Abstract
Purpose of Review
Precision medicine approaches, that tailor medications to specific individuals has made paradigm-shifting improvements for patients with certain cancer types.
Recent Findings
Such approaches, however, have not been implemented for patients with diabetic kidney disease. Precision medicine could offer new avenues for novel diagnostic, prognostic and targeted therapeutics development. Genetic studies associated with multiscalar omics datasets from tissue and cell types of interest of well-characterized cohorts are needed to change the current paradigm.
Summary
In this review, we will discuss precision medicine approaches that the nephrology community can take to analyze tissue samples to develop new therapeutics for patients with diabetic kidney disease.
Keywords: Precision medicine, Genetic, Genomics, Epigenetics, GWAS, eQTL, Kidney biopsy, Diabetic kidney disease
Introduction: What Is Precision Medicine?
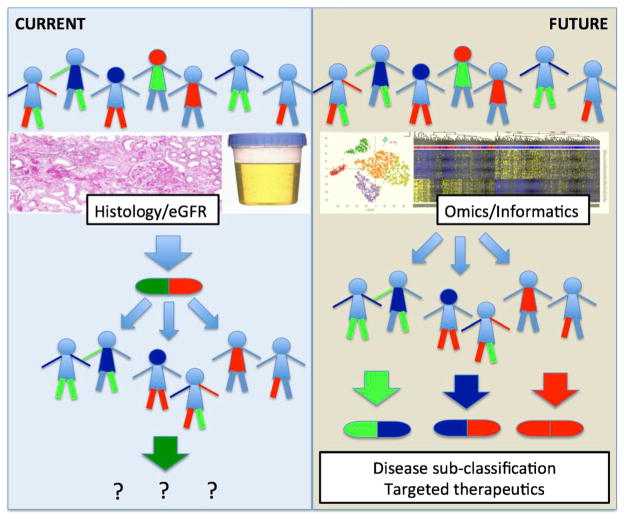
In his 2015 State of the Union address, President Obama announced the Precision Medicine Initiative, proposing a broad $215 million grant to support the Million American Genome Initiative, a national study initiative for precision medicine involving the health records and DNA of one million volunteers. The National Institute of Health defines precision medicine as follows (Fig. 1):
“Precision Medicine refers to the tailoring of medical treatment to the individual characteristics of each patient. It does not literally mean the creation of drugs or medical devices that are unique to a patient, but rather the ability to classify individuals into subpopulations that differ in their susceptibility to a particular disease, in the biology and/or prognosis of those diseases they may develop, or in their response to a specific treatment. Preventive or therapeutic interventions can then be concentrated on those who will benefit, sparing expense and side effects for those who will not. Although the term ‘Personalized Medicine’ is also used to convey this meaning, that term is sometimes misinterpreted as implying that unique treatments can be designed for each individual.”
Fig. 1.
Precision medicine approaches for diabetic kidney disease
Need for Patient-Oriented Precision Medicine Approaches
Diabetic kidney disease (DKD) is the most common cause of chronic kidney disease (CKD) in the USA and most of the world [1]. Histopathological characterization of kidney tissue samples remains the gold standard diagnostic criteria for DKD. Glomerular basement membrane (GBM) thickening (measured by point counting method of electron microscopical images; EM) and mesangial expansion observed on light microscopical analysis defines DKD [2••]. Nodular sclerosis is a highly specific, but not fully sensitive diagnostic criteria. DKD is also characterized by arterial hyalinosis and tubulointerstitial fibrosis. Early studies established an association between clinical presentations, such as albuminuria, cardiovascular disease, death, and histological lesions [3•, 4].
Progress in diabetic kidney research leading to new therapeutics development has been limited. Indeed, no new medicines indicated for the treatment of chronic kidney disease (CKD) have been approved since the AT1 receptor blocker (ARB) became the standard of care nearly 15 years ago. Several factors explain the limited progress, including but not limited to: (a) animal and cell culture models do not recapitulate human DKD; (b) DKD remains a clinical diagnosis with imprecise diagnostic criteria, and that (c) the clinical course of DKD is highly variable [5].
Laboratory mice have served as an invaluable tool for understanding the human disease development [6]. Mouse genetic tools have been developed enabling temporal- and cell-type-specific gene manipulation. The development and characterization of genetically engineered mouse disease models have helped to identify key disease associated pathways for a variety of conditions. Unfortunately, mouse models do not recapitulate human DKD because most animals only develop early morphological changes of human diabetic nephropathy, including mesangial expansion and early functional changes such as mild albuminuria [6]. Hallmarks of progressive DKD, such as arterial hyalinosis and tubulointerstitial fibrosis on histology, as well as functional changes, as evidenced by declining glomerular filtration rate (GFR), have not been described in most mouse models [2••].
In addition, human and murine kidneys show significant differences in gene expression and functional level [7]. Such differences may also explain the lack of translatability of pharmacological approaches aimed at treating DKD. DKD research must shift towards patient-centered and translational research approaches. Similar pivots in cancer, immunology, and neurodegenerative research have already led to prognosis-altering discoveries.
