Abstract
Genetic intratumor heterogeneity is associated with tumor occurrence, development and overall outcome. The present study aims to explore the association between mutant-allele fraction (MAF) heterogeneity and patient overall survival in lung cancer. Somatic mutation data of 939 non-small cell lung cancer (NSCLC) cases were obtained from The Cancer Genome Atlas. Entropy-based mutation allele fraction (EMAF) score was used to describe the uncertainty of individual somatic mutation patterns and to further analyze the association with patient overall survival. Results indicated that association between EMAF and overall survival was significant in the discovery set [hazard ratio (H)R=1.62; 95% confidence interval (CI): 1.08–2.41; P=0.018] and replication set (HR=1.63; 95% CI: 1.11–2.37; P=0.011). In addition, EMAF was also significantly different in lung adenocarcinoma and squamous cell carcinoma. Furthermore, a significant difference was indicated in early-stage patients. Results from c-index analysis indicated that EMAF improved the model predictive performance on the 3-year survival beyond that of traditional clinical staging, particularly in early-stage patients. In conclusion, EMAF successfully reflected MAF heterogeneity among patients with NSCLC. Additionally, EMAF improved the predictive performance in early-stage patient prognosis beyond that of traditional clinical staging. In clinical application, EMAF appears to identify a subset of early-stage patients with a poor prognosis and therefore may help inform clinical decisions regarding the application of chemotherapy after surgery.
Keywords: mutant-allele fraction heterogeneity, non-small cell lung cancer, overall survival, information entropy
Introduction
Lung cancer, predominantly non-small cell lung cancer (NSCLC), is the most commonly diagnosed malignancy and is a leading cause of cancer-related deaths worldwide (1,2). Diagnosis often occurs in late-stage disease, when most patients have missed the optimal window for surgery, so prognosis is usually poor. However, genomic profiling of tumor tissues can identify biomarkers for early diagnosis of NSCLC and its therapy. Early-diagnosed patients have considerably favorable prognosis, although divergence still exists among patients with similar clinical characteristics (3). This phenomenon indicates the importance of improved understanding of genetic and molecular heterogeneity among these patients.
Intratumor heterogeneity has been shown using somatic mutations and DNA copy number alterations among several cancers, including lung cancer (4,5), and is associated with worse clinical outcomes (6–10). Several methods have been proposed to explore tumor heterogeneity (11–14). However, most previous investigations are small-scale studies or single cell analyses, which are difficult to extend to large populations. Recently, a study has proposed a new method, mutant-allele tumor heterogeneity (MATH), that has been successfully applied in head and neck squamous cell carcinoma (HNSC) populations to differentiate patient prognosis (15–18). However, MATH is not generalizable to lung cancer using data from The Cancer Genome Atlas (TCGA), potentially due to different distribution patterns of mutational fractions they cannot fully reflect in lung cancer populations.
Therefore, we propose a measurement to describe mutant-allele fraction (MAF) heterogeneity that was evaluated based on whole exome sequencing of tumor and matched normal DNA (19) in lung cancer populations from TCGA (20). The proposed statistic successfully measures tumor heterogeneity and appears to be a novel prognostic biomarker for NSCLC.
Materials and methods
Study population
Clinical and tumor characteristic information and tumor-specific somatic mutation data of lung cancer were obtained from TCGA on June 23, 2016 including lung adenocarcinomas (LUAD) and squamous cell carcinomas (LUSC). Somatic mutations were identified from whole exome sequencing data by the TCGA Broad Institute Team, and followed by standard quality control processed (21–23) and VarScan algorithm (24). Patients with missing follow-up information were excluded. Total TCGA data included 939 NSCLC cases with both clinical information and mutation data. We randomly divided them into two datasets equally: Discovery set and replication set.
