Abstract
Cancer cell metabolism is currently considered to be context dependent, and metabolic reprogramming is being widely investigated. It is known that ovarian cancer often metastasizes to the omentum. Given that the omentum itself contains a high concentration of adipocytes, ovarian cancer is thought to be a good model for research into metabolic reprogramming (particularly the shift to lipid metabolism). The present study investigated the switch to lipid metabolism in the metabolic reprogramming of ovarian cancer cells. The present study first considered the possibility of epigenetic involvement. Using an open database (GSE 85293 and GSE2109), the methylation status and gene expression patterns of the primary tumor site (ovary) and the metastatic tumor site (omentum) were compared. However, no evidence was obtained regarding the involvement of epigenetics (at least in terms of DNA methylation). The influence of suspension in ascites on metabolism was then considered, and a suspension culture was used as an in vitro model. It was demonstrated that ovarian cancer cells that are detached from the primary site and suspended in ascites have enhanced lipid metabolism. Additionally, it was demonstrated that these cells express high levels of the cancer stem cell (CSC) marker cluster of differentiation 44 and c-kit in a balanced manner as they approach the omentum. Accordingly, these cells activate the mammalian target of rapamycin pathway, which is thought to be advantageous for cancer cell metastasis. In conclusion, the present study proposed one explanation for why ovarian cancer cells are likely to disseminate to the peritoneal cavity, and in particular to the omentum.
Keywords: metabolic reprogramming, ovarian cancer, lipid metabolism, cancer stem cells (CSCs), spheroid
Introduction
It has long been considered that cancer cells utilize glycolysis for their metabolism; they enhance glycolysis even in the presence of adequate oxygen to meet the high energy demands arising from their high proliferative status (1). This phenomenon, called the Warburg effect, was proposed almost one hundred years ago (1). Additionally, many studies concerning cancer metabolism have arisen from the development of metabolome analysis. In other words, researchers have reported the presence of metabolic heterogeneity even in the same tumor, and metabolic reprogramming can occur in a context-dependent manner (2–8).
Ovarian cancer is a leading cause of death worldwide, in part because most cases are identified late, when the cancer has reached advanced stages (9). Patients in advanced stages almost always have metastatic lesions in the omentum (9). The omentum itself contains a high concentration of adipocytes. The omentum is only one site of metastasis in the peritoneal cavity; however, it is reasonable to consider that ovarian cancer could utilize fatty acids as an energy resource and switch to lipid metabolism. Indeed, one research group elegantly demonstrated that co-culturing ovarian cancer cells and adipocyte tissues resulted in the enhancement of fatty acid metabolism in cancer cells (10). However, it remains unclear what type of cellular and molecular processes specifically causes the metabolic reprogramming.
We investigated the switch to lipid metabolism in the metabolic reprogramming of ovarian cancer cells.
We first considered the involvement of epigenetics. Using an open database, we compared the methylation status between the primary tumor site (ovary) and the metastatic tumor site (omentum). However, we found no evidence of the involvement of epigenetics (at least in terms of DNA methylation).
Then, we considered the influence of suspension in ascites on the metabolism, and we performed a suspension culture for the in vitro model (8). We demonstrated that culturing cells in suspension caused an increase in the expression of enzymes related to lipid metabolism. Additionally, we found that those cells showed decreased expression of cancer stem cell (CSC) markers, such as CD44 and c-kit. However, we also found that the expression of CD44 and c-kit remained considerably higher after those cells were exposed to starvation and IL-6 (which is thought to be secreted by mesenchymal cells in the peritoneal cavity), which might result in increased phosphorylation of S6, the downstream product of mammalian target of rapamycin (mTOR). In other words, we considered that i) suspension in ascites, ii) exposure to starvation and iii) exposure to the microenvironment (including IL-6) harmonically changed the metabolic status of ovarian cancer cells and CSC markers, and we hypothesized that the combination of these effects explained why ovarian cancer cells were likely to disseminate to the peritoneal cavity. The microenvironment in the body is likely to be more complex; nonetheless, we proposed one process of ovarian cancer metastasis. We suggest that ovarian cancer may be a good model for the investigation of metabolic reprogramming.
