Abstract
Piwi-interacting RNAs (piRNAs/piRs) are small non-coding RNAs that can serve important roles in genome stability by silencing transposable genetic elements. piR651, one of these novel piRNAs, regulates a number of biological functions, as well as carcinogenesis. Previous studies have reported that piR651 is overexpressed in human gastric cancer tissues and in several cancer cell lines, including non-small cell lung cancer (NSCLC) cell lines. However, the role of piRNAs in carcinogenesis has not been clearly defined. In the present study, a small interfering RNA inhibitor of piR651 was transfected into the NSCLC A549 and HCC827 cell lines to evaluate the effect of piR651 on cell growth. The association between piR651 expression and apoptosis was evaluated by flow cytometry and western blot analysis. Wound-healing and Transwell migration and invasion assays were used to determine the effect of piR651 on the migration and invasion of NSCLC cell lines. The results revealed that inhibition of piR651 inhibited cell proliferation and significantly increased the apoptotic rate compared with the negative control (NC), as well as altering the expression of apoptosis-associated proteins. There were fewer migrating and invading cells in the piR651-inhibited group than in the NC group in the Transwell assays. Furthermore, in the wound-healing assay, the wound remained wider in the piR651 inhibitor group, suggesting decreased cell migration compared with that in the NC group. The results of the present study demonstrate that piR651 potentially regulates NSCLC tumorigenic behavior by inhibiting cell proliferation, migration and invasion and by inducing apoptosis. Therefore, piR651 is a potential cancer diagnosis marker.
Keywords: piwi-interacting RNA, non-coding RNA, molecular diagnosis, non-small cell lung cancer, carcinogenesis
Introduction
Lung cancer is the leading cause of cancer-associated mortality in numerous countries, and the incidences of morbidity and mortality associated with the disease are increasing; lung cancer was responsible for >160,000 mortalities in the United States in 2010 (1). Among all the types of lung cancer, non-small cell lung cancer (NSCLC) is the most common histological subtype, accounting for 80–90% of all cases (2). Although multiple effective treatments, including radiotherapy, chemotherapy and immunotherapy, have recently become available for the management of locally confined NSCLC, these treatments have been unable to reduce the high mortality rate among patients with advanced-stage NSCLC (1). Thus, it has become increasingly important to identify methods to diagnose early-stage lung cancer with high sensitivity and predict clinical outcome.
In recent years, numerous studies have focused on the association between carcinogenesis and small non-coding regulatory RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and piwi-interacting RNAs (piRNAs) (3–5). Small non-coding regulatory RNAs may have key roles in carcinogenesis. The most extensively researched small non-coding regulatory RNAs are miRNAs, which can act as either oncogenes or tumor suppressors, according to the function of their target genes (6–8). Previous studies have confirmed that the up- or downregulation of certain miRNAs may contribute to human carcinogenesis and cancer progression (9,10). However, research regarding piRNAs, which are a novel type of small non-coding RNA with lengths of 26–31 nucleotides, remains insufficient (11,12). piRNAs serve biological roles through their specific associations with the piwi proteins (13,14). piRNAs aid the maintenance of DNA integrity, epigenetic regulation, germ line stem cell differentiation, embryonic development and disease occurrence and development. Previous studies have confirmed that piRNAs can also serve similar roles to miRNAs, acting as oncogenes or tumor suppressors in several types of cancer (15–17), including those of the cervix (18), bladder (19), lung (20), gastrointestinal tract (21), breast (22,23) and liver (24). It was previously reported that the piRNA piR651 is overexpressed in several types of human cancer tissue, including gastric, lung, colon, breast, and multiple myeloma cancer tissues, compared with paired adjacent normal tissues (21,22). Furthermore, piR651 expression levels in gastric cancer tissues are associated with tumor-node-metastasis (TNM) stage (25,26). The overexpression of piR651 has been demonstrated in several cancer cell lines, including those of the lung, gastric, mesothelium, cervix, breast and liver (21). These findings indicate that piR651 may serve an oncogenic role in carcinogenesis. However, the mechanism by which piR651 regulates carcinogenesis is unclear. Therefore, the present study aimed to elucidate the mechanisms of action of piR651 in NSCLC.