Lack of Reliable Diagnostic Biomarker
Even though histopathology remains the gold standard diagnostic criteria, very few patients with suspected DKD undergo kidney biopsy procedure. Instead, non-invasive diagnostic parameters are recommended to ascertain DKD diagnosis. Our society guidelines, state that in patients with CKD in the presence of diabetes and albuminuria, the diagnosis of DKD can safely be made. These recommendations were developed based on observations originating back to Mogensen et al. in the 1970–1980s [3•, 4, 8, 9]. Mogensen classified DKD as a progressive disease (stages I–V). Stage I and II were mainly preclinical stages representing hyperfiltration and then glomerular lesions in the absence of clinical disease, respectively. Stage III represented “incipient diabetic nephropathy” with the presence of microalbuminuria (30–300 mg/day) while stage IV represented “overt diabetic nephropathy” with macroalbuminuria (>0.5 g/day) and progressive GFR decline with ultimate end-stage renal disease, or stage V. Studies from the 1980s indicated that early DKD can be diagnosed based on microalbuminuria [10]. Indeed, studies show statistically significant increased risk (3.6–4.8) between ESRD in patients with DM and microalbuminuria. This indicates that microalbuminuria can identify subjects with increased risk of irreversible GFR decline and ESRD.
Recent studies, however suggest that microalbuminuria is a poor diagnostic marker of DKD. Many patients, especially those with type 2 diabetes, do not manifest excessive urinary albumin loss, yet still develop CKD and ESRD. Indeed, of the 28% in the United Kingdom Prospective Diabetes Study (UKPDS) cohort who developed moderate to severe renal impairment, half did not have preceding albuminuria. And of the 11% of patients with type 1 diabetes in the Diabetes Control and Complications Trial (DCCT) who developed an eGFR < 60 ml/min/1.73 m2, 40% never had experienced overt proteinuria [11]. In addition, the majority of patients with microalbuminuria do not exhibit a progressive increase in urinary albumin excretion as described by the classic course. Both treatment-induced and spontaneous ‘remission’ of albuminuria [12, 13] seem to occur. Consequently, individuals with microalbuminuria may be better regarded as being at increased risk of developing progressive renal disease, as well as cardiovascular disease and other diabetic complications, rather than as having DKD per se.
These observations suggest that disease manifestation could have changed over the last several decades, or observations made in a small cohort of patients with type1 diabetes are not fully generalizable to large cohorts of patients with type 2 diabetes. Current observational cohorts indicate that albuminuria and GFR might be independent manifestations of DKD. Some patients present with albuminuria while others only show GFR decline in the absence of albuminuria. Treatment with blockers of the renin angiotensin aldosterone system, that is now standard of care, significantly reduces albuminuria and might contribute to these changes in presentation; however, it does not explain it. According to the review of MacIsaac et al., somewhere between 20 and 70% of patients with DKD have reduced GFR with normoalbuminuria (depending on the cohort), most of this is not related to RAAS blockade.
DKD in the absence of albuminuria appears to be a novel clinical presentation of DKD. Patients with type 1 diabetes, normoalbuminuria, and low GFR appear to show classic changes of DKD by histopathological analysis. On the other hand, type 2 patients appear to have heterogeneous histological changes. The Fioretto group showed that some subjects have normal or near-normal histology, some present with typical diabetic nephropathy, some with atypical histology with disproportionately severe interstitial/tubular/vascular damage, while others have no or only mild diabetic glomerular changes [14, 15].
The gold standard, histopathological diagnosis, is recommended for patients who show atypical presentation and those with diabetes and positive serological indicators of non-diabetic kidney disease (such as low complement, positive HIV or protein electrophoresis screen). The Columbia Renal Pathology group recently analyzed all biopsy samples obtained from patients with diabetes [16••] to understand the value of these diagnostic recommendations. Among 2642 native kidney biopsies examined in 2011, they found that 620 (23.5%) came from patients with diabetes [16••]. On histological diagnosis, 37% of patients had DKD alone, 36% had non-diabetic renal disease (NDRD) alone, and 27% had DKD plus NDRD. In multivariate analyses, longer duration of DM was associated with a greater likelihood of DKD and a lower likelihood of NDRD: each added year of diabetes reduced the odds of NDRD by 5% (odds ratio, 0.95; 95% confidence interval, 0.91 to 0.98; P = 0.004). DM duration ≥12 years was the best predictor (58% sensitivity, 73% specificity) of DKD alone. In contrast, serological screens with low complement levels (C3 and/or C4), M-spike in either serum or urine, positive viral serologies for hepatitis, HIV, and active urine sediment did not predict the presence or absence of histopathological DKD.
DKD remains a histopathological diagnosis. While initial studies indicated that albuminuria could be used as an important diagnostic biomarker, the diagnostic value of albuminuria has recently come under spotlight. Novel precision medicine-based diagnostic biomarkers are desperately needed for DKD. These novel diagnostic markers, however, will need to be calibrated against the current gold standard: histopathology, highlighting the critical need for biopsy tissue samples from patients with diabetes.
