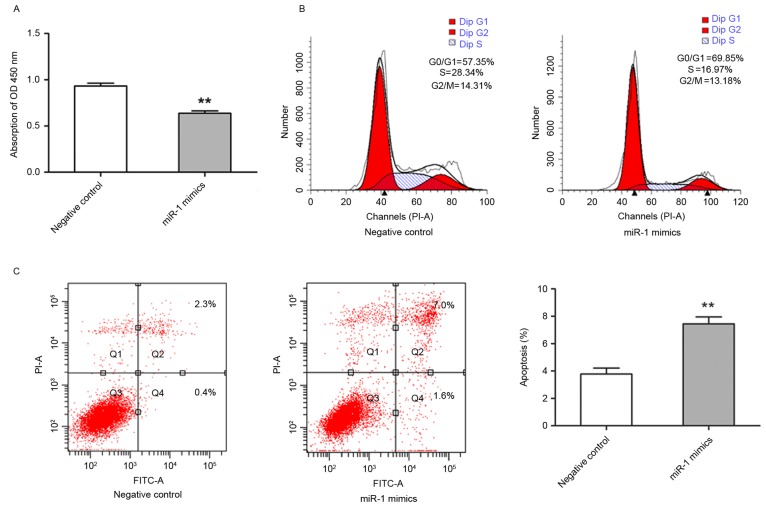
Exogenous expression level of miR-1 inhibited growth of TE-1 cells
It was demonstrated that miR-1 expression was downregulated in ESCC, therefore, subsequent study aimed to establish the biological effect of this change in expression on cell growth. TE-1 cells were transfected with miR-1 mimics, and the effects on cell proliferation were analyzed. TE-1 cell growth was significantly inhibited in miR-1-transfected cells compared with control miRNA-transfected cells (P<0.01; Fig. 3A). To further characterize miR-1-mediated inhibition of cell proliferation, cell cycle distribution and apoptotic rate were evaluated by flow cytometry. The cells transfected with miR-1 had an increased percentage of G0/G1 phase cells and a decreased percentage of G2/M phase cells in comparison with the cells transfected with control miRNA. The percentage of S phase cells was also markedly decreased in miR-1-expressing cells (Fig. 3B). In addition, the cells transfected with miR-1 exhibited significantly increased apoptosis, including early and late apoptosis, compared with the control group (P<0.01; Fig. 3C). These results suggested that miR-1 was able to inhibit growth of ESCC cells by modulating apoptosis and cell cycle progression.
Figure 3.
Exogenous expression of miR-1 inhibited growth of TE-1 cells. TE-1 cells were transiently transfected with miR-1 mimics (50 nM) or the negative control. (A) Cell proliferation was determined by cell counting kit-8 assay at 48 h post-transfection. (B) The cells were harvested for cell cycle analysis by flow cytometry at 48 h post-transfection. (C) The cells were harvested for apoptosis analysis by flow cytometry at 48 h post-transfection. Data are presented as the mean ± standard error from three independent experiments. **P<0.01 vs. negative control. miR-1, microRNA-1; OD, optical density; FITC, fluorescein isothiocyanate; PI, propidium iodide.