Materials and methods
Cell culture
The human NSCLC A549 and HCC827 cell lines were purchased from the Fourth Affiliated Hospital of Harbin Medical University (Harbin, China) and the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Science (Shanghai, China), respectively. HCC827 and A549 cells were cultured in RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and high-glucose Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.), respectively, supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin, and 10% fetal bovine serum (FBS; Tianhang Biotechnology Co., Hangzhou, China) in a humidified atmosphere with 5% CO2 at 37°C.
Cell transfection and detection of transfection rate
To investigate the possible effects of piRNAs on tumorigenesis, an siRNA inhibitor of piR651 was transfected into NSCLC cells. The piR651 inhibitor and negative control (NC) siRNA were synthesized by Shanghai GenePharma Co. (Shanghai, China). The sequence of the piR651 inhibitor was 5′-GACGCUUUCCAAGGCACGGGCCCCUCUCU-3′. The sequence of the NC was 5′-CAGUACUUUUGUGUAGUACAA-3′. The inhibitor and NC were transfected into A549 and HCC827 cells using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Finally, fluorescence microscopy and flow cytometry were utilized to determine the transfection rate. Cells were seeded into 6-well plates 1 day before transfection. When cells reached between 50 and 60% confluence, 100 pmol piR651 inhibitor with fluorescent marker [6-carboxyfluorescein (FAM)], or 100 pmol NC with FAM was transfected using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The transfection rate was determined using a fluorescence microscope (magnification, ×200), by observing the number of cells, and a flow cytometer (BD LSRII; BD Biosciences, Franklin Lakes, NJ, USA), by detecting the degree of fluorescence in the piRNA inhibitor and the NC.
MTT assay
MTT assays were utilized to ascertain the cell growth rate following transfection with the piR651 inhibitor. Five replicate wells were seeded for each group in this assay. A549 and HCC827 cells were cultured in 96-well plates for 24 h at 37°C, and transfected with either the piR651 inhibitor or the NC. After 12, 24, 48 and 72 h, 20 µl MTT (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) solution was added to each well. Cells were incubated at 37°C for 4 h in the dark and subsequently the supernatant was discarded and 150 µl dimethyl sulfoxide (Thermo Fisher Scientific, Inc.) was added to dissolve the crystallized formazan product. The plate was agitated mildly for 10 min. The optical density (OD) value was detected for each well at 490 nm using a Wellscan reader (Labsystems, Santa Fe, NM, USA). Growth proliferation was calculated as a percentage using the following formula: (OD experiment/OD control) ×100. Independent MTT experiments were performed three times.
Apoptosis analysis
To evaluate the association between piR651 and apoptosis, HCC827 and A549 cells were collected and washed with PBS 24 h after transfection. Subsequently, 106 cells/ml were stained using an Annexin V-fluorescein isothiocyanate apoptosis detection kit (cat. no. CX1001-2; Beijing Biosea Biotechnology Co., Ltd., Beijing, China), according to the manufacturer's protocol. The cells were subsequently suspended in binding buffer (Beijing Biosea Biotechnology Co., Ltd.) and analyzed using a flow cytometer (BD FACSCalibur; BD Biosciences). All tests were repeated three times.