Great Emphasis, but Limited Success with Predisposition DKD Biomarkers
Not everyone with diabetes develops kidney disease. The exact number of patients with diabetes and kidney disease remains elusive, due to diagnostic issues detailed above. Of the approximately 400 million people with type 2 diabetes worldwide, approximately half will have evidence of CKD. Approximately one in five adults with type 2 diabetes will have an eGFR of <60 ml/min/1.73 m2 and between 30 and 50% will have elevated urinary albumin excretion [17•]. The incidence of CKD in type 1 diabetes differs from that observed in type 2 diabetes. It is estimated that approximately one third of all people with type 1 diabetes will develop CKD over the course of their lifetime.
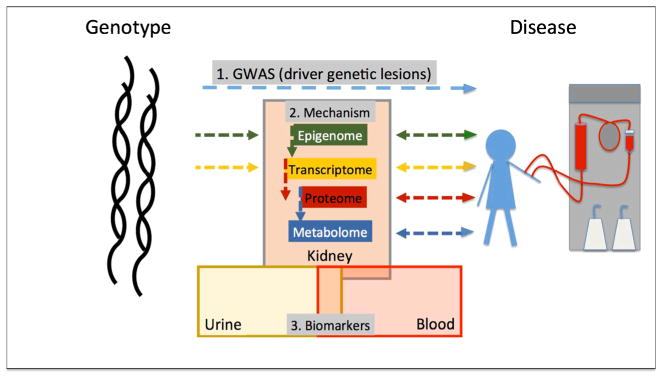
Poor glycemic and blood pressure control in addition to genetic predisposition remain the key factors that determine predisposition to DKD. Genetic studies have played a critical role in precision medicine and therapeutics development for multiple reasons. Recent advances in sequencing technology made genetic analysis significantly faster and easier. Furthermore, genetic analysis has a high yield of identifying causal genes and pathways as genetic variations are established before disease development (Fig. 2).
Fig. 2.
Integrated omics analysis to understand diabetic kidney disease
Human genetic studies have already made paradigm-shifting observations in relatively rare monogenic forms of kidney diseases (including polycystic kidney disease and focal segmental glomerulosclerosis). DKD, on the other hand, follows a complex polygenic pattern. Currently, the most powerful method to define the genetics of complex diseases such as DKD is genome-wide association studies (GWAS), where associations between polymorphisms and disease states are tested. To understand the genetics of DKD, GWAS have been performed to identify genetic variants that are significantly associated to DKD. For example, the GEnetics of Nephropathy: an International Effort (GENIE) consortium identified a significant locus for DKD subjects with type 1 diabetes (T1D) in a 12,000 cohort. The meta-analysis for DKD in T2D in the Family Investigation of Nephropathy and Diabetes (FIND) study discovered one locus at chromosome 6q25.2 in American Indians [18] and one locus was discovered at chr2 in T2DM-ESRD in African Americans [19]. The Finnish Diabetic Nephropathy (FinnDiane) Study found association on chromosome 2q31.1 [20]. The inconsistency between published DKD and GWAS reflects that each study has different disease criteria; for example, the FIND study did not include microalbuminuric participants, choosing to use only individuals with advanced nephropathy with modest sample size. The CKDGen consortium has been a bit more successful in identification of genetic variants associated with kidney function in a large (>100,000 samples) mostly European population. They have identified close to 70 loci associated with kidney function and disease in diabetic and non-diabetic populations [21].
Unfortunately, the biology explaining the GWAS association studies remains elusive. More than 90% of CKD-associated variants are localized to non-coding regions. Non-coding variants present special challenges as there are no simple tools that would allow prediction of functional consequences of such variations. Identification of target genes and target cell types that are affected by these genetic variants is necessary to explain the associations between genetic variants and phenotype [22]. Such studies require large collection of human tissue samples from disease-relevant organs.
The expression of quantitative trait loci (eQTL) method examines correlation between genetic variants and gene expression levels. eQTL is able to identify genetic variants that are associated with the heritability of gene expression. The integration of eQTL studies with GWAS analysis provides a powerful analytical framework to identify variants associated with disease development (GWAS) and target gene expression (eQTL) (Fig. 2). There is a large NIH funded consortium (GTEx, Genotype-Tissue Expression) working to correlate genetic variations and gene expression changes in 1000 patients and more than 50 tissue samples. Unfortunately, the GTEX consortium has only been able to collect a limited number of kidney samples [23]. To fill this gap, our group has generated one of the first eQTL maps for human kidney samples from people of European descent. Combining the eQTL and GWAS datasets, we were able to identify genes for which expression is associated with the GWAS identified genetic variants in human kidney tissue samples. These genes can serve as potential targets for the GWAS studies. This integrative approach highlighted that mannosidase beta is a likely CKD target gene. Validation studies using the zebrafish model system confirmed the role of MANBA in kidney development (Ko and Susztak unpublished). These studies highlight the critical need for human tissue samples and genetic approaches in understanding disease predisposition.
