Abstract
The second messenger, bis-(3′,5′)-cyclic dimeric guanosine monophosphate (cyclic di-GMP), is involved in the control of multiple bacterial phenotypes, including those that impact host–pathogen interactions. Bioinformatics analyses predicted that Mycobacterium leprae, an obligate intracellular bacterium and the causative agent of leprosy, encodes three active diguanylate cyclases. In contrast, the related pathogen Mycobacterium tuberculosis encodes only a single diguanylate cyclase. One of the M. leprae unique diguanylate cyclases (ML1419c) was previously shown to be produced early during the course of leprosy. Thus, functional analysis of ML1419c was performed. The gene encoding ML1419c was cloned and expressed in Pseudomonas aeruginosa PAO1 to allow for assessment of cyclic di-GMP production and cyclic di-GMP-mediated phenotypes. Phenotypic studies revealed that ml1419c expression altered colony morphology, motility and biofilm formation of P. aeruginosa PAO1 in a manner consistent with increased cyclic di-GMP production. Direct measurement of cyclic di-GMP levels by liquid chromatography–mass spectrometry confirmed that ml1419c expression increased cyclic di-GMP production in P. aeruginosa PAO1 cultures in comparison to the vector control. The observed phenotypes and increased levels of cyclic di-GMP detected in P. aeruginosa expressing ml1419c could be abrogated by mutation of the active site in ML1419c. These studies demonstrated that ML1419c of M. leprae functions as diguanylate cyclase to synthesize cyclic di-GMP. Thus, this protein was renamed DgcA (Diguanylate cyclase A). These results also demonstrated the ability to use P. aeruginosa as a heterologous host for characterizing the function of proteins involved in the cyclic di-GMP pathway of a pathogen refractory to in vitro growth, M. leprae.
Keywords: Mycobacterium leprae, diguanylate cyclase, Cyclic di-GMP, second messenger, gene function
Introduction
The bacterial metabolite bis-(3′,5′)-cyclic dimeric guanosine monophosphate (cyclic di-GMP) serves as a signal-transducing second messenger (Römling et al., 2013). Cyclic di-GMP has been described in a broad range of bacterial species and is associated with regulation of a variety of bacterial activities including survival (Bharati et al., 2012), virulence (Kulasakara et al., 2006), cell differentiation (Abel et al., 2011) and biofilm formation (Hickman et al., 2005; Irie et al., 2012; Borlee et al., 2010). Pathogen-produced cyclic di-GMP has also been found to stimulate the host innate immune response (Karaolis et al., 2007). The formation of cyclic di-GMP is catalysed via diguanylate cyclase (DGC), an enzyme that utilizes GTP as the substrate and possesses a conserved GG(D/E)EF motif that is part of the active-site domain (Chan et al., 2004; Wassmann et al., 2007). Approximately 50% of proteins with characterized DGC activity (Römling et al., 2013) are regulated via allosteric control and the binding of cyclic di-GMP to a conserved inhibitory site (I-site) motif (RxxD) directly upstream of the GGDEF motif (Christen et al., 2006; Wassmann et al., 2007). Phosphodiesterase (PDE) proteins that possess EAL or HD-GYP domains are responsible for the depletion of cyclic di-GMP and conversion into 5′-phosphoguanylyl-(3′,5′)-guanosine (pGpG) or two molecules of guanosine monophosphate (GMP), respectively (Ryan et al., 2006; Schmidt et al., 2005). Recently, oligoribonuclease (Orn) has also been identified to participate in the conversion of pGpG into GMP in Pseudomonas aeruginosa (Cohen et al., 2015; Orr et al., 2015). The activities of GGDEF and EAL domains can be incorporated into a single bifunctional protein or separated on individual proteins. More importantly, sensory input domains typically located in the N-terminus of proteins containing DGC or PDE domains receive environmental signals that activate the DGC or PDE activity (Römling et al., 2013; Sondermann et al., 2012). The potential combinatorial diversity of various sensory input domains coupled to individual DGC or PDE activity or bifunctional proteins allows organisms such as Pseudomonas spp. to utilize cyclic di-GMP in the regulation of multiple activities such as biofilm formation, motility and virulence (Hickman et al., 2005; Kulasakara et al., 2006; Merritt et al., 2007). The genome of P. aeruginosa PAO1 encodes 41 proteins predicted to metabolize intracellular cyclic-di-GMP levels. This includes 17 GGDEF domain-containing proteins, 5 EAL domain-containing proteins, 16 proteins that contain composite GGDEF-EAL domains and 3 proteins with HD-GYP domains (Kulasakara et al., 2006; Ryan et al., 2009; Römling et al., 2013; Mills et al., 2011).
In mycobacteria, the production and regulatory activity of cyclic di-GMP has been investigated in Mycobacterium tuberculosis, the causative agent of tuberculosis (Gengenbacher & Kaufmann, 2012). This bacterium encodes a single bifunctional DGC-PDE protein (Rv1354c) and both domains have been shown to be active (Gupta et al., 2010). Disruption of Rv1354c PDE activity decreases the pathogenicity and dormancy in M. tuberculosis (Hong et al., 2013). In addition, studies of cyclic di-GMP PDE activity of Rv1357c in the closely related Mycobacterium bovis BCG Pasteur 1173P2 demonstrated that cyclic di-GMP was associated with the regulation of lipid production and pellicle growth and promoted resistance to nitrosative stress (Flores-Valdez et al., 2015). Protein interaction studies have also suggested that cyclic di-GMP production in M. tuberculosis is involved in regulation of rhamnose biosynthesis, a key sugar in the formation of the mycobacterial cell wall (Deng et al., 2014). Moreover, studies of the Mycobacterium smegmatis homologue of rv1354c demonstrated that the cyclic di-GMP was involved in colony morphology and long-term survival during nutrient starvation (Sharma et al., 2014; Bharati et al., 2012; Gupta et al., 2015).