Materials and methods
Cell line and cell culture
The cancer cell line SKOV3 was obtained from ATCC (USA). Cells were cultured in DMEM (cat. no. 043-30085; Wako, Japan), supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) and 100 U/ml penicillin/100 µg/ml streptomycin (Wako), and sub-cultured via 0.25% trypsin/EDTA (Wako) detachment. The FBS lot number was 1706567. For the starvation culture, low-glucose DMEM without glutamine (cat. no. 044-33555; Wako) that was not supplemented with FBS was used. All cells were grown in a humidified atmosphere at 37°C with 5% CO2.
Adherent culture and suspension culture
Dissociated single cells (1×105 cells/ml) were seeded into conventional adherent plates (Nalge Nunc International; Thermo Fisher Scientific, Waltham, MA, USA) or ultra-low attachment plates (Corning Inc., Acton, MA, USA) and cultured for 48 h.
GSE 85293 and GSE 2109
Data analyses were performed by Subio, Inc., (Kagoshima, Japan) with Subio Platform.
GSE 85293 contained 16 samples (ovary, 4; omentum, 4; ascites, 4; normal, 4). Probe sets that were not changed (|beta-value|<0.3) compared to normal were excluded. This gave us 153,045 probe sets. After that, the omentum probe sets that were changed compared to the ovary samples (|beta-value|>0.3) were extracted.
GSE 2109 contained 2158 samples, including ovary (86 cases) and omentum (38 cases). The probe sets in which the ABS call value was A (Absent) in more than 90% of cases were excluded. This gave us 38877 probe sets. This set was called the QC1 list. Then, the probe sets in which the processed signal average ranged from −0.1 to 0.1 both in omentum and ovary samples were excluded from the QC1 list. This gave us 16649 probe sets. This set was called the QC2 list. From the QC2 list, the genes that differed significantly in the omentum and ovary were extracted. The criteria were i) |fold change| ≥1.5 and ii) P-value less than 0.1 according to the Mann-Whitney U-test.
Reagent
IL-6 was purchased from Wako. The final concentration of IL-6 was 50 ng/ml.
Western blotting
The same amount of protein from whole cell lysates was subjected to SDS-polyacrylamide gel (Bio-Rad, Berkeley, CA, USA) electrophoresis and then electrotransferred onto polyvinylidene difluoride membranes (Bio-Rad). Blotting was performed with Trans-Blot Turbo (Bio-Rad) according to the manufacturer's instructions. From blocking to the secondary antibody reaction, iBind Western System (Invitrogen) was used according to the manufacturer's instructions. The secondary antibodies were detected using Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore, Billerica, MA, USA) according to the manufacturer's instructions.
Antibodies
Anti-PGC1α antibody, anti-ACOX1 antibody, anti-CPT1A antibody, anti-CD44 antibody and anti-c-kit antibody were purchased from Abcam (cat. no. ab54481, ab184032, ab128568, ab51037 and ab32363, respectively). Anti-EHHADH antibody and anti-ACSS2 antibody were purchased from SantaCruz (cat. no. SC-393123 and SC-398559, respectively). Anti-p-HSL (Ser 660) antibody, anti-p-AMPK (Thr172) antibody and anti-S6 antibody were purchased from Cell Signaling Technology (cat. no. 4126, 2535 and 2217, respectively). Anti-LDHA antibody was purchased from Proteintech (cat. no. 19987-1-AP). Anti-α-tubulin antibody was purchased from Millipore (cat. no. CP06).
Results
DNA methylation might not be involved in metabolic reprogramming during metastasis
It has been noted that epigenetics are involved in the metabolic shift toward lipid metabolism (11). Thus, we speculated that epigenetic regulation was also related to metabolic reprogramming in ovarian cancer between the primary site (ovary) and the metastatic site (omentum). GSE 85293 was the dataset of DNA methylation status from sites including the ovary and omentum. GSE 2109 was the dataset of gene expressions (microarray or RNA-sequencing) from many sites, including the ovary and omentum. Using these datasets, we investigated and compared the methylation status and gene expression patterns, focusing on the PPAR signaling pathway, a representative lipid metabolism pathway from the Kyoto Encyclopedia of Genes and Genomes (KEGG). As shown in Table I, we could not find a correlation between DNA methylation and gene expression, although the investigation was limited because the datasets used were from different patients and studies.
Table I.