Western blot analysis
Western blot analysis was performed to detect changes in apoptosis-associated proteins following transfection of NSCLC cells with the piR651 inhibitor. In brief, HCC827 and A549 cells transfected with the piR651 inhibitor or the NC were lysed with radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China) after 46 h. The lysates were separated by centrifugation at 12,000 × g for 15 min at 4°C. The supernatants were boiled (10 min) and quantified using the bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology), according to the manufacturer's protocol. Subsequently, protein was separated using SDS-PAGE (12 and 6% gels; Beyotime Institute of Biotechnology) and transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). The membrane was subsequently blocked in Tris-buffered saline-Tween-20 (TBST), containing 5% skimmed milk powder (TBSTM), for 1 h at room temperature with gentle agitation. Membranes were incubated at 4°C overnight with the following primary antibodies against: Antibodies B-cell lymphoma 2 (Bcl-2; cat. no. ZS-492; dilution, 1:100), Bcl-2-associated X protein (Bax; cat. no. ZS-526; dilution, 1:100), β-actin (cat. no. TA-09; dilution, 1:1,000; all from OriGene Technologies, Beijing, China), caspase-3-p17 (cat. no. 25546-1-AP; dilution, 1:200; ProteinTech Group, Inc., Wuhan, China) and poly(ADP-ribose) polymerase 1 (PARP1; cat. no. 13371-1-AP; dilution, 1:200; ProteinTech Group, Inc.). Subsequently, membranes were washed with TBST (four times for 15 min each) and incubated at room temperature with horseradish peroxidase (HRP) conjugated affinipure goat anti-mouse immunoglobulin G (IgG) (cat. no. ZB-2305; dilution, 1:5,000; OriGene Technologies, Inc., Beijing, China) or HRP conjugated affinipure goat anti-rabbit IgG (cat. no. ZB2301; dilution, 1:5,000; OriGene Technologies, Inc.) secondary antibodies for 1 h. Following incubation, members were washed with TBST three times for 15 min each. Subsequently, membranes were incubated in Pierce ECL Western Blotting Substrate (cat. no. 32106; Thermo Fisher Scientific, Inc.) for 5 min at room temperature. Visualization was achieved using the X-OMAT BT Film (cat. no. 03690336810396; Carestream Health, Inc., Rochester, NY, USA) and the Medical Intensifying Screen (cat. no. 2013004; Jianmin Medical Instrument Co., Nanchang, China). All experiments were performed three times.
Transwell migration and invasion assays
Cell migration and invasion assays were performed using Transwell chambers with 8-mm membrane pores (Corning Incorporated, Corning, NY, USA). For the migration assay, 7–8×104 transfected cells per well were seeded in serum-free medium in the upper chamber with a non-coated membrane. In the invasion assays, 7–8×104 cells per well were seeded in serum-free medium in the upper chamber with a membrane that was pre-coated with Matrigel (BD Biosciences). For each assay, the contents of the lower chambers were the same as those of the upper chamber. After incubation for 48 h, the membranes were removed from the plate, and the non-migrated cells were removed from the upper surface with a cotton swab. The cells on the underside of the membrane were fixed in 100% methanol and stained with 0.5% crystal violet (diluted in 100% methanol). The cells on the lower surface were counted under an inverted light microscope and images were captured. Cells were counted in six randomly selected fields and the average number was recorded. The relative migration/invasion (%) was calculated as follows: (Average number/100) ×100. Three independent experiments were performed.
Wound-healing assays
For the wound-healing assays, cells were grown in 6-well plates to near confluence. A linear scratch was carefully made with a 10-µl pipette tip, and any cell debris was gently washed away with phosphate-buffered saline (PBS). The HCC827 or A549 cells were then incubated with serum-free RPMI-1,640 or DMEM, respectively. Images were captured of the cells 48 h after the scratch was created using an inverted light microscope (Olympus Corporation, Tokyo, Japan). The distance between the cells which migrated the furthest on each side to calculate the relative wound closure (%).