The production and regulatory role of cyclic di-GMP is unknown in Mycobacterium leprae, which is an acid-fast bacterium and the causative agent of leprosy or Hansen’s disease. Leprosy remains a public health concern in several low- and middle-income countries, with over 200 000 new cases reported each year (WHO, 2014). M. leprae cannot be cultured in vitro and is characterized by a degenerative genome that possesses a large number of pseudogenes (Cole et al., 2001). Nevertheless, this bacterium has the ability to adapt and survive within different intracellular environments of its human host. This includes infection of the upper respiratory tract, skin and peripheral nerves (Walker & Lockwood, 2007). More importantly, M. leprae can maintain its viability across a spectrum of disease pathology defined by two poles: tuberculoid leprosy that is typified by a dominant Th1 immune response and paucibacillary disease and lepromatous leprosy that presents with a high bacterial load and a nonprotective but robust Th2 immune response (Ridley & Jopling, 1966; Walker & Lockwood, 2007). The chronic nature of leprosy and the ability of M. leprae to adapt to various host environments is contradictory to a bacterium with a degenerative genome, as well as a reduced number of transcription factors and regulatory proteins (Cole et al., 2001). Bioinformatics analyses of the annotated M. leprae genome revealed three putative DGC proteins, which all retain the conserved GGDEF domain. One of the putative DGC coding sequences of M. leprae, ML1419c, was previously shown to be expressed when M. leprae was experimentally infected in the mouse footpad (Williams et al., 2009). More importantly, immunological studies revealed a strong antigen-specific T cell response to peptides of ML1419c in paucibacillary patients and in the household contacts of multibacillary patients (Spencer et al., 2005). This demonstrated that M. leprae produces ML1419c in the early stages of leprosy; however, the physiological function of this protein was unknown.
The inability to culture M. leprae in vitro or subject it to genetic manipulation is a major impediment in characterizing the physiological function of M. leprae proteins. To overcome these limitations, we chose to express ml1419c in an alternative heterologous host, P. aeruginosa PAO1, in which the effects of increasing and decreasing levels of cyclic di-GMP are well documented and control a range of phenotypes (Borlee et al., 2010; Hickman & Harwood, 2008; Hickman et al., 2005; Irie et al., 2012; Kulasakara et al., 2006; Merritt et al., 2007; Lee et al., 2007; Mills et al., 2011). Previously published studies have specifically shown that overexpression of the DGC encoded by tpbB (PA1120) increases cyclic di-GMP concentrations and corresponding biofilm formation in P. aeruginosa (Hickman & Harwood, 2008; Kulasakara et al., 2006). This study demonstrated the DGC activity of ML1419c from M. leprae in P. aeruginosa and provides strong evidence that M. leprae is capable of producing the second messenger cyclic di-GMP.
Methods
Bacterial strains, genomic DNA and growth conditions.
P. aeruginosa PAO1 and recombinant strains were grown at 37 °C in either Lennox LB medium or Vogel–Bonner minimal medium (VBMM) with l-arabinose (Gold Biotechnology) for inducible expression experiments. Escherichia coli strain BL21(DE3) (Invitrogen) was grown in Miller LB medium at 37 °C. Gentamicin (100 µg ml−1) (Gold Biotechnology) was used for selection of recombinant P. aeruginosa strains. Kanamycin (50 µg ml−1) (Sigma Aldrich) and gentamicin (10 µg ml−1) were used for selection of recombinant E. coli strains. Genomic DNA from M. leprae strains NHDP63 and Thai-53 was obtained from Biodefense and Emerging Infections Research Resources Repository.
Construction of plasmids and site-directed mutagenesis of ml1419c.
Heterologous gene expression in P. aeruginosa PAO1 was accomplished by using the arabinose-inducible vector pJN105 containing PBAD promoter (Newman & Fuqua, 1999). M. leprae ml1419c was amplified by PCR from genomic DNA of M. leprae with Q5 high-fidelity DNA polymerase (New England BioLabs), the forward primer 5′-GGAATTCGAGGAGGATATTCGTGTTGGAGACGGTGCGTAG-3′ and the reverse primer 5′-GGACTAGTTCAGCTAGGTTGTTGGTTGAACGTG-3′. Underlined sequences represent EcoRI and SpeI sites in the forward and reverse primers, respectively. Boldfaced sequence represents inclusion of an optimized ribosome binding site (Wolfgang et al., 2003). The 1692-bp ml1419c fragment was cloned into pJN105 (Newman & Fuqua, 1999) using the EcoRI and SpeI sites to generate pMRLB105.
To express ml1419c in E. coli for protein purification, we performed PCR amplification using M. leprae genomic DNA, as well as the forward primer 5′-AAACATATGTTGGAGACGGTGCGTAGCG-3′ and the reverse primer 5′-TTAAGCTTGCTAGGTTGTTGGTTGAACG-3′. Underlined sequences represent the NdeI and HindIII sites, respectively. The resulting PCR product was cloned into the expression vector pET-28a (+) at NdeI and HindIII sites to generate pMRLB109.
Site-directed mutagenesis of ml1419c was accomplished with the QuikChange Lightning Site-Directed Mutagenesis (Agilent Technologies) following the manufacturer’s recommendations. The forward primer 5′-GTGGTGGGTAGGTTCGTCGCTCTGATCCTG-3′ and the reverse primer 5′-CAGGATCAGAGCGACGAACCTACCCACCAC-3′ were used to generate ML1419c sequences encoding proteins with a deletion of the 472GGDEF476 motif (ML1419cΔGGDEF). The ml1419cΔGGDEF construct was digested with EcoRI and SpeI endonucleases and cloned into the pJN105 to generate pMRLB108. The plasmid pJN1120 (Hickman & Harwood, 2008) that expresses tpbB (PA1120), a well-characterized DGC from P. aeruginosa, was used as a positive control (Kulasakara et al., 2006; Ueda & Wood, 2009) and pJN105 was used as a negative control. All constructs were confirmed by nucleotide sequencing. Plasmids used in this study are shown in Table S1 (available in the online Supplementary Material).
Protein and whole cell lysate isolation.