Of the environmental factors hyperglycemia, probably as a measure of metabolic dysfunction, seems to be essential for DKD development, as no DKD is observed in patients in the absence of diabetes. Furthermore, natural history studies (mostly in subjects with type1 DM) indicate that the degree of hyperglycemia correlates with DKD incidence. Based on these observations, investigators have tried to control glycemia almost to the degree of non-diabetic levels in the ACCORD, ADVANCE, and VA-DT clinical trials [24]. Surprisingly, these studies failed to show lower mortality and complication rates. Several researchers propose that a “metabolic memory” effect might be responsible for these unexpected results. For example, metabolic dysregulation results in epigenetic alterations causing not only acute but sustained differences in gene expression. These differences are maintained during cell division over several decades. Several experimental and human studies support the metabolic memory theory in DKD. Epigenetic differences have been observed in endothelial cells cultured in high glucose medium; these differences are maintained even after the cells are returned to “normal” glucose solution. Epigenetic differences may explain the increased inflammatory gene expression in DKD. Several studies describe cytosine methylation differences in blood samples of patients with DKD compared to controls. As the epigenome is cell type specific, the study by Ko et al. is especially important. It indicates that cytosine methylation changes can be observed in kidneys of patients with CKD [25•]. Furthermore, cytosine methylation changes correlated with gene expression differences supporting their functional role in DKD.
The clinical use of predisposition markers remains elusive as we do not have clear therapeutic options besides good glycemic and BP control for subjects with DM. Genetic and epigenetic markers, however, can provide critically needed insight into disease pathogenesis.
Prognostic Biomarkers
Therapeutic interventions for DKD are aimed at reducing death and ESRD. The greatest emphasis is therefore placed on prognostic DKD biomarkers that can be used to identify patients who will reach these endpoints. A critical issue has been that the progression of diabetic kidney disease is highly variable and potentially confounded by an age-associated decline in kidney function [26•]. Data from large observational cohorts indicate that decline in GFR does not always follow a linear course and also depends on baseline kidney function. Such studies contributed to emphasizing patients termed as “rapid progressors”. Although there is not a clear consensus as to this definition, many studies define rapid progressors as patients with greater than 3 ml/year GFR decrease but alternative cut-points as rapid as 10 cm3/year have also been used. Identification and clinical characterization of rapid progressors became the focus of several large-scale efforts as these are the patients who would likely benefit from intensive clinical management [27]. Recent post hoc analyses of the Irbesartan Type II Diabetic Nephropathy Trial and Reduction of Endpoints in NIDDM with the Angiotensin II Antagonist Losartan studies also showed that clinical trial outcomes are mostly driven by a small number of subjects with unusually rapid progressive GFR decline.
The clinical and histological characteristics of subjects with rapid vs. slow progression are not well understood. Albuminuria remains one of the strongest risk factors for progression. Some of the latest studies indicate that using a 4- or a 6-variable model that includes albuminuria, age, sex, serum phosphate, serum calcium, and serum albumin has a C-statistics score of 0.84–0.91 to predict progression and ESRD [28, 29••].
During the last few years, several additional new bio-markers have been described that can potentially also identify patients at increased risk for rapid loss of kidney function. For example, blood and urinary levels of Kidney Injury Molecule-1 (KIM1) shows promise to identify patients who are at risk for kidney function decline. Recently, Krolewski et al. showed that circulating levels of tumor necrosis factor receptor 1 and 2 (TNFR1 and TNFR2) can identify patients with rapidly declining renal function [6, 30, 31]. According to studies published by the Joslin Diabetes Center, the cumulative incidence of ESRD for patients in the highest TNFR1 quartile was 54% after 12 years but only 3% for the other quartiles (P < 0.001). On the other hand, subjects with higher TNFR1 and TNFR2 also had lower baseline GFR and higher albuminuria values. Therefore, at present it is not clear whether serum TNFR1 and TNFR2 can predict progressive DKD independently of albuminuria and kidney function. Unfortunately, these findings are only validated in few independent studies. In addition, studies have not been performed to examine the correlation between TNFR1 or TNFR2 and histological manifestation of DKD to show the specificity of these markers for DKD [32].
In clinical practice, kidney biopsy is often performed to evaluate the degree of fibrosis as it is perceived that fibrosis and glomerulosclerosis are the best indicators of progression. Due to the lack of samples, few studies, however, have tested this hypothesis formally. This likely reflects that the current standard of care does not require renal biopsy for diagnostic or prognostic purposes. In addition, samples from patients without significant renal disease would also be needed to develop correlation analysis. Recently, Mise et al., reported that in patients with diabetes who underwent renal biopsy for clinical purpose, renal fibrosis score correlated with hazard rate of ESRD (HR 2.31) and improved current models for progression prediction [33]. Menn-Josephy et al. analyzed 434 consecutive renal biopsies at a single center and showed that for patients less than 70 years of age, greater than 50% interstitial fibrosis was predictive of progression to dialysis (C = 0.866) and when added to a full-prediction model (including known risk factors) it improved upon the predictive ability of the model [34]. However, this was not true for those over age 70 years old, as well as those with histological evidence of diabetic nephropathy. In summary, additional studies are needed to identify novel biomarkers of risk of progression. These studies should also include histopathological diagnostic and prognostic parameters.