Methylation and expression status (omentum/ovary) of the genes related to lipid metabolism.
| Genes | Methylation (GSE 85293) | Expression (GSE 2109) |
|---|---|---|
| FATP | ↑ | – |
| ME1 | ↑ | – |
| LPL | – | ↑ |
| CD36 | – | ↑ |
| ACS | ↓ | – |
| EHHADH | ↑ | – |
| LCAD | ↑ | – |
| ACOX1 | ↑ | – |
| Perlipin | – | ↑ |
| FABP4 | – | ↑ |
| ADIPO | – | ↑ |
| SORBS1 | – | ↑ |
| MMP-1 | – | ↑ |
We did not find a correlation between DNA methylation and gene expression, although our investigation was limited by the fact that the two studied datasets came from different patients and studies. ↑, hyper-methylated or up-regulated; ↓, hypo-methylated or down-regulated; -, not significant; FATP, fatty acid transporter; ME1, malic enzyme 1; LPL, lipoprotein lipase; CD36, CD36 molecule; ACS, acyl-CoA synthetase long-chain family member 1; EHHADH, enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase; LCAD, long-chain-acyl-CoA dehydrogenase; ACOX1, acyl-CoA oxidase 1; Perlipin, perilipin-2; FABP4, fatty acid binding protein 4; ADIPO, adiponectin; SORBS1, sorbin and SH3 domain containing 1, MMP-1, matrix metallopeptidase 1.
Cells cultured in suspension show enhanced lipid metabolism and relatively less sensitivity to starvation compared with cells cultured in adherent plates
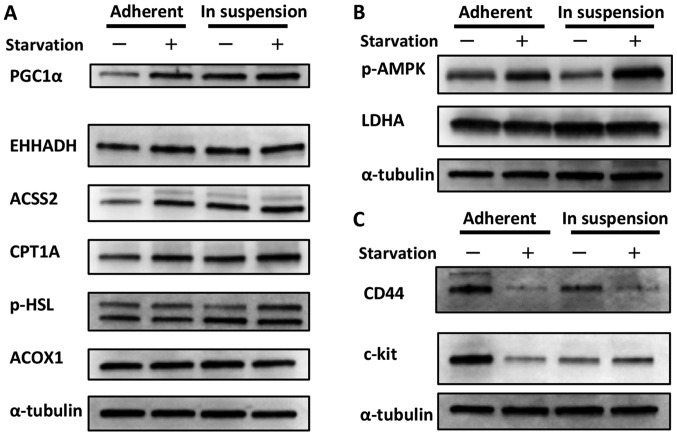
We could not obtain evidence that epigenetics (at least DNA methylation) were involved in metabolic reprogramming in ovarian cancer. We speculated that a more direct phenomenon was related to the metabolic shift toward lipid metabolism; in other words, detachment from the primary site and suspension in ascites itself affected metabolism (through morphological changes, for instance). Indeed, we have already demonstrated that cancer cells cultured in suspension are more likely to show an activated TCA cycle than cells cultured in adherent plates (8). In the present study, we focused on lipid metabolism. As shown in Fig. 1A, cells cultured in suspension showed increased expression of PGC1α, a sensor for lipid metabolism, compared with cells cultured in adherent plates (12). Additionally, the expression of some types of enzymes related to fatty acid metabolism was also increased, although the level of increase depended on the enzyme. While the expressions of these enzymes were increased in cells cultured in adherent plates when they were exposed to starvation, it seemed that cells cultured in suspension showed less sensitivity to starvation. This could be interpreted to indicate that cells in suspension already utilize fatty acid metabolism and maintain a relative advantage against starvation. Conversely, we considered the possibility that the phosphorylation of AMPK, a sensor for glycolysis, was decreased in cells cultured in suspension compared with cells cultured in adherent plates, as shown in Fig. 1B (13). The expression of LDHA, the indicator of glycolysis, was thought to be unchanged between the different cell culture statuses. Taken with the result from Fig. 1A, this finding suggests that cells cultured in suspension showed a metabolic shift from glycolysis toward lipid metabolism. Although the phosphorylation of AMPK was increased in cells cultured in suspension and exposed to starvation, as shown in Fig. 1B, it is possible that these cells were preparing for impending nourishment.
Figure 1.