Recombinant ML1419c was purified from E. coli BL21(DE3). E. coli transformed with pMRLB109 was cultured to an OD600 of ~0.4–0.6 and induced with 0.5 mM IPTG (EMD Millipore) for 3 h. Cells were lysed using an ultrasonic processor (Vibra-cell VC750) with an amplitude setting of 30 % with six 20 s pulses and a 59 s pause between the pulse cycles in lysis buffer [PBS (pH 7.4) with 1× proteinase inhibitor (Roche), 50 µg ml−1 DNase I (Sigma Aldrich) and 10 µg ml−1 RNase A (Sigma Aldrich)]. The majority of the E. coli recombinant protein was produced as inclusion bodies. Thus, protein inclusion bodies were collected by centrifugation and suspended in binding buffer [50 mM Tris/HCl (pH 8.0), 300 mM NaCl, 10 mM imidazole and 8 M urea]. Protein purification was achieved by immobilized metal affinity chromatography with Ni-NTA agarose resin (Qiagen). The purified protein was eluted with binding buffer containing 150 mM imidazole and was dialysed against 50 mM Tris/HCl (pH 8.0) and 150 mM NaCl with a gradual reduction of urea. Protein concentrations were determined using the bicinchoninic acid (BCA) assay (Smith et al., 1985). The purified recombinant ML1419c was provided to Lampire Biological Laboratories for the production of rabbit anti-ML1419c polyclonal serum.
P. aeruginosa PAO1 was transformed with pMRLB105, pMRLB108, pJN1120 or pJN105 by electroporation (Choi et al., 2006). Overnight cultures of P. aeruginosa strains were diluted (1 : 100) into fresh Lennox LB broth containing 100 µg ml−1 gentamicin and 0.2 % l-arabinose and grown to log-phase (OD600 of ~0.6–0.7). Cells were collected by centrifugation and suspended in lysis buffer and lysed as described for E. coli. The soluble proteins were isolated from whole cell lysates by centrifugation at 14 000 g 4 °C for 15 min. Protein concentrations were determined using the BCA assay.
SDS-PAGE and Western blot analysis.
Purified E. coli recombinant ML1419c (500 ng or 25 ng), P. aeruginosa whole cell lysates (5 µg), soluble fractions (5 µg) and insoluble fractions of proteins (5 µg) were resolved under denaturing conditions on NuPAGE 4–12 % Bistris polyacrylamide gels (Invitrogen). Proteins were visualized by staining with Coomassie G-250 stain (Invitrogen). For Western blots, proteins were transferred to PVDF membranes and probed with rabbit anti-ML1419c polyclonal serum (1 : 200 000). The secondary antibody was anti-rabbit IgG conjugated to HRP (1 : 20 000) (Promega). Chemiluminescence was used to visualize reactive proteins by incubating membrane with luminol-based enhanced chemiluminescence HRP substrate (Thermo Scientific).
Phenotypic assays.
Colony morphology and dye binding: P. aeruginosa strains were grown on VBMM agar containing 40 µg ml−1 Congo red (Sigma Aldrich), 15 µg ml−1 Coomassie brilliant blue (Sigma Aldrich) and 100 µg ml−1 gentamicin and in the presence or absence of 1 % l-arabinose at 30 °C for 48 h. The colony morphology was observed under a Leica MZ9.5 stereomicroscope (Leica Microsystems).
Motility assays: Swimming and twitching motility were assessed as previously described (Déziel et al., 2001; Darzins, 1993) with minor modifications. Swimming motility was assayed by stab inoculating 1 µl of a P. aeruginosa overnight culture into low-viscosity Lennox LB agar (0.3 % Bacto agar) containing with 100 µg ml−1 gentamicin and 0.2 % l-arabinose. The diameters of the swimming zone were measured after growth at 37 °C for 24 h. Twitching motility was assessed by stab inoculating a colony of P. aeruginosa through Lennox LB agar containing 100 µg ml−1 gentamicin and 0.2 % l-arabinose and cultured at 37 °C for 48 h. The migration of bacteria attached on the polystyrene plate surface was visualized by staining with 0.1 % crystal violet and the diameters were measured.
Biofilm formation: Biofilm formation was assessed as previously described (O'Toole, 2011) with modifications. The log-phase P. aeruginosa cultures (OD600 of ~0.6–0.7) were adjusted to an OD600 of ~0.1 in VBMM containing 100 µg ml−1 gentamicin and 0.2 % l-arabinose. Aliquots (150 µl) of diluted cultures were added to 96-well polystyrene plates (Nunc Microwell 96-well microplates (#243656), Thermo Scientific) in replicates of six and incubated at 37 °C in a sealed bag for 24 h. The 96-well plates were washed twice with water, stained with 0.1 % crystal violet for 10 min and washed twice with water. Bound crystal violet was solubilized with 30 % acetic acid and the absorbance was measured at 590 nm.
Quantitative analysis of cyclic di-GMP by liquid chromatography–mass spectrometry (LC-MS).
Overnight cultures of P. aeruginosa were diluted with VBMM (1 : 100) containing 100 µg ml–1 gentamicin and 0.2 % l-arabinose. An aliquot (2 ml) of P. aeruginosa culture grown to OD600 of ~0.6–0.7 was extracted with 100 µl of 0.6 M (final concentration) perchloric acid (Sigma Aldrich) (Hickman & Harwood, 2008) spiked with 100 nM [13C]adenosine (Omicron Biochemicals). The precipitate of the perchloric acid extraction was used for determination of protein concentration by the BCA assay and sample normalization (Irie & Parsek, 2014) and the extracts were neutralized with 20 µl of 2.5 M potassium bicarbonate. The neutralized supernatants were stored at −80 °C.
Cyclic di-GMP extracts (10 µl) were applied to an Agilent 1200 HPLC system coupled to an Agilent 6520 quadrupole time-of-flight (Q-TOF) mass spectrometer. Samples were resolved on an Atlantis T3 C18 column (3 µm particle size, 2.1×150 mm, Waters) at a flow rate of 350 µl min−1 at 30 °C. The gradient consisted of 100 % solvent A (0.1 % acetic acid, 10 mM ammonium acetate in water) for 1 min followed by a 1 min linear gradient to 10 % solvent B (methanol), a 1.4 min linear gradient to 20 % buffer B and a 1 min linear gradient to 100 % buffer B and held for 1 min. The Q-TOF mass spectrometer was operated in positive ion mode at 2 GHz extended dynamic range, an m/z range from 100 to 1700 and at a scan rate of one spectrum per second. MS data were collected in profile and centroid mode. Electrospray ionization source parameters were 2.5 kV, 350 °C gas temperature, drying gas flow rate of 11 l min–1 and a nebulizer flow rate of 45 psi. The cyclic di-GMP peak was confirmed by LC-tandem mass spectrometry (MS/MS) using collision energy of 19.8 eV. The cyclic di-GMP standard was purchased from BIOLOG Life Science Institute. The relative abundance of cyclic di-GMP was calculated as cyclic di-GMP peak area normalized to total protein (mg) and divided by the peak area of the [13C]adenosine internal standard.