Precision Medicine and Targeted Therapeutics
The primary goal of precision medicine is to improve survival and prevent ESRD. While novel diagnostic and prognostic biomarkers are essential to reach this goal, the ultimate goal is to develop targeted and impactful therapeutics. For therapeutic target identification, pathways that are causal to disease development are essential. While patient studies are very effective in establishing association between disease traits and prognosis, it is difficult to establish causality in patient-oriented studies. In general, the Hill’s criteria are used to establish causality including; effect size, reproducibility, specificity, temporality, biological gradient, plausibility, and coherence in addition to experiments and analogy. This is particularly difficult in fields where mouse models do not faithfully recapitulate the disease condition, given experimental studies are still considered gold standard to define causality. Lack of reliable experimental systems lead to increasing reliance on temporal relationships to infer causality and genetic variation to prove the causal relationship. Indeed, the best precision medicine examples come from large effect size genetic variants observed in patients with cancer. In certain types of cancer, genetic studies have made paradigm-shifting observations by identifying cancer driving mutations. Targeting these mutations such as EGFR in lung cancer and B-Raf in melanoma resulted in novel targeted therapeutics. Similarly, gain and loss of function variants in PCSK9 resulted in novel targets for hyperlipidemia. These “genetically targeted” therapeutics not only made major impact on patients’ lives but also remarkably increased the speed of drug development and approval. Several other ‘omics’ approaches have been applied to identify causal targetable pathways for disease development. These include epigenetics, transciptomics, proteomics, and metabolomics. These methods are appealing as these analytes are closer to phenotype development (Fig. 2). To take these studies to their full potential, it is essential that they are performed on tissue samples obtained from healthy and diseased human samples. Comprehensive metabolome-wide association studies and proteomics have been performed for CKD and future functional decline [35]. As of yet, no metabolite and protein markers has been identified that could consistently improve upon existing prognostic models. Furthermore, blood protein and metabolite changes do not fully translate to kidney specific differences making it exceedingly difficult to develop a coherent and targetable hypothesis based on these markers.
Transcriptomics and epigenetic profiling of human diabetic kidney disease samples have been performed and published for a limited number of samples [36]. These studies identified the dysregulation of several key pathways. Inflammation and metabolic pathway associated genes represent the largest gene group functions showing association with kidney function (eGFR) or structural damage (fibrosis). This association appears strong, coherent, and reproducible in multiple studies. The association between inflammatory gene expression such as CCL2, CCL5, and DKD development could also be replicated in mice, and genetic or pharmacological intervention studies indicated that the increase in cytokine levels contribute to DKD development [37]. Despite the strong preclinical data, human phase II studies only indicated modest effect size for CCL2 and CCL5 blockers for DKD development. Transcription factor targeting (such as Jak) seemed to be more effective, albeit associated with side effects. The transcriptomics data also indicated a decrease in expression of genes associated with fatty acid oxidation and mitochondrial content [38]. Clinical studies show a modest effect size in preserving kidney function with the use of PPARA agonist fenofibrate [39]. While transcriptomic studies are very powerful to demonstrate the dysregulation of specific pathways, they are confounded by the massive amount of secondary changes, making it challenging to identify a single causal gene or protein. In addition, cells and organs mostly exist as networks and any small differences in expression levels will trigger compensatory changes.
Many researchers are proposing collection of samples from patients with early stage disease or even at healthy state (before disease development) to address issues around temporality and causality. This is an interesting and important proposal and such approaches will likely reduce secondary changes induced data noise. Unfortunately, given the small expected effect size and the inherent variability of the transcriptomic studies, it would require a very large sample size to compensate for these issues. As of now, we are not aware of any studies that would have the power to successfully identify therapeutic targets using such approaches.
Following this logic, collecting “control or healthy” tissue samples with associated genetic data appears to be the most powerful method for the identification of causal pathways. We know GWAS studies can establish the genotype-phenotype association (and disease risk development). Furthermore, genetic variations have been selected over millions of years and thereby fully tested by nature for tolerability. Network level analysis and transcript profiling at baseline and following system perturbation is also essential to identify critical genes and nodes in DKD development. This framework would then further benefit from other omics analysis such as proteomics and metabolite level characterization. In summary, tissue level omics characterization is then essential to complement genetic and biomarker studies to understand disease mechanism and therapeutics targeting.
Conclusions: a Vision for the Future
There is a critical need for precision medicine-based approaches for diagnostic, prognostic, and therapeutics development in DKD. It is increasingly clear that these approaches rely on using tissue samples from well-phenotyped patient cohorts. As histopathological diagnosis remains the gold standard criteria for DKD, all future diagnostic markers will need to be calibrated to the current gold standard approach. The demand for better diagnostic approaches is vast as it seems that the clinical manifestation of disease is different from those originally observed by Mogensen in the 1980s.