MAF
To identify genomic loci that had tumor-specific mutations based on tumor-normal pairs, the number of mutant reads and reference allele reads at each mutant locus was obtained from whole exome sequencing data of tumor and adjacent normal tissues. MAF, also called variant allele frequency (VAF), was calculated as:
As reported previously, MAF was influenced by presence of sub-clonal mutations and copy number alterations, which are higher when a locus is mutated earlier in a clonal evolution or undergoes allele-specific amplification (17). The patients usually had a number of mutant loci, leading to different MAF values (most of them were hundreds) within each patient. As well, the distribution of MAF values within each patient was unique and differed from the others.
Further, we referred the theory of information entropy to describe MAF heterogeneity from the MAF values. Entropy measures a quantity of uncertainty (25), and is originally defined by a discrete random variable (26):
In case of continuous MAF, the value was categorized into bins by length ∆ (∆→0). Thus, the entropy for a continuous variable was:
Where f(x) is the probability density function of x. We named it entropy-based MAF (EMAF).
To estimate it, we smoothed MAF distribution by kernel function θ (27), which measures ‘similarity’ between pairs of samples Xn and Xn. Kernel density estimation is of the form:
Where θ is the default step kernel [θ (x>0)=0, θ (x≤0)=1], |xn-xn'| represents distance between paired samples, and r is kernel width.
MATH method
MATH is a simple method that calculates the variance of MAF values. In MATH, the median absolute deviation (MAD) of MAF values was calculated first: MAD = (|xi -median(x)|). MATH was calculated as 100xMAD/median. Further, calculation of MAD followed with values scaled by a constant factor (1.4826) so that the expected MAD of a sample from a normal distribution equals the standard deviation (15).
Statistical analysis
Continuous variables were described as mean ± SD and compared by student's t-test, while categorized variables were summarized by frequency (n) and compared by Fisher's exact test. General linear model was used to compare EMAFs with other characteristics. Associations between EMAF and overall survival were evaluated by Cox proportional hazard models with adjustment for common clinical variables (age, gender, smoking status, clinical stage, T classification, N classification and histology type). Survival curves were drawn with the Kaplan-Meier method and were compared among subgroups using log-rank tests. C-index was used for evaluating overall adequacy of risk prediction procedures with censored survival data (28).
Statistical analyses were performed using R v.3.2.2 (The R Foundation). P-values were two-sided and P<0.05 was considered to indicate a statistically significant difference.
Results
Demographic and clinical characteristics
The 939 lung cancer cases were equally divided into the discovery set and replication set (Table I). The discovery set (n=469) had an average age of 65.82±9.72 years, ranging from 33–86 years, and 111 (23.0%) individuals were followed until death. Of them, 80.4% had early stage disease (stage I–II). Among the 470 cases in the replication set, they had an average age of 66.29±9.06 years, ranging from 38–90 years, and 127 (27.0%) individuals were followed until death. 81.0% had early stage disease in the replication set. The comparisons of baseline information in the two sets were all non-significant (P>0.05).
Table I.
Demographic and clinical characteristics of lung cancer patients in The Cancer Genome Atlas.