Statistical analysis.
P values were calculated by one-way ANOVA followed by Tukey comparison using GraphPad Prism version 6.0 (GraphPad Software). Data were expressed as mean values±sd. The P value <0.05 was considered as statistically significant.
Bioinformatics analyses.
The annotated proteins containing either GGDEF, EAL or HD-GYP domains of M. leprae strain TN were identified from the NCBI Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (Marchler-Bauer et al., 2015) using the following domain accession numbers: GGDEF, cd01949; EAL, cd01948; and HD-GYP, cd00077 and SMART database (http://smart.embl-heidelberg.de/) (Letunic et al., 2015) using the following domain accession numbers: GGDEF, SM00267; EAL, SM00052; and HD-GYP, SM00471. M. leprae proteins containing GGDEF, EAL or HD-GYP motifs that were identified in both databases were used for further analyses. ML1419c haem-binding sites were identified from the NCBI CDD. Alignment of M. leprae strain TN protein sequences possessing GGDEF and/or EAL motifs was performed with the T-Coffee alignment tool (Di Tommaso et al., 2011). Evaluation of putative transmembrane domains was performed with TMHMM program (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al., 2001).
Results
Bioinformatics analyses and identification of cyclic di-GMP-related proteins in M. leprae
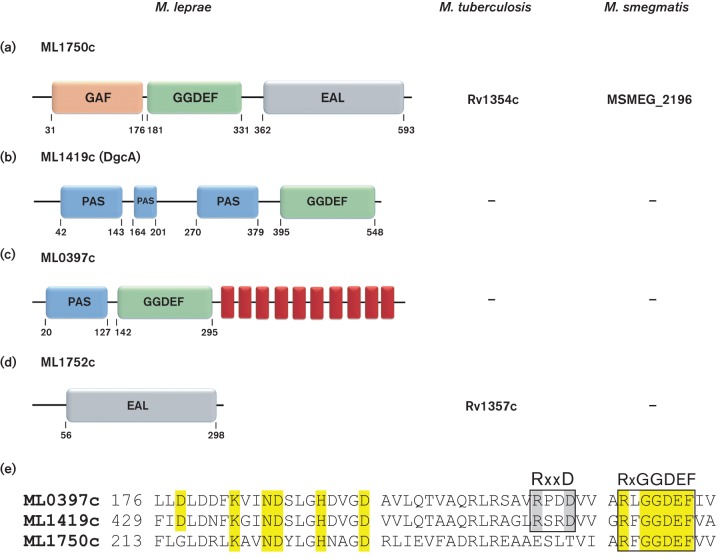
Identification of putative DGC proteins (GGDEF) or cyclic di-GMP PDE proteins (EAL or HD-GYP) encoded in the genome of M. leprae strain TN was achieved by interrogation of the NCBI CDD and the SMART databases. Bioinformatics analyses revealed that M. leprae harbours one putative hybrid DGC-PDE protein (ML1750c), two putative DGC proteins (ML1419c and ML0397c) and one putative PDE protein with an EAL domain (ML1752c) (Fig. 1). No proteins with a HD-GYP motif were identified. Results from these bioinformatics analyses of M. leprae strain TN agree with the large-scale census of cyclic di-GMP-related proteins (Römling et al., 2013).
Fig. 1.
Bioinformatics analyses of putative DGC and PDE of M. leprae. (a) ML1750c (623 aa) (gi|15827936|NP_302199) is a hybrid protein containing both GGDEF and EAL motifs and an N-terminal GAF sensory domain. (b) ML1419c (563 aa) (gi|15827746|NP_302009) contains a GGDEF motif and three consecutive PAS sensory domains upstream to GGDEF domain. (c) ML0397c (602 aa) (gi|15827122|NP_301385) possesses a GGDEF motif, an N-terminal PAS sensor domain and 10 transmembrane α-helices (red rectangles). (d) ML1752c (302 aa) (gi|15827938|NP_302201) has a single EAL motif and lacks a sensory domain. Homologues of ML1750c are produced in both M. tuberculosis and M. smegmatis and a homologue of ML1752c is identified in M. tuberculosis. Numbers indicate amino acid positions as reported by CDD NCBI. (e) Alignment of conserved DGC domains of M. leprae proteins ML0397c, ML1419c and ML1750c. The conserved I-site, RxxD motif of ML0397c and ML1419c is highlighted in grey. The conserved A-site, RxGGDEF motif, is present in all proteins. Conserved amino acids involved in enzymatic activity are highlighted in yellow.
The hybrid DGC-PDE protein ML1750c possesses an N-terminal GAF sensor domain, as well as GGDEF and EAL domains (Fig. 1a). This protein is homologous to M. smegmatis MSMEG_2196 with 64.39 % identity and M. tuberculosis Rv1354c with 62.30 % identity, both of which were experimentally defined as possessing active GGDEF and EAL domains (Gupta et al., 2010; Bharati et al., 2012; Hong et al., 2013). Unexpectedly, M. leprae with its reduced genome encoded two additional DGC proteins, ML1419c and ML0397c (Fig. 1b, c), that are not encoded by M. smegmatis or M. tuberculosis. ML1419c possesses three sequential PAS-signalling domains N-terminal to the GGDEF domain. Two predicted haem-binding sites were also identified within the N-terminal region of the ML1419c PAS-signalling domains. ML0397c harbours a single N-terminal PAS sensor domain linked to a GGDEF domain and 10 C-terminal transmembrane domains as predicted by analysis with the TMHMM program. A single predicted PDE protein (ML1752c) is homologous to Rv1357c of M. tuberculosis (Gupta et al., 2010; Römling et al., 2013), but it has no homology to proteins encoded by M. smegmatis (Fig. 1d).