Change in the metabolic status and CSC marker expression of cells in adherent plates vs. suspension. (A) Change in lipid metabolism. Cells cultured in suspension showed increased expression of PGC1α, which is thought to be a sensor for lipid metabolism, compared with cells cultured in adherent plates. Additionally, the expression of some types of enzymes related to fatty acid metabolism were also increased, although the level of increase depended on the enzyme. The expressions of these enzymes were increased in cells cultured in adherent plates when they were exposed to starvation; however, the cells cultured in suspension showed less sensitivity to starvation. This could be interpreted to indicate that cells in suspension already utilize fatty acid metabolism and maintain a relative advantage against starvation. (B) Change in glycolysis. The phosphorylation of AMPK, a marker for glycolysis, was decreased in cells cultured in suspension compared with cells cultured in adherent plates. The expression of LDHA, the indicator for glycolysis, was nearly unchanged between the two groups. Although the phosphorylation of AMPK was increased in cells cultured in suspension and exposed to starvation, it is possible that these cells were preparing for upcoming nourishment. (C) Change in CSC marker expression. Cells cultured in suspension showed decreased expression of CD44 and c-kit compared with cells cultured in adherent plates. Interestingly, the response during starvation differed between the two culture groups. While the expression of CD44 was decreased during starvation in cells cultured in both suspension and adherent plates, the expression of c-kit increased in cells cultured in suspension but decreased in cells cultured in adherent plates.
The expression of CSC markers changes according to the cell status
At present, it is well-documented that CSCs are related to metastasis (14). To add to this knowledge, we investigated how CSC markers changed according to the cell status. We considered CD44 and c-kit as CSC markers for ovarian serous carcinoma, as previously described (14). As shown in Fig. 1C, cells cultured in suspension showed decreased expression of CD44 and c-kit compared with cells cultured in adherent plates. Interestingly, the response during starvation differed between the two culture environments. While the expression of CD44 was decreased during starvation in both cells cultured in suspension and those cultured in adherent plates, the expression of c-kit increased in cells cultured in suspension but decreased in cells cultured in adherent plates. This phenomenon was considered advantageous for metastasis overall, as discussed below.
Cells cultured in suspension show balanced high expression of CD44 and c-kit and possible mTOR pathway activation after exposure to IL-6
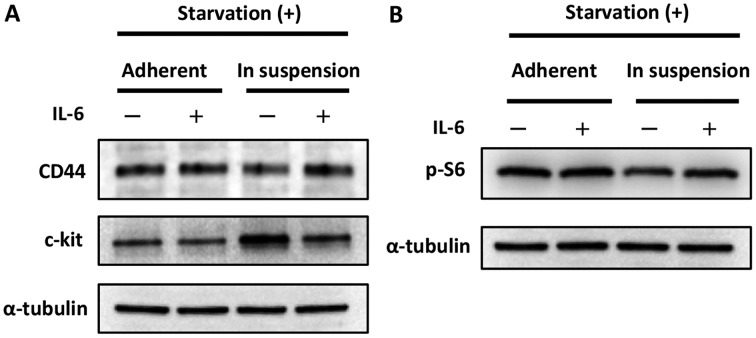
Above, we describe an in vitro model that mimics the condition of being detached from the primary site and suspended in ascites. Then, we performed an experiment that mimicked the status of ‘cells suspended in ascites and approaching the omentum’. We focused on IL-6, which is thought to be secreted from the omentum or other mesenchymal cells and is known to have some impact on cancer (stem) cells (9). In other words, we investigated the behavior of cancer cells in the presence or absence of IL-6. As shown in Fig. 2A, the expression of CD44 was increased in cells cultured in suspension when exposed to IL-6, while the expression of c-kit was decreased. These results seem paradoxical; however, they could be interpreted to indicate that the cells expressed CD44 and c-kit in relatively high but balanced manner. Indeed, the phosphorylation of S6, the indicator of mTOR pathway activation, was increased in cells cultured in suspension when exposed to IL-6, as shown in Fig. 2C.
Figure 2.
Impact of IL-6 on the maintenance of CSC-like status. (A) Impact of IL-6 on CSC markers. The expression of CD44 increased in cells cultured in suspension when they were exposed to IL-6. In contrast, the expression of c-kit was decreased in cells cultured in suspension when they were exposed to IL-6. These results seemed paradoxical and are discussed in the Discussion section. (B) Impact of IL-6 on the mTOR pathway. The phosphorylation of S6, the indicator of mTOR pathway activation, increased in cells cultured in suspension when they were exposed to IL-6.