Statistical analysis
Data were presented as the mean ± standard error of the mean, of three independent experiments. MTT data was processed using two-factor analysis of variance followed by Bonferroni's correction and the remaining tests were analyzed by a two-sample t-test using SPSS software (version 17.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
piR651 promotes cell proliferation in a time-dependent manner and inhibits apoptosis in NSCLC cells
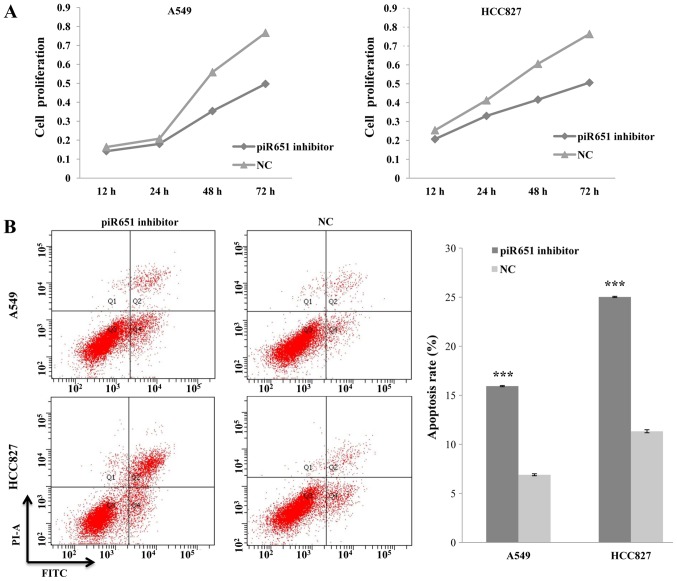
A previous study found piR651 to be overexpressed in several cancer cell lines, including those of lung cancer (21). Thus, the present study reduced the endogenous expression of piR651 by transfecting the NSCLC A549 and HCC827 cell lines with a piR651 inhibitor. The rate of cell proliferation decreased, compared with the NC, in response to transfection with the piR651 inhibitor in the two cell lines. In addition, the piR651 inhibitor regulated the cell proliferation rate in a time-dependent manner (P<0.001; Fig. 1A).
Figure 1.
Association between piR651 and cell proliferation and apoptosis. (A) In A549 cells, the proliferation rate decreased in a time-dependent manner following transfection with the piR651 inhibitor compared with the NC (P<0.001). The results were similar in HCC827 cells (P<0.001). (B) The piR651 inhibitor increased A549 and HCC827 cell apoptosis, as detected by flow cytometry. The apoptosis rate of cells transfected with the piR651 inhibitor was significantly higher than that of the NC in A549 and HCC827 cells. ***P<0.001 vs. corresponding NC group. piR651, piwi-interacting RNA-651; NC, negative control; PI-A, propidium iodide; FITC, fluorescein isothiocyanate.
A previous study indicated that apoptosis serves an important role in regulating carcinogenesis (27). Therefore, we hypothesized that piR651 regulates tumorigenesis by altering the rate of NSCLC cell apoptosis. An Annexin V-PI kit was used to detect apoptosis in A549 and HCC827 cells following transfection with the piR651 inhibitor or the NC. The apoptotic rates of the piR651 inhibitor-transfected groups were higher than those of the respective the NC-transfected groups (t=145.31, P=0.001; t=135.5, P=0.001; Fig. 1B). On the basis of these results, it was concluded that piR651 may affect tumorigenesis by promoting cell proliferation and inhibiting apoptosis in NSCLC cells.