Multiple alignments of the M. leprae proteins containing GGDEF and EAL domains (Figs 1e and S1a) were performed with T-Coffee (Di Tommaso et al., 2011). This demonstrated conservation of key amino acids in putative active sites. The three predicted DGCs of M. leprae (ML1750c, ML1419c and ML0397c) all contain the A-site sequence, RxGGDEF (Ryjenkov et al., 2005). Thus, these proteins are expected to act as functional DGCs producing cyclic di-GMP. In addition, ML1419c and ML0397c possess an I-site, RxxD motif (Wassmann et al., 2007), that is located directly upstream of the A-site. Multiple alignment analysis for the proteins with predicted PDE activity and EAL domains revealed that the conserved residues of the EAL active site are present in ML1750c and ML1752c. This conservation included appropriately spaced residues of E, N, E, E, D, K and E, except for the last E residue of ML1752c that is replaced with a K residue (Römling et al., 2013) (Fig. S1b). These in silico data indicate that M. leprae has a greater capacity than M. tuberculosis or M. smegmatis for cyclic di-GMP production. Based on immunological data, one of the putative DGCs (ML1419c) is known to be produced in vivo by M. leprae (Spencer et al., 2005; Williams et al., 2009), and thus, it was selected for further assessment of DGC activity.
Conditionalexpression of ml1419c in P. aeruginosa PAO1
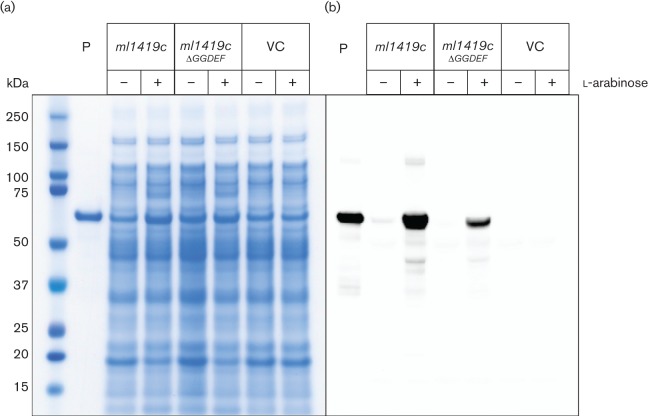
M. leprae cannot be cultured in vitro; thus, protein functions for this bacterium are typically studied in model organisms (Monot et al., 2009). The production and function of cyclic di-GMP has been extensively studied in P. aeruginosa, where phenotypes and mutants associated with this second messenger molecule are well described. In this study, ml1419c and a mutated construct of this gene, ml1419cΔGGDEF, were conditionally expressed in P. aeruginosa PAO1 under the control of the l-arabinose-responsive PBAD promoter (Newman & Fuqua, 1999). Recombinant protein production was assessed by SDS-PAGE and Western blot of whole cell lysates of P. aeruginosa strains (Fig. 2). Protein of the expected size (approximately 61 kDa) that reacted with anti-ML1419c polyclonal serum was observed in the whole cell lysates of P. aeruginosa expressing ml1419c or ml1419cΔGGDEF when grown in the presence of arabinose (Fig. 2b). Moreover, ml1419c and ml1419cΔGGDEF were expressed in P. aeruginosa and produced soluble protein (Fig. S2). In the absence of arabinose, no or low levels of recombinant protein production was observed. No products reactive to the anti-ML1419c polyclonal serum were observed for P. aeruginosa containing the pJN105 vector control regardless of the presence or absence of arabinose.
Fig. 2.
Expression of ml1419c from M. leprae in P. aeruginosa PAO1. Recombinant P. aeruginosa PAO1 containing ml1419c, ml1419cΔGGDEF or the pJN105 vector (VC) were grown in the presence (+) or in the absence (−) of 0.2 % l- arabinose. Whole cell lysates (5 µg) of the recombinant strains were analysed by SDS-PAGE with Coomassie blue staining (a) and Western blot (b). Purified recombinant ML1419c produced in E. coli was used as a positive control (lane P) for Coomassie blue staining (500 ng) and Western blot (25 ng).
ml1419c alters P. aeruginosa colony morphology
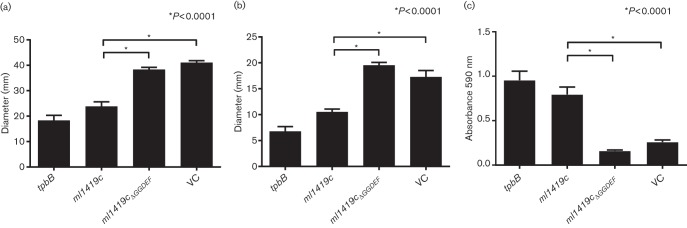
To provide an initial assessment of whether expression of ml1419c resulted in cyclic di-GMP production in P. aeruginosa PAO1, we investigated the colony morphology of recombinant P. aeruginosa strains on VBMM Congo red and brilliant blue agar plates (Fig. 3). P. aeruginosa PAO1 typically forms round colonies with smooth surfaces and regular borders; however, increased intracellular cyclic di-GMP levels induced formation of small colonies with wrinkly or rugose colony morphology and increased Congo red and brilliant blue binding that is correlated to the increase of exopolysaccharide production (Starkey et al., 2009; Hickman et al., 2005). The recombinant P. aeruginosa PAO1 conditionally expressing ml1419c in the presence of arabinose resulted in small and wrinkled colonies exhibiting rugose morphology. These colonies were similar in appearance to the positive control of P. aeruginosa expressing tpbB, a well-studied DGC. Colony morphology of recombinant P. aeruginosa PAO1 harbouring the arabinose-inducible ml1419c expression plasmid is similar to wild-type colony morphology when arabinose is omitted from the growth medium (Fig. S3). The first two glycine residues of the GGDEF motif confirmed DGCs participate in binding of the GTP substrate and the glutamic acid binds Mg2+ that is necessary for DGC activity (Chan et al., 2004; Wassmann et al., 2007). Thus, it was expected that alteration of P. aeruginosa colony morphology by ml1419c expression would be abrogated by an in-frame deletion of 472GGDEF476 (ml1419cΔGGDEF). P. aeruginosa expressing ml1419cΔGGDEF resulted in larger colonies that resembled the smooth colony morphology of wild-type P. aeruginosa PAO1 or vector control (Fig. 3). These data indicated that recombinant ML1419c functions as a DGC and the GGDEF domain of this protein is essential for this activity.