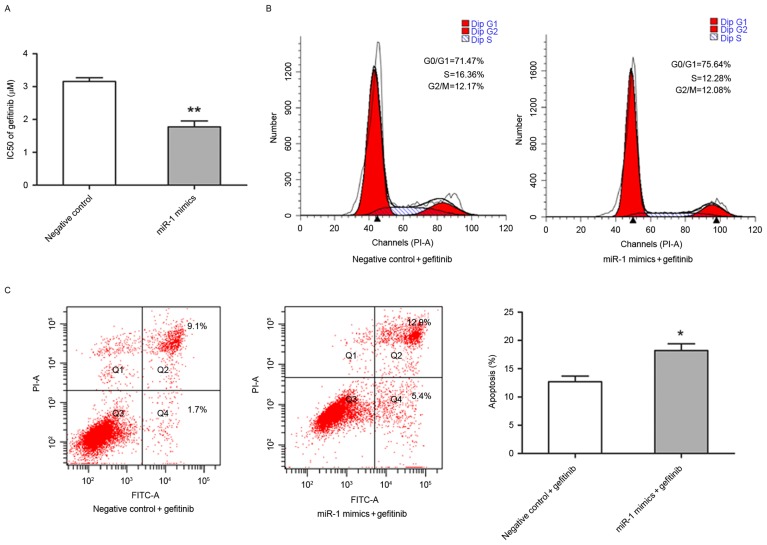
miR-1 increased sensitivity to gefitinib in TE-1 cells
The present study also investigated whether miR-1 is able to affect the sensitivity of TE-1 cells to gefitinib. The half-maximal inhibitory concentration value of gefitinib was significantly lower in miR-1-expressing TE-1 cells (1.77±0.18 µM) compared with the control TE-1 cells (3.16±0.11 µM; P<0.01; Fig. 4A). Flow cytometry was performed to determine whether the increased sensitivity to gefitibnib was due to alternation of cell cycle progression and apoptotic rate. Notably, that the percentage of TE-1 cells in different stages of the cell cycle were distinct in miR-1-expressing cells compared with control cells following exposure to 3.0 µM gefitinib for 24 h (Fig. 4B). In the presence of gefitinib, the apoptotic rate of TE-1 cells transfected with miR-1 mimics was significantly higher compared with TE-1 cells transfected with the control miRNA (P<0.05; Fig. 4C).
Figure 4.
Exogenous expression of miR-1 enhanced sensitivity to gefitinib in TE-1 cells. TE-1 cells were transiently transfected with miR-1 mimics (50 nM) or the negative control. (A) The cells following transfection were treated with various concentrations of gefitinib (0, 0.01, 0.1, 1 or 10 µM) for 48 h, and the IC50 for gefitinib were calculated by cell counting kit-8 assay. (B) The cells following transfection were treated with gefitinib (3 µM) for 24 h and were harvested for cell cycle analysis by flow cytometry. (C) The cells following transfection were treated with gefitinib (3 µM) for 24 h and were harvested for apoptosis analysis by flow cytometry. Data are presented as the mean ± standard error from three independent experiments. **P<0.01 vs. negative control; *P<0.05 vs. negative control plus gefitinib. IC50, half-maximal inhibitory concentrations; miR, microRNA; FITC, fluorescein isothiocyanate; PI, propidium iodide.
Discussion
The carcinogenesis of ESCC is a multi-stage process involving a variety of changes in gene expression and physiological structure. miRNA expression is aberrant in ESCC, suggesting that miRNAs serve an important role in ESCC progression (33). Recently, it was confirmed that miR-1 functions as a tumor suppressor in carcinogenesis (21–29,34,35). The present study revealed that miR-1 was significantly downregulated in ESCC tissues compared with normal esophageal tissues. Therefore, miR-1 may act as a tumor suppressor in ESCC.
Previous studies by the present authors revealed that miR-1 expression was significantly lower in patients with lung cancer compared with normal tissues, and the low-miR-1 expression group exhibited a significantly higher recurrence rate compared with those in the moderate-miR1 expression level group (28,35). Therefore, detection of miR-1 expression may be a valuable tool to evaluate invasion and metastasis of human NSCLC. Similar to the results obtained for patients with NSCLC, the present study demonstrated that the level of miR-1 expression was associated with clinical stage and lymph node metastasis in patients with ESCC, suggesting that low miR-1 expression may be associated with ESCC progression. Functional studies in the TE-1 ESCC cell line confirmed that upregulation of miR-1 may inhibit growth via increased apoptosis and/or cell cycle arrest in the G0/G1 phase.