The need for novel precision medicine-based targeted therapeutics cannot be greater. This requires the generation of large multiscalar datasets using human tissue samples and associated genetic data to increase the statistical power to identify causal genes and pathways. As current society guidelines do not necessitate the kidney biopsy procedure for DKD, there is a significant challenge to obtain such critical tissue samples. Research biopsies on a small cohort (<100) samples have been performed by the Nelson and Mauer groups. These groups have reported low complications rates in their single center studies. Use of nephrectomies can also represent an important source of tissue material. While we cannot exclude increased heterogeneity due to ischemia that occurs during the surgery, it can also provide critically needed control samples from patients with diabetes and/or hypertension in absence of kidney disease. Our group has collected more than 1600 samples with associated clinical information and associated genetic, epigenetic, and genomics datasets [25•, 36, 40]. As a first step in precision medicine approaches, collecting and analyzing all clinically indicated biopsies from patients with diabetes will likely be essential. Our initial assessment indicates that patients with rapidly progressive GFR decline are overrepresented in the biopsy group. Capturing this group is critically needed for future therapeutics development. TRIDENT; transformative research in diabetic nephropathy is a novel phase 0 study aiming to recruit 300 patients with diabetes and clinically indicated kidney biopsies, and perform histological, genetic, genomics, and epigenetic analysis of these samples. The aim is to identify genes and pathways associated with rapid functional loss; this phenotype is enriched in the clinically indicated biopsy group. Furthermore, the NIDDK has put forward an ambitious proposal to collect kidney tissue samples and perform genetic, genomics studies for novel target identification for chronic and acute kidney disease. These new developments should have a significant impact and will hopefully be able to turn the corner in diabetic kidney disease and lead to much needed new diagnostics and therapeutics.
Footnotes
Compliance with Ethical Standards
Conflict of Interest Caroline Gluck and Yi-An Ko declare that they have no conflict of interest.
Katalin Susztak reports grants from Biogen, ONO Pharma, Boehringer Ingelheim, Regeneron, GSK, Celgene, NIH, and JDRF.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Breyer MD, Coffman TM, Flessner MF, Fried LF, Harris RC, Ketchum CJ, Kretzler M, Nelson RG, Sedor JR, Susztak K Kidney Research National D. Diabetic nephropathy: a national dialogue. Clin J Am Soc Nephrol. 2013;8(9):1603–5. doi: 10.2215/CJN.03640413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2••.Tervaert TW, Mooyaart AL, Amann K, Cohen AH, Cook HT, Drachenberg CB, Ferrario F, Fogo AB, Haas M, de Heer E, Joh K, Noel LH, Radhakrishnan J, Seshan SV, Bajema IM, Bruijn JA, Renal PS. Pathologic classification of diabetic nephropathy. J Am Soc Nephrol. 2010;21(4):556–63. doi: 10.1681/ASN.2010010010. Important paper described a new classification system for DKD. [DOI] [PubMed] [Google Scholar]
- 3•.Mogensen CE, Christensen CK. Predicting diabetic nephropathy in insulin-dependent patients. N Engl J Med. 1984;311(2):89–93. doi: 10.1056/NEJM198407123110204. Original publication describing the clinical presentation of DKD. [DOI] [PubMed] [Google Scholar]
- 4.Mogensen CE, Christensen CK, Vittinghus E. The stages in diabetic renal disease: with emphasis on the stage of incipient diabetic nephropathy. Diabetes. 1983;32(Suppl 2):64–78. doi: 10.2337/diab.32.2.s64. [DOI] [PubMed] [Google Scholar]
- 5.Breyer MD, Susztak K. Developing treatments for chronic kidney disease in the 21st century. Semin Nephrol. 2016a;36(6):436–47. doi: 10.1016/j.semnephrol.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius FC, 3rd, Alpers CE, Bottinger EP, Breyer MD, Coffman TM, Gurley SB, Harris RC, Kakoki M, Kretzler M, Leiter EH, Levi M, McIndoe RA, Sharma K, Smithies O, Susztak K, Takahashi N, Takahashi T. Animal models of diabetic complications C. Mouse models of diabetic nephropathy. J Am Soc Nephrol. 2009;20(12):2503–12. doi: 10.1681/ASN.2009070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Si H, Banga RS, Kapitsinou P, Ramaiah M, Lawrence J, Kambhampati G, Gruenwald A, Bottinger E, Glicklich D, Tellis V, Greenstein S, Thomas DB, Pullman J, Fazzari M, Susztak K. Human and murine kidneys show gender- and species-specific gene expression differences in response to injury. PLoS One. 2009;4(3):e4802. doi: 10.1371/journal.pone.0004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen CK, Mogensen CE. The course of incipient diabetic nephropathy: studies of albumin excretion and blood pressure. Diabet Med. 1985;2(2):97–102. doi: 10.1111/j.1464-5491.1985.tb00608.x. [DOI] [PubMed] [Google Scholar]