Fig. 3.
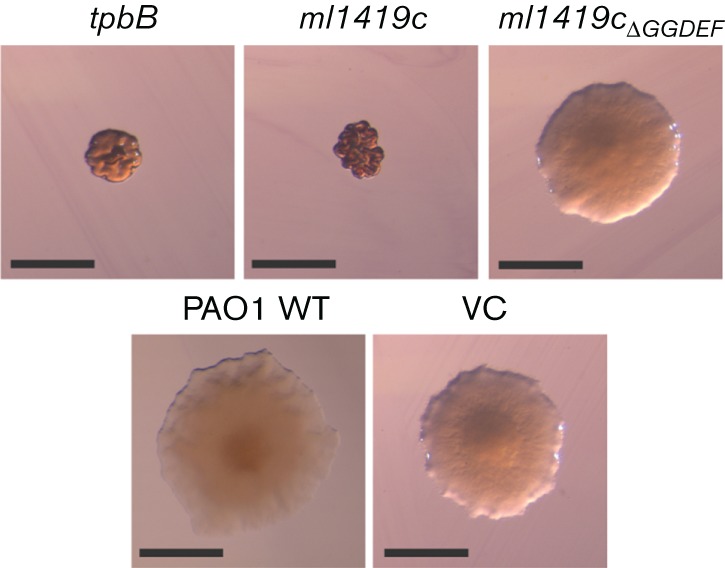
Colony morphology of P. aeruginosa PAO1 and recombinant strains. Strains were grown on VBMM agar containing Congo red, brilliant blue and 1 % l-arabinose. Rugose colonies were observed in P. aeruginosa expressing tpbB and ml1419c. PAO1 wild-type, P. aeruginosa expressing ml1419cΔGGDEF and the vector control (VC) form round colonies with smooth surfaces. Scale bar corresponds to 1 mm.
ml1419c expression provides for quantifiable phenotypic differences associated with DGC activity
To provide better assessment of potential cyclic di-GMP production by ML1419c and the impact of this production on P. aeruginosa, we measured several quantifiable phenotypes (twitching motility, swimming motility and biofilm formation). Swimming was assessed by quantitatively measuring the swim zone diameter formed by bacteria from the point of inoculation in low-viscosity agar (O'Toole & Kolter, 1998). In contrast, twitching motility was quantified by measuring the migration of bacteria that were inoculated at a single point between the agar and the polystyrene petri dish (Darzins, 1993; Déziel et al., 2001). Similar to P. aeruginosa expressing tpbB, the expression of ml1419c suppressed swimming and twitching motility (Figs 4a, b and S4a, b). This is consistent with increased production of cyclic di-GMP and corresponding inhibition of bacterial flagella and type IV pili function (Merritt et al., 2007; Simm et al., 2004). In concordance with decreased DGC activity, alteration of the GGDEF domain in ML1419c (ML1419cΔGGDEF) resulted in swimming and twitching motility that was similar to wild-type PAO1 and the vector control (Figs 4a, b and S4a, b). The suppression of the swimming and twitching phenotypes of P. aeruginosa expressing ml1419c was significant when compared to wild-type PAO1 and strains expressing the ΔGGDEF mutation in ml1419c (Fig. 4a, b). It was also noted that suppression of motility was only observed when P. aeruginosa expressing ml1419c was grown in the presence of arabinose.
Fig. 4.
P. aeruginosa expressing ml1419c suppresses motility and enhances biofilm formation. (a) Swimming and (b) twitching motility were suppressed in P. aeruginosa expressing tpbB and ml1419c as compared to the wild-type PAO1 strain and vector control (VC) in the presence of 0.2 % l-arabinose. Twitching and swimming motility were restored in P. aeruginosa expressing mutated ml1419c (ml1419cΔGGDEF). The diameters (mm) of (a) swim zones and (b) twitching zones were measured for four replicates of each strain and the mean was determined, *P<0.0001. (c) Biofilm formation as measured by crystal violet binding was increased in P. aeruginosa expressing tpbB and ml1419c. Biofilm formation was abrogated in P. aeruginosa expressing ml1419cΔGGDEF. The mean was determined from six replicates. *P<0.0001.
Another well-documented activity of cyclic di-GMP in P. aeruginosa is the induction of biofilm formation (Hickman et al., 2005; Lee et al., 2007). Elevated cellular levels of cyclic di-GMP increase the production of biofilm matrix components in P. aeruginosa including the Pel and Psl polysaccharides and a biofilm-associated adhesin (Hickman et al., 2005; Hengge, 2009; Borlee et al., 2010). Under conditions of arabinose induction, P. aeruginosa expressing ml1419c or tpbB produced significantly more biofilm as compared to wild-type PAO1 and the vector control (Fig. 4c). As observed with the other phenotypic assays, P. aeruginosa biofilm formation was significantly reduced when the GGDEF motif was deleted from ML1419c (ML1419cΔGGDEF) (Fig. 4c). The effect of ml1419c on multiple phenotypes of P. aeruginosa PAO1 associated with increased levels of cyclic di-GMP production provides strong evidence that the M. leprae ML1419c functions as a DGC. Additionally, the deletion of GGDEF motif of ML1419c reduced or eliminated ml1419c induction of phenotypes associated with elevated cellular levels of cyclic di-GMP.