The PI3K/Akt signaling pathway serves a critical role in esophageal cancer pathogenesis (32). The involvement of the PIK3CA gene, which encodes the PI3K protein p110α catalytic subunit, is activated by a series of cell surface tyrosine kinase receptors, including platelet-derived growth factor receptor and insulin growth factor receptor (38). Upon activation of these receptors, PIK3CA binds to its heterodimer p85 and promotes Akt phosphorylation at Thr308 and/or Ser473. P-Akt then activates a series of processes that drive tumor progression, including cell growth, proliferation, survival and motility (39,40). Previously, PIK3CA was identified as a target of miR-1 in the NSCLC A549 cell line by the present authors (35). miR-1 overexpression in A549 cells suppressed tumorigenic properties via PIK3CA repression and constitutive suppression of the PI3K/Akt/survivin signaling pathway (28,35). The present study revealed that the level of PIK3CA expression was higher in ESCC tissues compared with normal tissues and was inversely correlated with the level of miR-1 expression. Upregulation of miR-1 may also inhibit growth of ESCC cells via downregulation of the PI3K/Akt/survivin signaling pathway. On the basis of these findings, the present study suggested that decreased miR-1 expression modulates PIK3CA signaling in the PI3K/Akt/survivin pathway and promotes cell growth. Therefore, increasing miR-1 expression may be a novel approach for the treatment of ESCC.
High expression of EGFR may correlate with a poor response to therapy, development of cytotoxic drug resistance, disease progression and poor survival in various types of human cancer (41–43). Therefore, blockade of EGFR signal transduction appears to be a promising strategy for cancer therapy. Gefitinib is an orally active EGFR tyrosine kinase inhibitor. The antitumor activity of gefitinib has been confirmed in vitro and in vivo in various types of human cancer, including NSCLC, and colorectal, breast and head and neck cancer (41–44). Approximately 40–70% of patients with ESCC demonstrated high expression levels of EGFR (45,46). Numerous previous studies revealed that gefitinib had strong antitumor activity against ESCC in vitro and in vivo (7–9). However, a large recent clinical trial involving patients with esophageal cancer revealed that the use of gefitinib as a second-line treatment in unselected patients does not improve overall survival compared with the placebo (47). Another phase II study demonstrated that gefitinib was well tolerated by patients with recurrent or metastatic adenocarcinoma or squamous cell carcinoma of the esophagus or gastroesophageal junction, but had limited efficacy (48). It has been revealed that the PI3K/Akt signaling pathway confers gefitinib resistance independent of EGFR (49). Gefitinib in combination with specific inhibitors of the PI3K/Akt signaling pathway may cause additional cytotoxic effects in ESCC cell lines. Previous studies indicated that certain miRNAs may alter the sensitivity of cancer cells to therapeutic agents (50,51). The present study investigated whether exogenous miR-1 is able to alter sensitivity of ESCC cells to gefitinib. It was revealed that miR-1 blocked the activation of the PI3K/Akt signaling pathway and increased the sensitivity of ESCC cells to gefitinib. Therefore, the results of the present study suggested a role for microRNAs in chemosensitivity of ESCC cells. The present study suggested that a combination of miR-1 and gefitinib may be a successful therapeutic strategy for ESCC. It would be of interest to elucidate the underlying molecular mechanisms for miR-1-mediated gefitinib-induced antitumor activity. Although efficacy and tolerability of gefitinib and miR-1 needs to be extensively tested in preclinical models, the results of the present study provided a novel promising approach to improving chemotherapeutic efficacy.
To conclude, the present study demonstrated that miR-1 was downregulated in ESCC and is able to function as a tumor suppressor in this type of cancer. Overexpression of miR-1 inhibited growth, increased apoptosis and induced cell cycle arrest in the G0/G1 cell cycle phase, possibly by suppressing the PI3K/Akt/survivin signaling pathway. Additionally, miR-1 may increase the sensitivity of ESCC cells to gefitinib. Therefore, miR-1 appears to be a promising therapeutic target for ESCC treatment.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81172217), the Medical Important Talents of Jiangsu Province (grant no. RC201157) and the Project of Oncology Translational Medicine Central of Jiangsu Province (grant no. BL2012008).