| Characteristic | Discovery set (n=469) | Replication set (n=470) | P-value |
|---|---|---|---|
| Median survival time (months) | 45.30 | 41.33 | 0.326 |
| Censored rate (%) | 76.33 | 72.97 | 0.260 |
| Age (year) | 65.82±9.72 | 66.29±9.06 | 0.443 |
| Gender | |||
| Male | 268 | 295 | 0.083 |
| Female | 201 | 175 | |
| Race | 0.851 | ||
| White | 370 | 348 | |
| American Indian/Alaska native | 1 | 0 | |
| Asian | 8 | 8 | |
| Black/African American | 28 | 31 | |
| Missing | 62 | 83 | |
| Tobacco history | 0.776 | ||
| Never smoke/quit >15 y | 142 | 146 | |
| Current smoker/quit <15 y | 310 | 318 | |
| Missing | 17 | 16 | |
| Histology type | 0.896 | ||
| Adenocarcinoma | 231 | 229 | |
| Squamous cell carcinoma | 238 | 241 | |
| T classification | 0.657 | ||
| T1 | 140 | 123 | |
| T2 | 251 | 273 | |
| T3 | 54 | 53 | |
| T4 | 22 | 20 | |
| Missing or not available | 2 | 1 | |
| N classification | 0.148 | ||
| N0 | 309 | 290 | |
| N1 | 108 | 102 | |
| N2 | 44 | 61 | |
| N3 | 2 | 5 | |
| Missing or not available | 6 | 12 | |
| M classification | 0.903 | ||
| M0 | 350 | 349 | |
| M1 | 16 | 14 | |
| Missing or not available | 103 | 107 | |
| Clinical stage | 0.826 | ||
| I | 243 | 236 | |
| II | 134 | 130 | |
| III | 74 | 84 | |
| IV | 18 | 20 |
MATH in NSCLC cases
We applied MATH method to NSCLC cases to determine if the method was applicable to cancers beyond HNSC. Using multivariable Cox regression model adjusted for age, gender, smoking status, histology type and clinical stage, MATH showed non-significant associations with survival for either discovery set (HR=1.17; 95% CI: 0.80–1.72; P=0.409) or replication set (HR=0.85; 95% CI: 0.51–1.40; P=0.533) cases. The results may reveal MATH is not generalizable in lung cancer.
MAF and EMAF profiles
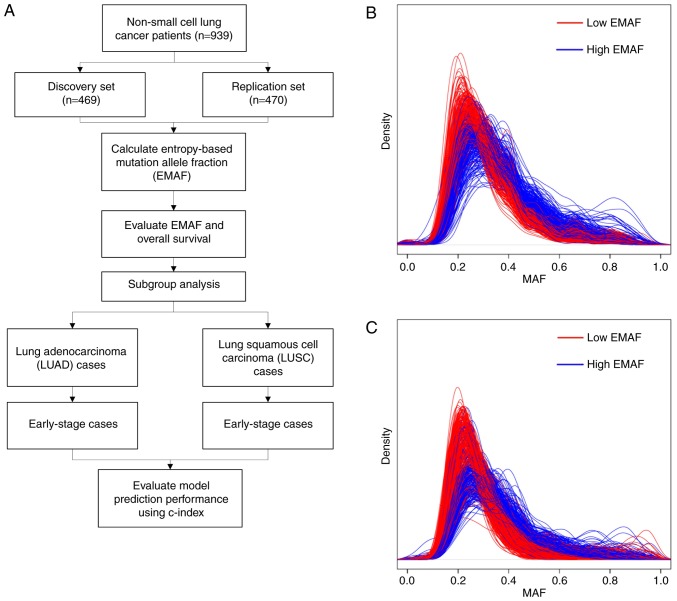
We categorized the EMAF values into high- and low-EMAF group by the median value within each dataset. Kernel smoothed distributions of MAF values of discovery and replication cases were shown in Fig. 1B and C. The distributions of cases with lower EMAF scores tended to have a smaller uncertainty. In the discovery set, EMAF ranged from 1.87 to 3.02, with a mean of 2.77 and a median of 2.79. The relation between EMAF and clinical stage was not statistically significant (β=−0.003, P=0.248). In the replication set, EMAF ranged from 1.94 to 3.02, with a mean of 2.78 and a median of 2.81. EMAF also showed no relationship with the clinical stage (β=0.001, P=0.859). This might indicate that EMAF was independent from clinical stage.
Figure 1.
(A) Flow chart of our study design. (B and C) MAF distribution of the discovery set (A) and replication set (B) cases smoothed by kernel density function with a bandwidth of 0.3. Patients were divided into low-EMAF (red) and high-EMAF (blue) groups by the median value. Distribution of MAF values within each patient was represented as a smooth line in the figures.