- 9.Schmitz O, Hansen HE, Orskov H, Mogensen CE, Posborg PV. End-state renal failure in diabetic nephropathy: pathophysiology and treatment. Blood Purif. 1985;3(1–3):120–39. doi: 10.1159/000169405. [DOI] [PubMed] [Google Scholar]
- 10.Newman DJ, Mattock MB, Dawnay AB, Kerry S, McGuire A, Yaqoob M, Hitman GA, Hawke C. Systematic review on urine albumin testing for early detection of diabetic complications. Health Technol Assess. 2005;9(30):iii–vi. xiii–163. doi: 10.3310/hta9300. [DOI] [PubMed] [Google Scholar]
- 11.Nathan DM, Cleary PA, Backlund JY, Genuth SM, Lachin JM, Orchard TJ, Raskin P, Zinman B, Diabetes C. Complications trial/epidemiology of diabetes I, complications study research G. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N Engl J Med. 2005;353(25):2643–53. doi: 10.1056/NEJMoa052187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perkins BA, Ficociello LH, Roshan B, Warram JH, Krolewski AS. In patients with type 1 diabetes and new-onset microalbuminuria the development of advanced chronic kidney disease may not require progression to proteinuria. Kidney Int. 2010;77(1):57–64. doi: 10.1038/ki.2009.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perkins BA, Aiello LP, Krolewski AS. Diabetes complications and the renin-angiotensin system. N Engl J Med. 2009;361(1):83–5. doi: 10.1056/NEJMe0904293. [DOI] [PubMed] [Google Scholar]
- 14.Ekinci EI, Jerums G, Skene A, Crammer P, Power D, Cheong KY, Panagiotopoulos S, McNeil K, Baker ST, Fioretto P, Macisaac RJ. Renal structure in normoalbuminuric and albuminuric patients with type 2 diabetes and impaired renal function. Diabetes Care. 2013;36(11):3620–6. doi: 10.2337/dc12-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caramori ML, Fioretto P, Mauer M. Low glomerular filtration rate in normoalbuminuric type 1 diabetic patients: an indicator of more advanced glomerular lesions. Diabetes. 2003;52(4):1036–40. doi: 10.2337/diabetes.52.4.1036. [DOI] [PubMed] [Google Scholar]
- 16••.Sharma SG, Bomback AS, Radhakrishnan J, Herlitz LC, Stokes MB, Markowitz GS, D’Agati VD. The modern spectrum of renal biopsy findings in patients with diabetes. Clin J Am Soc Nephrol. 2013;8(10):1718–24. doi: 10.2215/CJN.02510213. Recent publication describing pathological changes in kidney biopsies of patients with diabetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Thomas MC, Brownlee M, Susztak K, Sharma K, Jandeleit-Dahm KA, Zoungas S, Rossing P, Groop PH, Cooper ME. Diabetic kidney disease. Nat Rev Dis Primers. 2015;1:15018. doi: 10.1038/nrdp.2015.18. Excellent review summarizing DKD, both clinical and basic science updates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyengar SK, Sedor JR, Freedman BI, et al. Genome-wide association and trans-ethnic meta-analysis for advanced diabetic kidney disease: Family Investigation of Nephropathy and Diabetes (FIND) PLoS Genet. 2015;11(8):e1005352. doi: 10.1371/journal.pgen.1005352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer ND, McDonough CW, Hicks PJ, et al. A genome-wide association search for type 2 diabetes genes in African Americans. PLoS One. 2012;7(1):e29202. doi: 10.1371/journal.pone.0029202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandholm N, McKnight AJ, Salem RM, Brennan EP, Forsblom C, Harjutsalo V, Makinen VP, McKay GJ, Sadlier DM, Williams WW, Martin F, Panduru NM, Tarnow L, Tuomilehto J, Tryggvason K, Zerbini G, Comeau ME, Langefeld CD, Godson C, Hirschhorn JN, Maxwell AP, Florez JC, Groop PH Consortium F, FinnDiane Study G. The GC. Chromosome 2q31.1 associates with ESRD in women with type 1 diabetes. J Am Soc Nephrol. 2013;24(10):1537–43. doi: 10.1681/ASN.2012111122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pattaro C, Teumer A, Gorski M, et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun. 2016;7:10023. doi: 10.1038/ncomms10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamatoyannopoulos JA. What does our genome encode? Genome Res. 2012;22(9):1602–11. doi: 10.1101/gr.146506.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Germain M, Pezzolesi MG, Sandholm N, McKnight AJ, Susztak K, Lajer M, Forsblom C, Marre M, Parving HH, Rossing P, Toppila I, Skupien J, Roussel R, Ko YA, Ledo N, Folkersen L, Civelek M, Maxwell AP, Tregouet DA, Groop PH, Tarnow L, Hadjadj S. SORBS1 gene, a new candidate for diabetic nephropathy: results from a multi-stage genome-wide association study in patients with type 1 diabetes. Diabetologia. 2015;58(3):543–8. doi: 10.1007/s00125-014-3459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Ko YA, Mohtat D, Suzuki M, Park AS, Izquierdo MC, Han SY, Kang HM, Si H, Hostetter T, Pullman JM, Fazzari M, Verma A, Zheng D, Greally JM, Susztak K. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013;14(10):R108. doi: 10.1186/gb-2013-14-10-r108. Epigenetic changes in kidney samples of patients with diabetic kidney disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Perkins BA, Nelson RG, Ostrander BE, Blouch KL, Krolewski AS, Myers BD, Warram JH. Detection of renal function decline in patients with diabetes and normal or elevated GFR by serial measurements of serum cystatin C concentration: results of a 4-year follow-up study. J Am Soc Nephrol. 2005;16(5):1404–12. doi: 10.1681/ASN.2004100854. CKD development in patients with diabetes in absence of albuminuria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsa A, Kao WH, Xie D, Astor BC, Li M, Hsu CY, Feldman HI, Parekh RS, Kusek JW, Greene TH, Fink JC, Anderson AH, Choi MJ, Wright JT, Jr, Lash JP, Freedman BI, Ojo A, Winkler CA, Raj DS, Kopp JB, He J, Jensvold NG, Tao K, Lipkowitz MS, Appel LJ Investigators AS, Investigators CS. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013;369(23):2183–96. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levey AS, Tangri N, Stevens LA. Classification of chronic kidney disease: a step forward. Ann Intern Med. 2011;154(1):65–7. doi: 10.7326/0003-4819-154-1-201101040-00012. [DOI] [PubMed] [Google Scholar]
- 29••.Tangri N, Stevens LA, Griffith J, Tighiouart H, Djurdjev O, Naimark D, Levin A, Levey AS. A predictive model for progression of chronic kidney disease to kidney failure. JAMA. 2011;305(15):1553–9. doi: 10.1001/jama.2011.451. Current prognostic biomarkers, 6 biochemical parameter can accurately predic functional decline in patients with CKD. [DOI] [PubMed] [Google Scholar]
- 30.Gohda T, Niewczas MA, Ficociello LH, Walker WH, Skupien J, Rosetti F, Cullere X, Johnson AC, Crabtree G, Smiles AM, Mayadas TN, Warram JH, Krolewski AS. Circulating TNF receptors 1 and 2 predict stage 3 CKD in type 1 diabetes. J Am Soc Nephrol. 2012;23(3):516–24. doi: 10.1681/ASN.2011060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niewczas MA, Gohda T, Skupien J, Smiles AM, Walker WH, Rosetti F, Cullere X, Eckfeldt JH, Doria A, Mayadas TN, Warram JH, Krolewski AS. Circulating TNF receptors 1 and 2 predict ESRD in type 2 diabetes. J Am Soc Nephrol. 2012;23(3):507–15. doi: 10.1681/ASN.2011060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavkov ME, Weil EJ, Fufaa GD, Nelson RG, Lemley KV, Knowler WC, Niewczas MA, Krolewski AS. Tumor necrosis factor receptors 1 and 2 are associated with early glomerular lesions in type 2 diabetes. Kidney Int. 2016;89(1):226–34. doi: 10.1038/ki.2015.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mise K, Hoshino J, Ueno T, Hazue R, Hasegawa J, Sekine A, Sumida K, Hiramatsu R, Hasegawa E, Yamanouchi M, Hayami N, Suwabe T, Sawa N, Fujii T, Hara S, Ohashi K, Takaichi K, Ubara Y. Prognostic value of tubulointerstitial lesions, urinary N-acetyl-beta-d-glucosaminidase, and urinary beta2-microglobulin in patients with type 2 diabetes and biopsy-proven diabetic nephropathy. Clin J Am Soc Nephrol. 2016;11(4):593–601. doi: 10.2215/CJN.04980515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menn-Josephy H, Lee CS, Nolin A, Christov M, Rybin DV, Weinberg JM, Henderson J, Bonegio R, Havasi A. Renal interstitial fibrosis: an imperfect predictor of kidney disease progression in some patient cohorts. Am J Nephrol. 2016;44(4):289–99. doi: 10.1159/000449511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekula P, Goek ON, Quaye L, Barrios C, Levey AS, Romisch-Margl W, Menni C, Yet I, Gieger C, Inker LA, Adamski J, Gronwald W, Illig T, Dettmer K, Krumsiek J, Oefner PJ, Valdes AM, Meisinger C, Coresh J, Spector TD, Mohney RP, Suhre K, Kastenmuller G, Kottgen A. A metabolome-wide association study of kidney function and disease in the general population. J Am Soc Nephrol. 2016;27(4):1175–88. doi: 10.1681/ASN.2014111099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60(9):2354–69. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breyer MD, Susztak K. The next generation of therapeutics for chronic kidney disease. Nat Rev Drug Discov. 2016b;15(8):568–88. doi: 10.1038/nrd.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stadler K, Goldberg IJ, Susztak K. The evolving understanding of the contribution of lipid metabolism to diabetic kidney disease. Curr Diab Rep. 2015;15(7):40. doi: 10.1007/s11892-015-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang HM, Ahn SH, Choi P, Ko YA, Han SH, Chinga F, Park AS, Tao J, Sharma K, Pullman J, Bottinger EP, Goldberg IJ, Susztak K. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med. 2015;21(1):37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han SH, Malaga-Dieguez L, Chinga F, Kang HM, Tao J, Reidy K, Susztak K. Deletion of Lkb1 in renal tubular epithelial cells leads to CKD by altering metabolism. J Am Soc Nephrol. 2016;27(2):439–53. doi: 10.1681/ASN.2014121181. [DOI] [PMC free article] [PubMed] [Google Scholar]