Detection of cyclic di-GMP in vivo by LC–MS
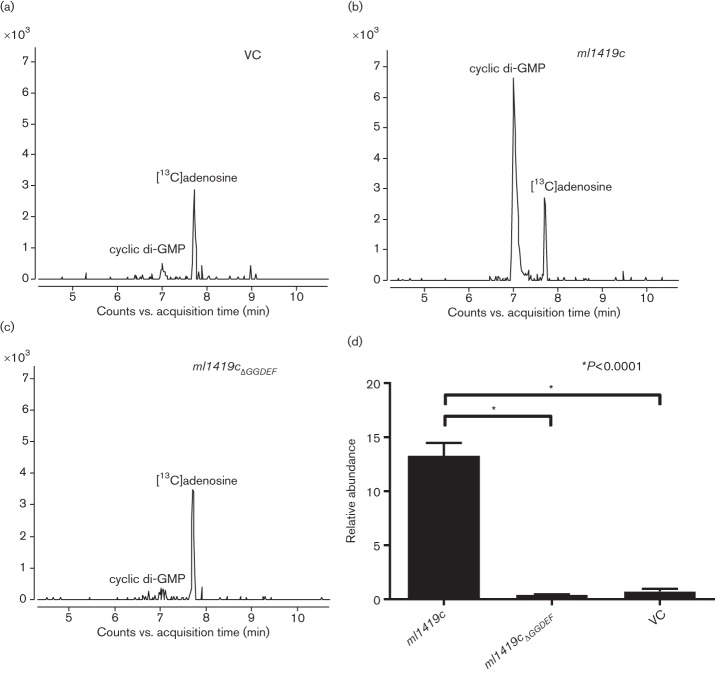
To directly assess the DGC function of ML1419c in the P. aeruginosa genetic background, we performed LC–MS to detect and measure the relative abundance of cyclic di-GMP in P. aeruginosa extracts. Initial analyses of cyclic di-GMP standard and [13C]adenosine (applied as an internal standard) demonstrated that these two products eluted with retention times of 7.061 and 7.702 min and yielded m/z values of 691.1021 and 269.1065, respectively. MS/MS fragmentation of cyclic di-GMP resulted in transition ions of m/z 152.0577, 248.0786 and 540.0566. These transition ions were used to confirm that the parent ion m/z 691.1021 represented cyclic di-GMP (Fig. S5).
LC–MS analyses of wild-type PAO1 and recombinant P. aeruginosa strains revealed the presence of cyclic di-GMP (Fig. 5a–c). The quantitative analyses of the relative cyclic di-GMP levels were based on the normalized peak area of cyclic di-GMP to [13C]adenosine. P. aeruginosa expressing ml1419c significantly increased the abundance (approximately ninefold) of cyclic di-GMP detected as compared to the vector control (Fig. 5d). However, the cyclic di-GMP abundance of P. aeruginosa expressing ml1419c was lower than that of P. aeruginosa expressing tpbB (Fig. S6). In comparison, there were no significant differences in cyclic di-GMP abundance between the vector control and P. aeruginosa expressing ml1419cΔGGDEF. These data confirm that ML1419c of M. leprae functions to induce cyclic di-GMP synthesis in P. aeruginosa and that the GGDEF motif is part of the active-site domain as shown in other DGCs (Ryjenkov et al., 2005).
Fig. 5.
Cyclic di-GMP detection and relative quantification of cyclic di-GMP in recombinant P. aeruginosa strains. The LC–MS extracted ion chromatogram of cyclic di-GMP (m/z 691.102) in extracts of recombinant P. aeruginosa strains; (a) vector control (VC), (b) ml1419c and (c) ml1419cΔGGDEF. [13C]Adenosine was applied as an internal standard (retention time 7.702 min, m/z 269.1065). (d) The relative quantification demonstrated a significant increase in the abundance of cyclic di-GMP produced in P. aeruginosa expressing ml1419c as compared to VC and P. aeruginosa expressing ml1419cΔGGDEF, *P<0.0001. Experiments were performed with three biological and three technical replicates.
Discussion
This current study demonstrated that ML1419c encoded on the genome of M. leprae possesses functional DGC activity resulting in the synthesis of cyclic di-GMP that can be measured by phenotypic or analytical assays. Thus, the M. leprae ML1419c protein was renamed as DgcA (Diguanylate cyclase A). Given the inability to grow or manipulate M. leprae in vitro, the use of P. aeruginosa as a heterologous expression host was critical for demonstrating the enzymatic activity of DgcA. In fact, the recombinant expression of the M. leprae dgcA was able to mimic the same phenotypes in P. aeruginosa that were induced by overexpression of a well-characterized P. aeruginosa encoded DGC, tpbB (Kulasakara et al., 2006; Ueda & Wood, 2009). DgcA from M. leprae possesses conserved DGC A-site (RFGGDEF) and I-site (RSRD) motifs. The consensus sequence of the A-site motif, RxGG(D/E)EF, has been well studied and is known to participate in protein dimerization, as well as substrate binding and catalytic activity (Wassmann et al., 2007; Chan et al., 2004). Thus, we hypothesized that deletion of the GGDEF domain would prevent enhanced production of cyclic di-GMP by recombinant ML1419c and the subsequent loss of P. aeruginosa phenotypes associated with increased DGC activity. Modification and expression of recombinant ML1419c, ml1419cΔGGDEF, resulted in decreased cyclic di-GMP levels as compared to that of P. aeruginosa expressing full-length ml1419c. Likewise, swimming and biofilm phenotypes associated with increased cyclic di-GMP production were altered.
Two of the DGC genes from M. leprae (dgcA and ml0397c) were previously found to be expressed during infection in animal models and leprosy patients (Williams et al., 2009; Spencer et al., 2005). These data indicate that, at various stages during infection, the M. leprae DGCs may play important roles in leprosy pathogenesis. The production of DgcA during the early stages of leprosy as described in previous studies (Williams et al., 2009; Spencer et al., 2005) and the ability of DgcA to induce a robust immune response (Geluk et al., 2011) indicate a potential role for this protein in the pathogen’s ability to sense and respond to environmental changes during the initial stages of infection. The putative sensing domain of DgcA from M. leprae possesses three PAS domains, and two of these PAS domains have conserved haem-binding sites. Thus, we hypothesize that DgcA likely responds to oxygen tension, nitric oxide and/or carbon monoxide (Henry & Crosson, 2011). Importantly, the environmental cues that are perceived by the PAS sensor domains of DgcA would be expected to alter downstream gene expression and protein function via the activity of DgcA-derived cyclic di-GMP. Previous research groups have overexpressed GGDEF proteins similar to DgcA in order to study protein activity in the absence of activating signal (Kulasakara et al., 2006; Hickman & Harwood, 2008), and it is likely that elevated protein levels facilitate the dimerization and activation of these DGCs (Hallberg et al., 2016). Thus, it is possible that the DGC activity demonstrated by the recombinant expression of M. leprae dgcA in P. aeruginosa was a result of protein abundance or interaction of the DgcA PAS domains with an environmental signal. Future structural studies of DgcA and targeted binding assays are required to define the ligands that bind to this protein and whether they induce or repress DGC activity.