piR651 regulates apoptosis by altering the expression of apoptosis-associated proteins in NSCLC cells
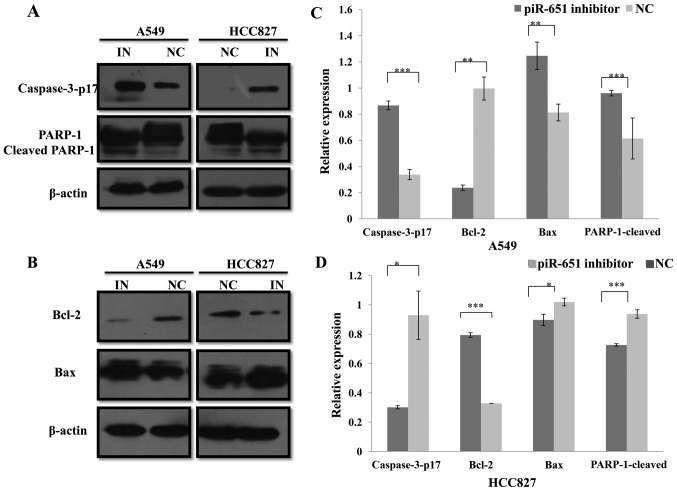
The previous experiment demonstrated that piR651 inhibited NSCLC cell apoptosis. Numerous studies have confirmed that several pathways can regulate tumor cell apoptosis, including the caspase pathway (28–31). Thus, classical apoptosis-associated proteins, including caspase-3, Bax, Bcl-2 and PARP1, were investigated. Using western blotting assays, it was confirmed that, compared with the NC in A549 and HCC827 cells, caspase-3-p17 (P<0.001 and P=0.022), Bax (P=0.004 and P=0.014) and cleaved-PARP-1 (P=0.014 and P<0.001) expression levels were increased, whereas Bcl-2 (P=0.003 and P<0.001) expression was decreased by the piR651, respectively (Fig. 2). Caspase-3 and Bax/Bcl-2 are proteins involved in the mitochondrial apoptosis pathway (30–32). In conclusion, the piR651 inhibitor induced apoptosis in A549 and HCC827 cells, most likely through a mitochondria-dependent pathway.
Figure 2.
piR651 regulates the expression of apoptosis-associated proteins in A549 and HCC827 cells. (A) Caspase-3-p17, PARP-1 and cleaved PARP1 protein levels in A549 and HCC827 cells were analyzed using western blot. β-actin was used as an internal control. (B) Bax and Bcl-2 protein levels in A549 and HCC827 cells were analyzed using western blot. β-actin was used as an internal control. (C) Compared with the NC, the expression of caspase-3-p17 (***P<0.001), Bax (**P=0.004) and cleaved PARP1 (*P=0.014) increased and the expression of Bcl-2 (**P=0.003;) decreased in response to the piR651 inhibitor in A549 cells. (D) In HCC827 cells, the expression of caspase-3-p17 (*P=0.022), Bax (*P=0.014) and cleaved PARP1 (***P<0.001) decreased and the expression of Bcl-2 (***P<0.001) increased in response to the piR651 inhibitor compared with the NC. piR651, piwi-interacting RNA-651; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein; PARP, poly(ADP-ribose) polymerase; IN, piR651 inhibitor; NC, negative control.
piR651 promotes NSCLC cell migration and invasion in vitro
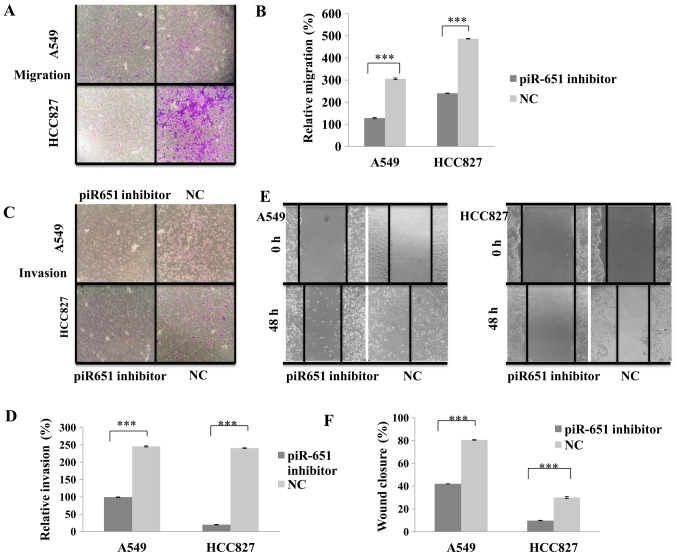
To determine whether the knockdown of endogenous piR651 affects NSCLC cell migration and invasion, the piR651 inhibitor was transfected into NSCLC A549 and HCC827 cell lines to downregulate the endogenous expression of piR651. As presented in Fig. 3A and B, compared with the NC, cells transfected with the piR651 inhibitor exhibited significantly decreased cell migration (P<0.001). The results of the wound-healing assay were similar to those of the migration assay; transfection with the piR651 inhibitor significantly decreased the wound-closure rate compared with the NC (P<0.001; Fig. 3C and D). The results of the cell invasion assay were also similar to those of the migration assay, with significantly fewer cells invading through the membrane in the piR651 inhibitor-transfected group compared with the NC group in the two cells lines (P<0.001; Fig. 3E and F). These data support the hypothesis that a reduction in endogenous piR651 expression may suppress NSCLC cell migration and invasion.