References
- 1.Smyth EC, Lagergren J, Fitzgerald RC, Lordick F, Shah MA, Lagergren P, Cunningham D. Oesophageal cancer. Nat Rev Dis Primers. 2017;3:17048. doi: 10.1038/nrdp.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng H, Zheng R, Zhang S, Zuo T, Xia C, Zou X, Chen W. Esophageal cancer statistics in China, 2011: Estimates based on 177 cancer registries. Thorac Cancer. 2016;7:232–237. doi: 10.1111/1759-7714.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Office for National Statistics, corp-author. Cancer survival in England: Patients diagnosed 2006–2010 and followed up to 2011. Newport: Office for National Statistics; 2012. [Google Scholar]
- 4.Hanawa M, Suzuki S, Dobashi Y, Yamane T, Kono K, Enomoto N, Ooi A. EGFR protein overexpression and gene amplification in squamous cell carcinomas of the esophagus. Int J Cancer. 2006;118:1173–1180. doi: 10.1002/ijc.21454. [DOI] [PubMed] [Google Scholar]
- 5.Mendelsohn J, Baselga J. Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol. 2003;21:2787–2799. doi: 10.1200/JCO.2003.01.504. [DOI] [PubMed] [Google Scholar]
- 6.Teraishi F, Kagawa S, Watanabe T, Tango Y, Kawashima T, Umeoka T, Nisizaki M, Tanaka N, Fujiwara T. ZD1839 (Gefitinib, ‘Iressa’), an epidermal growth factor receptor-tyrosine kinase inhibitor, enhances the anti-cancer effects of TRAIL in human esophageal squamous cell carcinoma. FEBS Lett. 2005;579:4069–4075. doi: 10.1016/j.febslet.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 7.Hara F, Aoe M, Doihara H, Taira N, Shien T, Takahashi H, Yoshitomi S, Tsukuda K, Toyooka S, Ohta T, Shimizu N. Antitumor effect of gefitinib (‘Iressa’) on esophageal squamous cell carcinoma cell lines in vitro and in vivo. Cancer Lett. 2005;226:37–47. doi: 10.1016/j.canlet.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 8.Wei Z, Ma W, Qi X, Zhu X, Wang Y, Xu Z, Luo J, Wang D, Guo W, Li X, et al. Pinin facilitated proliferation and metastasis of colorectal cancer through activating EGFR/ERK signaling pathway. Oncotarget. 2016;7:29429–29439. doi: 10.18632/oncotarget.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol. 2005;23:2445–2459. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- 10.Sutter AP, Höpfner M, Huether A, Maaser K, Scherübl H. Targeting the epidermal growth factor receptor by erlotinib (Tarceva) for the treatment of esophageal cancer. Int J Cancer. 2006;118:1814–1822. doi: 10.1002/ijc.21512. [DOI] [PubMed] [Google Scholar]
- 11.Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, Park F, Haley JD, Gibson N, Sliwkowski MX. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 12.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 13.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 15.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 16.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 17.Ma W, Yu J, Qi X, Liang L, Zhang Y, Ding Y, Lin X, Li G, Ding Y. Radiation-induced MicroRNA-622 inhibits radiosensitivity in colorectal cancer cells by targeting Rb. Oncotarget. 2015;6:15984–15994. doi: 10.18632/oncotarget.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu XJ, Qi X, Zheng DH, Ji LJ. Modelling cancer processes with CRISPR-Cas9. Trends Biotechnol. 2015;33:317–319. doi: 10.1016/j.tibtech.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Yan D, Dong Xda E, Chen X, Wang L, Lu C, Wang J, Qu J, Tu L. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Leone V, D'Angelo D, Rubio I, de Freitas PM, Federico A, Colamaio M, Pallante P, Medeiros-Neto G, Fusco A. MiR-1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF-1alpha. J Clin Endocrinol Metab. 2011;96:E1388–E1398. doi: 10.1210/jc.2011-0345. [DOI] [PubMed] [Google Scholar]