EMAF and clinical outcome
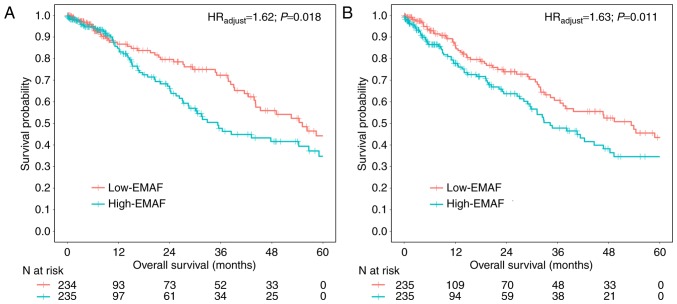
Univariate Cox regression showed a 1.50 times higher risk of death for the high-EMAF group compared to the low-EMAF group in the discovery set (HRunadjust=1.50; 95% CI: 1.03–2.18; P=0.035), and a 1.47 times in the replication set (HRunadjust=1.47; 95% CI: 1.04–2.09; P=0.031). Results retained statistical significance with further adjustment for covariates, including age, gender, smoking status, clinical stage, T classification, N classification and histology type for the discovery set (HRadjust=1.62; 95% CI: 1.08–2.41; P=0.018) (Fig. 2A) and replication set (HRadjust=1.63; 95% CI: 1.11–2.37; P=0.011) (Fig. 2B). We did find a relationship between MAF heterogeneity and clinical outcome (overall survival) (Table II).
Figure 2.
EMAF and NSCLC patients' overall survival. Survival curves for the discovery set (A) and replication set (B) cases, which were divided into low-EMAF (red) and high-EMAF (blue) groups by the median value. Hazard ratios (HRadjust) and P-values were estimated by Cox regression with adjustment for age, gender, smoking status, clinical stage, T classification, N classification and histology type l.
Table II.
Cox regression analysis of clinical characteristics and EMAF.
| Discovery set (n=469) | Replication set (n=470) | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariable | Multivariable | Univariable | Multivariable | |||||
| Characteristics | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
| High EMAF | 1.50 (1.03–2.18) | 0.035 | 1.62 (1.08–2.41) | 0.018 | 1.47 (1.04–2.09) | 0.031 | 1.63 (1.11–2.37) | 0.011 |
| Age (per year) | 0.99 (0.97–1.01) | 0.708 | 1.00 (0.98–1.03) | 0.660 | 1.01 (0.99–1.04) | 0.065 | 1.02 (1.00–1.04) | 0.034 |
| Gender (Female) | 1.01 (0.69–1.47) | 0.951 | 0.93 (0.62–1.39) | 0.726 | 0.92 (0.64–1.33) | 0.681 | 0.96 (0.64–1.46) | 0.878 |
| Clinical Stage (per stage) | 1.55 (1.28–1.89) | <0.001 | 1.34 (0.98–1.84) | 0.064 | 1.36 (1.15–1.62) | <0.001 | 0.98 (0.71–1.37) | 0.951 |
| Smoking status (Current smoker/quit <15 y) | 0.79 (0.65–0.96) | 0.021 | 0.82 (0.67–1.00) | 0.058 | 1.04 (0.87–1.24) | 0.615 | 0.96 (0.79–1.16) | 0.681 |
| T classification (per stage) | 1.34 (1.07–1.68) | 0.009 | 1.05 (0.80–1.39) | 0.702 | 1.56 (1.24–1.95) | <0.001 | 1.54 (1.15–2.06) | 0.003 |
| N classification (per stage) | 1.49 (1.18–1.87) | <0.001 | 1.16 (0.84–1.59) | 0.368 | 1.43 (1.16–1.77) | <0.001 | 1.35 (0.97–1.90) | 0.074 |
| Histology type (LUSC) | 0.75 (0.52–1.09) | 0.143 | 0.75 (0.50–1.13) | 0.179 | 1.14 (0.80–1.63) | 0.452 | 1.10 (0.72–1.69) | 0.637 |
EMAF, entropy-based mutation allele fraction; LUSC, lung squamous cell carcinoma; HR, hazard ratio; CI, confidence interval.