Figure 3.
piR651 promotes A549 and HCC827 cell migration and invasion. (A) Cell migration assays in A549 and HCC827 cells. (B) The number of cells migrated in the piR651 inhibitor was decreased, compared with that of the NC in A549 and HCC827 cell lines (***P<0.001). (C) Cell invasion assays in A549 and HCC827 cells. (D) The number of the cells invaded in the piR651 inhibitor was significantly decreased, compared with that of the NC in A549 and HCC827 cells (***P<0.001). (E) Wound healing assays in A549 and HCC827 cells. (F) The distance between the two cells in the piR651 inhibitor-transfected group was increased, compared with that in the NC group in the two cells lines (***P<0.001). ***P<0.001 piR651, piwi-interacting RNA-651; NC, negative control.
Discussion
Accumulating evidence suggests the presence of aberrant piRNA expression in numerous types of human cancer cells and tissues, including gastric carcinoma, hepatic carcinoma, bladder cancer, ovarian carcinoma and NSCLC, in which piRNAs may act as oncogenes or tumor suppressors (18–24,26). piR651, piR20365, piR4987, piR20485 and piR20582 are all overexpressed in breast cancer tissues and cell lines (22,23), whereas expression of piR823 is downregulated in gastric cancer tissues and cell lines (20). Although there are a growing number of studies concerning the association between piRNAs and carcinogenesis (18–25), the mechanism by which piRNAs regulate carcinogenesis remains inadequately understood.
In a previous study, the expression of piR651 in human gastric cancer tissues was found to be significantly increased compared with paired normal tissues and was associated with the TNM stage (20). Additionally, the expression levels of piR651 were higher in breast, colon, lung and gastric cancer tissues than in paired normal tissues (21). The upregulation of piR651 was also observed in several cancer cell lines, including those of NSCLC (20,21). However, the role of piR651 in carcinogenesis remains undefined. Therefore, in the present study, the potential roles of piR651 in NSCLC cells were investigated. The results of the present study revealed that that the knockdown of endogenous piR651 in NSCLC cells inhibited cell proliferation, induced apoptosis and inhibited cell migration and invasion. The proliferation rate of A549 and HCC827 cells decreased in a time-dependent manner following transfection with the piR651 inhibitor compared with the same cells transfected with the NC, whereas the apoptotic rate of these cells was significantly higher. These data suggest that piR651 may serve an oncogenic role in tumorigenesis.
Previous studies have demonstrated that mitochondria play an important role in the intrinsic apoptosis pathway, which is regulated by Bcl-2 family proteins through the pro-apoptotic protein Bax and the anti-apoptotic protein Bcl-2 (30,31). Analyses of mitochondria have confirmed their involvement in the production of reactive oxygen species (ROS), the downregulation of Bcl-2 family proteins, the upregulation of Bax, the release of cytochrome c, and the activation of caspase-3 in various cell lines (33–35). Therefore, we hypothesized that the knockdown of endogenous piR651 would increase the rate of apoptosis as well as the expression of cleaved caspase-3 and Bax, and would downregulate the expression of Bcl-2. Western blot analysis confirmed this hypothesis (Fig. 2).