- 23.Kojima S, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M, Seki N. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer. 2012;106:405–413. doi: 10.1038/bjc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson RS, Yi M, Esposito D, Watkins SK, Hurwitz AA, Yfantis HG, Lee DH, Borin JF, Naslund MJ, Alexander RB, et al. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res. 2012;40:3689–3703. doi: 10.1093/nar/gkr1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshino H, Enokida H, Chiyomaru T, Tatarano S, Hidaka H, Yamasaki T, Gotannda T, Tachiwada T, Nohata N, Yamane T, et al. Tumor suppressive microRNA-1 mediated novel apoptosis pathways through direct inhibition of splicing factor serine/arginine-rich 9 (SRSF9/SRp30c) in bladder cancer. Biochem Biophys Res Commun. 2012;417:588–593. doi: 10.1016/j.bbrc.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Migliore C, Martin V, Leoni VP, Restivo A, Atzori L, Petrelli A, Isella C, Zorcolo L, Sarotto I, Casula G, et al. MiR-1 downregulation cooperates with MACC1 in promoting MET overexpression in human colon cancer. Clin Cancer Res. 2012;18:737–747. doi: 10.1158/1078-0432.CCR-11-1699. [DOI] [PubMed] [Google Scholar]
- 27.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Zhao Q, Zhang B, Shao Y, Chen L, Wang X, Zhang Z, Shu Y, Guo R. Correlation between the expression levels of microRNA-1 and PIK3CA in non-small-cell lung cancer and their relationship with clinical characteristics and prognosis. Future Oncol. 2014;10:49–57. doi: 10.2217/fon.13.242. [DOI] [PubMed] [Google Scholar]
- 29.Nohata N, Sone Y, Hanazawa T, Fuse M, Kikkawa N, Yoshino H, Chiyomaru T, Kawakami K, Enokida H, Nakagawa M, et al. miR-1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget. 2011;2:29–42. doi: 10.18632/oncotarget.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 31.Chen H, Guan R, Lei Y, Chen J, Ge Q, Zhang X, Dou R, Chen H, Liu H, Qi X, et al. Lymphangiogenesis in gastric cancer regulated through Akt/mTOR-VEGF-C/VEGF-D axis. BMC Cancer. 2015;15:103. doi: 10.1186/s12885-015-1109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akagi I, Miyashita M, Makino H, Nomura T, Hagiwara N, Takahashi K, Cho K, Mishima T, Ishibashi O, Ushijima T, et al. Overexpression of PIK3CA is associated with lymph node metastasis in esophageal squamous cell carcinoma. Int J Oncol. 2009;34:767–775. doi: 10.3892/ijo_00000202. [DOI] [PubMed] [Google Scholar]
- 33.Yang YL, Chu JY, Luo ML, Wu YP, Zhang Y, Feng YB, Shi ZZ, Xu X, Han YL, Cai Y, et al. Amplification of PRKCI, located in 3q26, is associated with lymph node metastasis in esophageal squamous cell carcinoma. Genes Chromosomes Cancer. 2008;47:127–136. doi: 10.1002/gcc.20514. [DOI] [PubMed] [Google Scholar]
- 34.Wada S, Noguchi T, Takeno S, Kawahara K. PIK3CA and TFRC located in 3q are new prognostic factors in esophageal squamous cell carcinoma. Ann Surg Oncol. 2006;13:961–966. doi: 10.1245/ASO.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Yu QQ, Wu H, Huang X, Shen H, Shu YQ, Zhang B, Xiang CC, Yu SM, Guo RH, Chen L. miR-1 targets PIK3CA and inhibits tumorigenic properties of A549 cells. Biomed Pharmacother. 2014;68:155–161. doi: 10.1016/j.biopha.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Mönig SP, Hölscher AH. Clinical classification systems of adenocarcinoma of the esophagogastric junction. Recent Results Cancer Res. 2010;182:19–28. doi: 10.1007/978-3-540-70579-6_2. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Vanhaesebroeck B, Stein RC, Waterfield MD. The study of phosphoinositide 3-kinase function. Cancer Surv. 1996;27:249–270. [PubMed] [Google Scholar]