Subgroup analysis with histology type and clinical stage
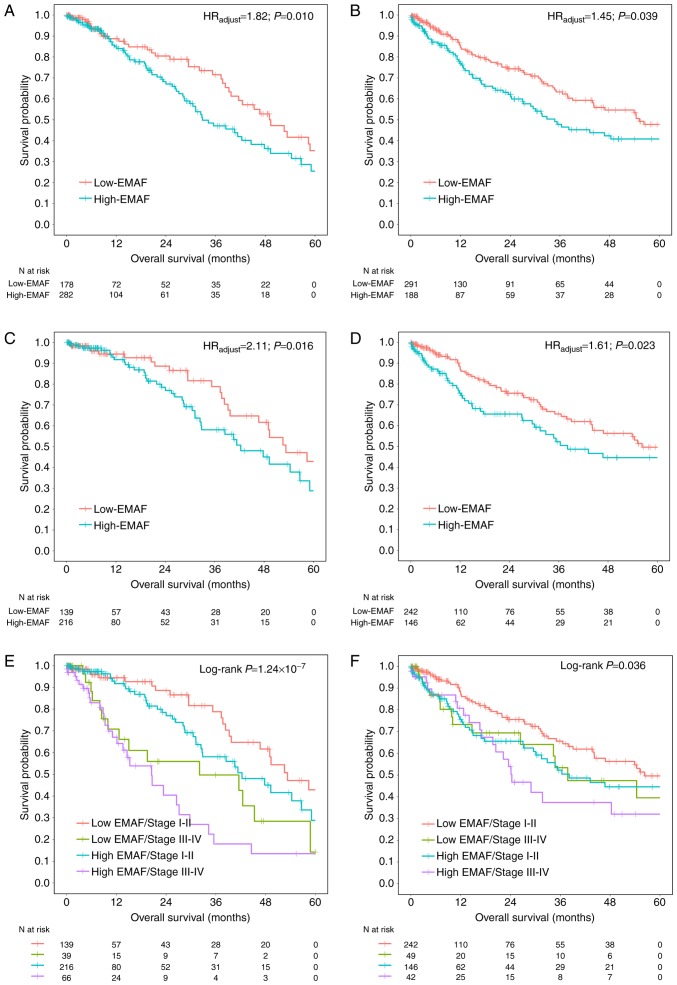
Further, we explored the relationship between EMAF and survival with different histology type. After adjustment for age, gender, smoking status, clinical stage, T classification, N classification, EMAF showed significance in both LUAD (HR=1.82; 95% CI: 1.15–2.87; P=0.010) and LUSC (HR=1.45; 95% CI: 1.02–2.05; P=0.039) cases (Fig. 3A and B).
Figure 3.
Subgroup analysis with histology type and clinical stage. Subgroup survival curves for 460 LUAD (A) and 479 LUSC (B) cases were analyzed. In addition, we also analyzed the early-stage (stage<3) patients in (C) LUAD and (D) LUSC. Further, survival curves were plotted depicting relation of EMAF and clinical stage to overall survival in (E) LUAD and (F) LUSC cases.
A biomarker for early-stage lung cancer is more important and urgent. Subgroup analysis among 355 early-stage (stage I–II) LUAD cases showed consistent results (HR=2.11; 95% CI: 1.15–3.90; P=0.016) (Fig. 3C), as well as positive results among 388 early-stage LUSC patients (HR=1.61; 95% CI: 1.06–2.43; P=0.023) (Fig. 3D). Besides, subgroup analyses by clinical stage (Fig. 3E and F) showed consistently significant results for LUAD (P=1.24×10−7) and LUSC (P=0.036).
Predict performance of EMAF on 3-year overall survival
Furthermore, the index of concordance (c-index) was used to evaluate the predict performance of EMAF on 3-year overall survival (Table III). Among LUAD cases, the c-index was 0.70 for clinical characteristics including age, gender, smoking status, TNM stage, T classification, and N classification, and was increased to 0.76 by adding on EMAF. Similarly, the c-index for LUSC cases was also improved from 0.58 (for clinical characteristics only) to 0.63 (by adding on EMAF score). Results were consistent among early-stage patients (LUAD: 0.65 to 0.73; LUSC: 0.58 to 0.64). Thus, EMAF appears to improve performance of prognostic prediction beyond clinical information.