It is estimated that ~90% of patients diagnosed with NSCLC succumb due to metastasis, rather than from the primary tumor. Owing to the high rates of recurrence and metastasis, NSCLC is associated with a strong tendency to metastasize to specific organs (32). Metastasis, which is a multi-step process involving the proliferation of cancer cells in the primary tumor, local invasion, invasion of lymphatic vessels and blood vessels, local engraftment and the formation of a new tumor (of the same tumor type) at a distal site (36,37), has been estimated to be associated with >70% of all cancer-associated mortalities (37). Therefore, the identification of molecular factors that predict tumor metastasis is clinically useful, and the identities of these factors may suggest effective therapeutic strategies against NSCLC. In the present study, the relative migration distance following transfection with a piR651 inhibitor was revealed to be less than that following transfection with the NC in wound-healing assays. Furthermore, the numbers of invasive or migratory cells on the underside of the Transwell membrane in the invasion and migration assays were lower in the piR651 inhibitor-transfected group compared with the NC group. These experiments thus confirmed that piR651 promotes cell migration and invasion.
In conclusion, the results of the present study results suggest the existence of a novel regulatory mechanism that drives NSCLC cell proliferation, apoptosis, invasion, and metastasis via the upregulation or downregulation of piR651 expression. The results of the current study confirm that the downregulation of piR651 inhibits proliferation, induces apoptosis and reduces invasion and migration in NSCLC. Therefore, piR651 may provide an opportunity to improve the clinical diagnosis and treatment of NSCLC.
Acknowledgements
The authors would like to thank Dr Pan (Hepatosplenic Surgery Center, The First Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang, China) for his advice with manuscript preparation.
References
- 1.Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: Epidemiology, etiology and prevention. Clin Chest Med. 2011;32:605–644. doi: 10.1016/j.ccm.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, Spitznagel EL, Piccirillo J. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: Analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006;24:4539–4544. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 3.Meister G. Argonaute proteins: Functional insights and emerging roles. Nat Rev Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 4.Peters L, Meister G. Argonaute proteins: Mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y, Ai X, Shen S, Lu S. NF-κB-mediated miR-124 suppresses metastasis of non-small-cell lung cancer by targeting MYO10. Oncotarget. 2015;6:8244–8254. doi: 10.18632/oncotarget.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Lin P, Yu S, Yang P. MicroRNA in lung cancer. Br J Cancer. 2010;103:1144–1148. doi: 10.1038/sj.bjc.6605901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kourtidis A, Anastasiadis PZ. Bringing together cell-to-cell adhesion and miRNA biology in cancer research. Future Oncol. 2016;12:1211–1214. doi: 10.2217/fon-2016-0012. [DOI] [PubMed] [Google Scholar]
- 10.Ling H, Krassnig L, Bullock MD, Pichler M. MicroRNAs in testicular cancer diagnosis and prognosis. Urol Clin North Am. 2016;43:127–134. doi: 10.1016/j.ucl.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 12.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 13.Kwon C, Tak H, Rho M, Chang HR, Kim YH, Kim KT, Balch C, Lee EK, Nam S. Detection of PIWI and piRNAs in the mitochondria of mammalian cancer cells. Biochem Biophys Res Commun. 2014;446:218–223. doi: 10.1016/j.bbrc.2014.02.112. [DOI] [PubMed] [Google Scholar]
- 14.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 15.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki R, Honda S, Kirino Y. PIWI expression and function in cancer. Front Genet. 2012;3:204. doi: 10.3389/fgene.2012.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. Elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y, Li C, Zhang K, Sun H, Tao D, Liu Y, Zhang S, Ma Y. Identification of piRNAs in Hela cells by massive parallel sequencing. BMB Rep. 2010;43:635–641. doi: 10.5483/BMBRep.2010.43.9.635. [DOI] [PubMed] [Google Scholar]