Discussion
Here, we proposed an explanation of why ovarian cancer cells are likely to disseminate to the peritoneal cavity, focusing on metabolic reprogramming and CSCs.
It is considered that cancer cells utilize glycolysis for metabolism even the presence of adequate oxygen to satisfy the high energy demands that arise from their high rate of proliferation. This phenomenon (aerobic glycolysis) is called the Warburg effect, and it was first documented in the 1920s (1). However, more precise and specific investigations of cancer cell metabolism have been performed since the development of metabolome analysis. Metabolic reprogramming is among these newly investigated processes, and the need to rethink the Warburg effect is widely recognized (2–8).
Meanwhile, ovarian cancer is known to frequently metastasize to the omentum (10). The omentum itself contains a high concentration of adipocytes, and ovarian cancer is thought to be a good research model for metabolic reprogramming (especially for lipid metabolism). Indeed, a previous report elegantly demonstrated that co-culturing ovarian cancer cells and adipocyte tissues resulted in the enhancement of fatty acid metabolism in ovarian cancer cells (10). However, an understanding of what specific cellular and molecular processes cause the metabolic reprogramming remains elusive. Collectively, we investigated the switch in the metabolic reprogramming of ovarian cancer cells to lipid metabolism.
While an unborn baby utilizes the glucose in blood for its nutrition before birth, it begins to utilize lipid metabolism during breast-feeding (i.e., upon exposure to fatty acids) (11). Epigenetics is known to be involved in the drastic shift (11). With this knowledge, we speculated that epigenetics was also in involved in the metabolic reprogramming during ovarian cancer. Using an open database, we compared the methylation status between the primary tumor site (the ovary) and the metastatic tumor site (the omentum). However, we did not obtain any evidence of the involvement of epigenetics (at least in terms of DNA methylation).
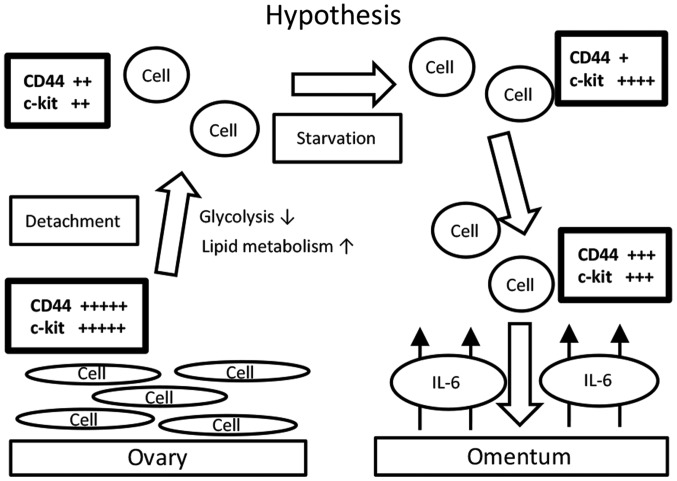
We then considered the influence of suspension in ascites on metabolism, and we performed a suspension culture for the in vitro model. A hypothetic schema of our results is shown in Fig. 3. Ovarian cancer cells that are detached from the primary site (the ovary) reprogram their metabolism from glycolysis to lipid metabolism. In the process, the expression levels of CD44 and c-kit decrease. During starvation, the expression level of CD44 decreases and that of c-kit increases. Upon exposure to IL-6, the expression level of CD44 further increase, but that of c-kit decreases. These changing patterns are believed to maintain an overall ‘CSC-like status’. Indeed, the mTOR pathway is reactivated accordingly. We propose that this is why ovarian cancer cells often metastasize to the omentum. Indeed, a previous report showed that CD44-positive and c-kit-positive cells isolated from the ascites of ovarian cancer patients presented activation of lipid metabolism (14). In the present study, we demonstrated that the reverse could also be true; that is, that the expression of CD44 and c-kit in ovarian cancer cells that are detached from the primary site and suspended seems to change significantly in a balanced manner as the cells approach the omentum. We do not know why detachment itself causes metabolic changes; however, cell polarity and focal adhesion might be responsible.
Figure 3.