Table III.
Performances of prognostic prediction on 3-year overall survival.
| Death rate, %(n/N) | Prognostic prediction performance, c-index (95%CI) | ||||
|---|---|---|---|---|---|
| Study population | Low-EMAF group | High-EMAF group | EMAF only | Clinical characteristics onlya | Both |
| LUAD | 20.6 (46/223) | 25.3 (60/237) | 0.60 (0.53,0.66) | 0.70 (0.63,0.78) | 0.76 (0.68,0.83) |
| Early-stage | 17.0 (33/193) | 22.2 (36/162) | 0.60 (0.51,0.68) | 0.65 (0.55,0.75) | 0.73 (0.63,0.83) |
| LUSC | 22.2 (57/257) | 33.8 (75/222) | 0.58 (0.53,0.63) | 0.58 (0.53,0.64) | 0.63 (0.57,0.68) |
| Early-stage | 20.7 (45/217) | 30.4 (52/171) | 0.60 (0.54,0.66) | 0.58 (0.51,0.65) | 0.64 (0.57,0.71) |
Clinical variables including age, gender, smoking status, clinical stage, T classification, and N classification. LUSC, lung squamous cell carcinoma; EMAF, entropy-based mutation allele fraction; LUAD, lung adenocarcinoma; CI, confidence interval.
Discussion
In this study, based on the theory that high genetic heterogeneity is associated with worse overall survival, we propose an information entropy-based score, EMAF, to evaluate the uncertainty of individual genome-wide mutational distribution patterns of tumor DNA, also described as MAF heterogeneity. Lung cancer patients with higher EMAF scores tend to have higher uncertainty of MAF distribution. Moreover, our study found that high EMAF scores correlate with poor clinical outcomes among NSCLC cases. Our hypothesis is that high EMAF indicates an early start and a high percentage of sub-clonal mutations, which make the tumor more aggressive (29) as well as representing a more disordered regulation mechanism. Both of them may have an adverse effect on the tumor progression and clinical outcome.
Mroz et al (17) reported a MATH score by MAD/median of mutation allele fraction distribution to describe the intratumor heterogeneity in HNSC data from TCGA. However, our results show that MATH scores failed to be applied in NSCLC data. A potential explanation may be that MATH used incomplete information of the complicated NSCLC MAF distribution, while EMAF scores proposed in this study consider overall MAF distribution comprehensively by information entropy and integral process.
EMAF appears to distinguish adequately NSCLC patient prognosis, and retains significant among early-stage patients. Besides, clinical information especially clinical stage is regarded as an efficient and common predictive factor to clinical outcome (30–32). Notably, EMAF provides additional distinguishing capability to survival in addition to clinical information. Thus, the combination of EMAF and clinical information could significantly improve the predictive performance for 3-year overall survival. Early-stage lung cancer patients are expected to have favorable clinical prognosis, although they actually have diverse survival outcomes (33), which may be due to timing for treatment after surgery (34). EMAF appears to identify a subset of early-stage patients with relatively poor prognosis, which may indicate an alternative preoperative chemotherapy or radiation strategy followed by surgery.
EMAF is based on next generation sequencing which will have a wide range of applications in the future. In addition, it is a quantitative measure as long as tumor and matched normal somatic mutations can be sequenced. Cancer consists of a quite large complex regulatory network, unlike some methods that need to select biological markers as the first step, EMAF is based on overall distribution of each person and is not restricted by a single locus. Further, due to the use of the kernel function estimation method, EMAF also is not restricted by distribution type and thus has wide applicability. Although information entropy with a continuous version cannot be used as a measure of amount of information, it can be used as a relative measure of uncertainty. We defined EMAF as continuous entropy of the patients, and it was based on distribution of MAF values that considered the uncertainty and ‘impurity’ of those values at the same time.