- 19.Chu H, Hui G, Yuan L, Shi D, Wang Y, Du M, Zhong D, Ma L, Tong N, Qin C, et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356:561–567. doi: 10.1016/j.canlet.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 21.Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. PiR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12–17. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15:563–568. doi: 10.1007/s12094-012-0966-0. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Ren Y, Xu H, Pang D, Duan C, Liu C. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg Oncol. 2013;22:217–223. doi: 10.1016/j.suronc.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, Lung RW, Chan AW, Chan TF, Wong N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58:1165–1173. doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 25.Cui L, Lou Y, Zhang X, Zhou H, Deng H, Song H, Yu X, Xiao B, Wang W, Guo J. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem. 2011;44:1050–1057. doi: 10.1016/j.clinbiochem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 26.UyBico SJ, Wu CC, Suh RD, Le NH, Brown K, Krishnam MS. Lung Cancer Staging Es-sentials: The new TNM staging system and potential imaging pitfalls. Radiographics. 2010;30:1163–1181. doi: 10.1148/rg.305095166. [DOI] [PubMed] [Google Scholar]
- 27.Shin MH, He Y, Marrogi E, Piperdi S, Ren L, Khanna C, Gorlick R, Liu C, Huang J. A RUNX2-mediated epigenetic regulation of the survival of p53 defective cancer cells. PLoS Genet. 2016;12:e1005884. doi: 10.1371/journal.pgen.1005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gherman C, Braicu OL, Zanoaga O, Jurj A, Pileczki V, Maralani M, Drigla F, Braicu C, Budisan L, Achimas-Cadariu P, Berindan-Neagoe I. Caffeic acid phenethyl ester activates pro-apoptotic and epithelial-mesenchymal transition-related genes in ovarian cancer cells A2780 and A2780cis. Mol Cell Biochem. 2016;413:189–198. doi: 10.1007/s11010-015-2652-3. [DOI] [PubMed] [Google Scholar]
- 29.Xu Z, Chen J, Shao L, Ma W, Xu D. Erratum to: Promyelocytic leukemia protein enhances apoptosis of gastric cancer cells through Yes-associated protein. Tumour Biol. 2016;37:2775. doi: 10.1007/s13277-015-4658-6. [DOI] [PubMed] [Google Scholar]
- 30.Malki A, Mohsen M, Aziz H, Rizk O, Shaban O, El-Sayed M, Sherif ZA, Ashour H. New 3-Cyano-2-substituted pyridines induce apoptosis in MCF 7 breast cancer cells. Molecules. 2016;21:E230. doi: 10.3390/molecules21020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamelos VA, Fisher N, Bamrah H, Voisey C, Price JC, Farrell WE, Redman CW, Richardson A. The BH3 mimetic obatoclax accumulates in lysosomes and causes their alkalinization. PLoS One. 2016;11:e0150696. doi: 10.1371/journal.pone.0150696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang JS, Su CY, Yu WH, Lee WJ, Liu YP, Lai TC, Jan YH, Yang YF, Shen CN, Shew JY, et al. GIT1 promotes lung cancer cell metastasis through modulating Rac1/Cdc42 activity and is associated with poor prognosis. Oncotarget. 2015;6:36278–36291. doi: 10.18632/oncotarget.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cryns V, Yuan J. Proteases to die for. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 34.Amar SK, Goyal S, Mujtaba SF, Dwivedi A, Kushwaha HN, Verma A, Chopra D, Chaturvedi RK, Ray RS. Role of type I and type II reactions in DNA damage and activation of caspase3 via mitochondrial pathway induced by photosensitized benzophenone. Toxicol Lett. 2015;235:84–95. doi: 10.1016/j.toxlet.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Hamacher-Brady A, Stein HA, Turschner S, Toegel I, Mora R, Jennewein N, Efferth T, Eils R, Brady NR. Artesunate activates mitochondrial apoptosis in breast cancer cells via iron-catalyzed lysosomal reactive oxygen species production. J Biol Chem. 2011;286:6587–6601. doi: 10.1074/jbc.M110.210047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Germanov E, Berman JN, Guernsey DL. Current and future approaches for the therapeutic targeting of metastasis. Int J Mol Med. 2006;18:1025–1036. [PubMed] [Google Scholar]
- 37.You J, He X, Ding H, Zhang T. BRMS1 regulates apoptosis in non-small cell lung cancer cells. Cell Biochem Biophys. 2015;71:465–472. doi: 10.1007/s12013-014-0226-8. [DOI] [PubMed] [Google Scholar]