Hypothetic schema of the data we obtained. Ovarian cancer cells that are detached from the primary site (the ovary) reprogram their metabolism from glycolysis to lipid metabolism. Accordingly, the expression levels of CD44 and c-kit decrease. During starvation, the expression level of CD44 further decreases, but that of c-kit increases. When ovarian cancer cells are exposed to IL-6 near the omentum, the expression level of CD44 further increases, but that of c-kit decreases. These changing patterns may contribute to maintaining an overall ‘CSC-like’ status. Indeed, the mTOR pathway is reactivated accordingly. This process was considered as an explanation of why ovarian cancer cells often metastasize to the omentum.
Although we only used one cell line and performed an in vitro experiment, we proposed a possible explanation for why ovarian cancer cells are likely to metastasize to the peritoneal cavity, especially to the omentum.
It is quite challenging to determine whether CSCs reprogram their own metabolism or the properties that could contribute to metabolic reprogramming are CSCs, and further investigations into this matter are expected. In that context, ovarian cancer is thought to be a good model for the investigation of metabolic reprogramming.
References
- 1.Dang CV. Rethinking the warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 2010;70:859–862. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altieri DC. Mitochondria on the move: Emerging paradigms of organelle trafficking in tumour plasticity and metastasis. Br J Cancer. 2017;117:301–305. doi: 10.1038/bjc.2017.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cliff TS, Dalton S. Metabolic switching and cell fate decisions: Implications for pluripotency, reprogramming and development. Curr Opin Genet Dev. 2017;46:44–49. doi: 10.1016/j.gde.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Fidoamore A, Cristiano L, Laezza C, Galzio R, Benedetti E, Cinque B, Cinque B, Antonosante A, d'Angelo M, Castelli V, et al. Energy metabolism in glioblastoma stem cells: PPARα a metabolic adaptor to intratumoral microenvironment. Oncotarget. 2017 doi: 10.18632/oncotarget.19086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawada K, Toda K, Sakai Y. Targeting metabolic reprogramming in KRAS-driven cancers. Int J Clin Oncol. 2017;22:651–659. doi: 10.1007/s10147-017-1156-4. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Han G, Li X, Kan Q, Fan Z, Li Y, Ji Y, Zhao J, Zhang M, Grigalavicius M, et al. Mitochondrial pyruvate carrier function determines cell stemness and metabolic reprogramming in cancer cells. Oncotarget. 2017;8:46363–46380. doi: 10.18632/oncotarget.18199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato M, Kawana K, Adachi K, Fujimoto A, Yoshida M, Nakamura H, Nishida H, Inoue T, Taguchi A, Takahashi J, et al. Spheroid cancer stem cells display reprogrammed metabolism and obtain energy by actively running the tricarboxylic acid (TCA) cycle. Oncotarget. 2016;7:33297–33305. doi: 10.18632/oncotarget.8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeung TL, Leung CS, Yip KP, Au Yeung CL, Wong ST, Mok SC. Cellular and molecular processes in ovarian cancer metastasis. A review in the theme: Cell and molecular processes in cancer metastasis. Am J Physiol Cell Physiol. 2015;309:C444–C456. doi: 10.1152/ajpcell.00188.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehara T, Kamei Y, Yuan X, Takahashi M, Kanai S, Tamura E, Tsujimoto K, Tamiya T, Nakagawa Y, Shimano H, et al. Ligand-activated PPARα-dependent DNA demethylation regulates the fatty acid β-oxidation genes in the postnatal liver. Diabetes. 2015;64:775–784. doi: 10.2337/db14-0158. [DOI] [PubMed] [Google Scholar]
- 12.Davidson B, Hadar R, Stavnes HT, Trope CG, Reich R. Expression of the peroxisome proliferator-activated receptors-alpha, -beta and -gamma in ovarian carcinoma effusions is associated with poor chemoresponse and shorter survival. Hum Pathol. 2009;40:705–713. doi: 10.1016/j.humpath.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 13.Zhang CS, Hawley SA, Zong Y, Li M, Wang Z, Gray A, Ma T, Cui J, Feng JW, Zhu M, et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature. 2017;548:112–116. doi: 10.1038/nature23275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastò A, Bellio C, Pilotto G, Ciminale V, Silic-Benussi M, Guzzo G, Rasola A, Frasson C, Nardo G, Zulato E, et al. Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation and resist glucose deprivation. Oncotarget. 2014;5:4305–4319. doi: 10.18632/oncotarget.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]