We acknowledge some limitations in this study. NSCLC is so complicated that could be affected by somatic mutation explored in this study as well as some other factors such as performance status, chemotherapy after surgery or relapse, which might cause bias. Besides, EMAF is limited by very small number of MAF values, which preclude determining an authentic distribution by kernel density estimation. In this study, one LUAD case was excluded due to the presence of only 2 mutant loci and EMAF was incalculable. Besides, EMAF is generated from sequencing data and influenced by sequencing depth. A low sequencing depth may result in an inaccurate MAF. Further, MAF heterogeneity is evaluated based on the whole genomic mutations, while in which probably only a small fraction relates to diseases. In addition, mutations in non-coding regions are gradually being cognized which warrant investigation in future (35–38).
In conclusion, the proposed entropy-based EMAF score can quantify MAF heterogeneity in NSCLC cases and is therefore suggested as a prognostic biomarker. In addition, EMAF differentiates a subgroup of early stage patents with an unfavorable prognosis, potentially providing clinical support for therapeutic decision-making.
Acknowledgements
The present study was partially supported by the National Natural Science Foundation of China (Nos. 81402764 to Y.W., 81373102 to Y.Z, 81402763 to R.Z., and 81473070 and 81530088 to F.C.) and the Natural Science Foundation of Jiangsu, China (No. BK20140907 to Y.W.). Y.W. was also partially supported by the Outstanding Young Teachers Training Program of Nanjing Medical University, and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions. Sponsors had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank the patients and investigators who participated in TCGA for providing the data.
Glossary
Abbreviations
- EMAF
entropy-based mutation allele fraction
- HR
hazard ratio
- HNSC
head and neck squamous cell carcinoma
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- MAF
mutant-allele fraction
- MATH
mutant-allele tumor heterogeneity
- NSCLC
non-small cell lung cancer
- TCGA
The Cancer Genome Atlas
- VAF
variant allele frequency
References
- 1.Ma H, Shu Y, Pan S, Chen J, Dai J, Jin G, Hu Z, Shen H. Polymorphisms of key chemokine genes and survival of non-small cell lung cancer in Chinese. Lung Cancer. 2011;74:164–169. doi: 10.1016/j.lungcan.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015;16:e342–e351. doi: 10.1016/S1470-2045(15)00077-7. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Jr, Wu YL, Paz-Ares L. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 4.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Håkansson L, Tropé C. On the presence within tumours of clones that differ in sensitivity to cytostatic drugs. Acta Pathol Microbiol Scand A. 1974;82:35–40. doi: 10.1111/j.1699-0463.1974.tb03825.x. [DOI] [PubMed] [Google Scholar]
- 7.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 8.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–895. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 9.Dexter DL, Kowalski HM, Blazar BA, Fligiel Z, Vogel R, Heppner GH. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 1978;38:3174–3181. [PubMed] [Google Scholar]
- 10.Heppner GH. Tumor heterogeneity. Cancer Res. 1984;44:2259–2265. [PubMed] [Google Scholar]