- 39.Andjelković M, Alessi DR, Meier R, Fernandez A, Lamb NJ, Frech M, Cron P, Cohen P, Lucocq JM, Hemmings BA. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 40.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 41.Bianco C, Tortora G, Bianco R, Caputo R, Veneziani BM, Caputo R, Damiano V, Troiani T, Fontanini G, Raben D, et al. Enhancement of antitumor activity of ionizing radiation by combined treatment with the selective epidermal growth factor receptor tyrosine kinase inhibitor ZD1839 (Iressa) Clin Cancer Res. 2002;8:3250–3258. [PubMed] [Google Scholar]
- 42.Qi X, Zhang L, Lu X. New insights into epithelial-to-mesenchymal transition in cancer. Trends Pharmacol Sci. 2016;37:246–248. doi: 10.1016/j.tips.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Sirotnak FM, Zakowski MF, Miller VA, Scher HI, Kris MG. Efficacy of cytotoxic agents against human tumor xenografts is markedly enhanced by coadministration of ZD1839 (Iressa), an inhibitor of EGFR tyrosine kinase. Clin Cancer Res. 2000;6:4885–4892. [PubMed] [Google Scholar]
- 44.Baselga J, Rischin D, Ranson M, Calvert H, Raymond E, Kieback DG, Kaye SB, Gianni L, Harris A, Bjork T, et al. Phase I safety, pharmacokinetic, and pharmacodynamic trial of ZD1839, a selective oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with five selected solid tumor types. J Clin Oncol. 2002;20:4292–4302. doi: 10.1200/JCO.2002.03.100. [DOI] [PubMed] [Google Scholar]
- 45.Iihara K, Shiozaki H, Tahara H, Kobayashi K, Inoue M, Tamura S, Miyata M, Oka H, Doki Y, Mori T. Prognostic significance of transforming growth factor-alpha in human esophageal carcinoma. Implication for the autocrine proliferation. Cancer. 1993;71:2902–2909. doi: 10.1002/1097-0142(19930515)71:10<2902::AID-CNCR2820711004>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 46.Ozawa S, Ueda M, Ando N, Abe O, Shimizu N. High incidence of EGF receptor hyperproduction in esophageal squamous-cell carcinomas. Int J Cancer. 1987;39:333–337. doi: 10.1002/ijc.2910390311. [DOI] [PubMed] [Google Scholar]
- 47.Dutton SJ, Ferry DR, Blazeby JM, Abbas H, Dahle-Smith A, Mansoor W, Thompson J, Harrison M, Chatterjee A, Falk S, et al. Gefitinib for oesophageal cancer progressing after chemotherapy (COG): A phase 3, multicentre, double-blind, placebo-controlled randomised trial. Lancet Oncol. 2014;15:894–904. doi: 10.1016/S1470-2045(14)70024-5. [DOI] [PubMed] [Google Scholar]
- 48.Adelstein DJ, Rodriguez CP, Rybicki LA, Ives DI, Rice TW. A phase II trial of gefitinib for recurrent or metastatic cancer of the esophagus or gastroesophageal junction. Invest New Drugs. 2012;30:1684–1689. doi: 10.1007/s10637-011-9736-z. [DOI] [PubMed] [Google Scholar]
- 49.Janmaat ML, Kruyt FA, Rodriguez JA, Giaccone G. Response to epidermal growth factor receptor inhibitors in non-small cell lung cancer cells: Limited antiproliferative effects and absence of apoptosis associated with persistent activity of extracellular signal-regulated kinase or Akt kinase pathways. Clin Cancer Res. 2003;9:2316–2326. [PubMed] [Google Scholar]
- 50.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 51.Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, Gillespie E, Slack FJ. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]