- 11.Cooke SL, Temple J, Macarthur S, Zahra MA, Tan LT, Crawford RA, Ng CK, Jimenez-Linan M, Sala E, Brenton JD. Intra-tumour genetic heterogeneity and poor chemoradiotherapy response in cervical cancer. Br J Cancer. 2011;104:361–368. doi: 10.1038/sj.bjc.6605971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumourevolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49:211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mroz EA, Tward AD, Pickering CR, Myers JN, Ferris RL, Rocco JW. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer. 2013;119:3034–3042. doi: 10.1002/cncr.28150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mroz EA, Tward AD, Hammon RJ, Ren Y, Rocco JW. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: Analysis of data from the cancer genome atlas. PLoS Med. 2015;12:e1001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocco JW. Mutant allele tumor heterogeneity (MATH) and head and neck squamous cell carcinoma. Head Neck Pathol. 2015;9:1–5. doi: 10.1007/s12105-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostrer H. Changing the game with whole exome sequencing. Clin Genet. 2011;80:101–103. doi: 10.1111/j.1399-0004.2011.01712.x. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broad Institute TCGA Genome Data Analysis Center, corp-author. Analysis Overview for Lung Adenocarcinoma (Primary solid tumor cohort) Broad Institute of MIT and Harvard; [Jan 28;2016 ]. 2016. [Google Scholar]
- 23.Cheng PF, Dummer R, Levesque MP. Data mining The cancer genome atlas in the era of precision cancer medicine. Swiss Med Wkly. 2015;145:w14183. doi: 10.4414/smw.2015.14183. [DOI] [PubMed] [Google Scholar]
- 24.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yee J, Kwon MS, Park T, Park M. A modified entropy-based approach for identifying gene-gene interactions in case-control study. PLoS One. 2013;8:e69321. doi: 10.1371/journal.pone.0069321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14:306–317. [PubMed] [Google Scholar]
- 27.Schreiber T. Measuring information transfer. Phys Rev Lett. 2000;85:461–464. doi: 10.1103/PhysRevLett.85.461. [DOI] [PubMed] [Google Scholar]
- 28.Uno H, Cai T, Pencina MJ, D'Agostino RB, Wei LJ. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med. 2011;30:1105–1117. doi: 10.1002/sim.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marquette D, Pichon E, Deschasse G, Lemaire B, Lemarie E, Diot P, Marchand-Adam S. Lung cancer in adults: Better prognosis of patients aged 45 and under related to good condition and lower TNM stage (a comparative and retrospective study) Presse Med. 2012;41:e250–e256. doi: 10.1016/j.lpm.2011.11.025. (In French) [DOI] [PubMed] [Google Scholar]
- 31.Liu M, Pan H, Zhang F, Zhang Y, Zhang Y, Xia H, Zhu J, Fu W, Zhang X. Identification of TNM stage-specific genes in lung adenocarcinoma by genome-wide expression profiling. Oncol Lett. 2013;6:763–768. doi: 10.3892/ol.2013.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Wang L, Lin XY, Wang J, Yu JH, Miao Y, Wang EH. The transcription factor DEC1 (BHLHE40/STRA13/SHARP-2) is negatively associated with TNM stage in non-small-cell lung cancer and inhibits the proliferation through cyclin D1 in A549 and BE1 cells. Tumour Biol. 2013;34:1641–1650. doi: 10.1007/s13277-013-0697-z. [DOI] [PubMed] [Google Scholar]
- 33.Yu KH, Zhang C, Berry GJ, Altman RB, Ré C, Rubin DL, Snyder M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erb CT, Su KW, Soulos PR, Tanoue LT, Gross CP. Surveillance practice patterns after curative intent therapy for stage I non-small-cell lung cancer in the medicare population. Lung Cancer. 2016;99:200–207. doi: 10.1016/j.lungcan.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao T, Xu J, Liu L, Bai J, Xu C, Xiao Y, Li X, Zhang L. Identification of cancer-related lncRNAs through integrating genome, regulome and transcriptome features. MolBiosyst. 2015;11:126–136. doi: 10.1039/c4mb00478g. [DOI] [PubMed] [Google Scholar]
- 36.Koch L. Cancer genomics: Non-coding mutations in the driver seat. Nat Rev Genet. 2014;15:574–575. doi: 10.1038/nrg3801. [DOI] [PubMed] [Google Scholar]
- 37.Piraino SW, Furney SJ. Beyond the exome: The role of non-coding somatic mutations in cancer. Ann Oncol. 2016;27:240–248. doi: 10.1093/annonc/mdv561. [DOI] [PubMed] [Google Scholar]
- 38.Tavare S. Data integration in cancer genomics: Non-coding mutations. Genet Epidemiology. 2015;585 [Google Scholar]