Comparison of the genomes of the two primary mycobacterial pathogens, M. leprae and M. tuberculosis, reveals that M. leprae has a significantly smaller genome, a relatively large number of pseudogenes, fewer functional proteins and fewer transcription factors (Cole et al., 2001). Consequently, M. leprae is refractory to in vitro growth and has evolved into an obligate intracellular pathogen. Given the narrow biological niche of M. leprae, it is intriguing that this pathogen harbours three coding sequences for known or predicted DGCs (dgcA, ml0397c and ml1750c), whilst M. tuberculosis has only one protein, Rv1354c, a homologue of ML1750c. This same coding sequence is also the only DGC found in the genome of M. smegmatis (msmeg_2196) (Bharati et al., 2012), a nonpathogenic saprophyte commonly used as a model to define gene function of mycobacterial pathogens (Reyrat & Kahn, 2001; Singh & Reyrat, 2009). Rv1354c and MSMEG_2196 have been confirmed to have DGC and PDE activity, and phenotypes have been associated with rv1354c and msmeg_2196.
During the course of our studies, we expressed ml1419c in M. smegmatis, which does not encode an ml1419c homologue. However, we found this to be an inadequate heterologous host system to elucidate the biochemical function of ML1419C. Expression of ml1419c in M. smegmatis under the control of an hsp60 promoter produced recombinant protein, which subsequently altered the colony morphology, pellicle formation and sliding motility of M. smegmatis (data not shown). However, we were unable to reproducibly detect cyclic di-GMP production with a reasonable amount of cells in this system. The requirement for 1–3 g of mycobacterial cells to conduct analytical measurement of cyclic di-GMP in M. tuberculosis or M. smegmatis (Bharati et al., 2012; Hong et al., 2013) does not provide a robust system to correlate cyclic di-GMP levels with phenotypes. Thus, given the ambiguity of cyclic di-GMP production in M. smegmatis and the reported cyclic di-GMP phenotype(s) of this bacterium (Bharati et al., 2012; Gupta et al., 2015), future efforts will focus on defining the function and significance of DGC activity in M. leprae, using a model system that allows for genetic analyses of individual DGCs. Towards this goal, all of the known and predicted DGCs of M. leprae (Fig. 1) are encoded in the genomes of Mycobacterium lepromatosis (another bacterium restricted to in vivo growth) and Mycobacterium haemophilum (an opportunistic pathogen). Homologues of ML1419c and ML1750c are also encoded in several environmental actinobacteria such as Mycobacterium rhodesiae, Mycobacterium chubuense, Mycobacterium chlorophenolicum, Mycobacterium rufum and Rhodococcus fascians. Evolutionary reduction of the number cyclic di-GMP signalling pathways is generally believed to be inversely correlated with a bacterium’s need and ability to adapt to rapidly changing environmental conditions (Römling et al., 2013). Therefore, the conservation of DGC genes in M. leprae and the presence of these genes in opportunistic pathogens and several environmental Mycobacterium spp. is indicative of the potential role for DGCs in the signalling response required for M. leprae to survive as an obligate intracellular pathogen. Interestingly, like M. leprae, M. haemophilum is a pathogen of the skin and displays optimal growth at 30 °C (Sompolinsky et al., 1978). Recently, it has been reported that M. haemophilum can be genetically manipulated to express foreign genes (Tufariello et al., 2015), and efforts are now under way to use this Mycobacterium sp. as a model to further define the physiological functions and signalling events associated with DgcA and the additional predicted PDE and DGCs of M. leprae.
Acknowledgements
This work was supported by National Institutes of Health grants, National Institute of Allergy and Infectious Diseases grant R01-AI022553 and National Institute of Arthritis and Musculoskeletal and Skin Diseases grant R01-AR040312. We thank Grace Borlee and Ashli Simone for technical assistance and advice in the completion of this work.
Supplementary Data
Abbreviations:
- BCA
bicinchoninic acid
- CDD
Conserved Domain Database
- DGC
diguanylate cyclase
- LC
liquid chromatography
- PDE
phosphodiesterase
- Q-TOF
quadrupole time-of-flight
- VBMM
Vogel–Bonner minimal medium
References
- Abel S., Chien P., Wassmann P., Schirmer T., Kaever V., Laub M. T., Baker T. A., Jenal U.(2011). Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell 43550–560. 10.1016/j.molcel.2011.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharati B. K., Sharma I. M., Kasetty S., Kumar M., Mukherjee R., Chatterji D.(2012). A full-length bifunctional protein involved in c-di-GMP turnover is required for long-term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology 1581415–1427. 10.1099/mic.0.053892-0 [DOI] [PubMed] [Google Scholar]
- Borlee B. R., Goldman A. D., Murakami K., Samudrala R., Wozniak D. J., Parsek M. R.(2010). Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75827–842. 10.1111/j.1365-2958.2009.06991.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C., Paul R., Samoray D., Amiot N. C., Giese B., Jenal U., Schirmer T.(2004). Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A 10117084–17089. 10.1073/pnas.0406134101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. H., Kumar A., Schweizer H. P.(2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: Application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64391–397. 10.1016/j.mimet.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Christen B., Christen M., Paul R., Schmid F., Folcher M., Jenoe P., Meuwly M., Jenal U.(2006). Allosteric control of cyclic di-GMP signaling. J Biol Chem 28132015–32024. 10.1074/jbc.M603589200 [DOI] [PubMed] [Google Scholar]
- Cohen D., Mechold U., Nevenzal H., Yarmiyhu Y., Randall T. E., Bay D. C., Rich J. D., Parsek M. R., Kaever V., et al. (2015). Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 11211359–11364. 10.1073/pnas.1421450112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T., Eiglmeier K., Parkhill J., James K. D., Thomson N. R., Wheeler P. R., Honoré N., Garnier T., Churcher C., et al. (2001). Massive gene decay in the leprosy bacillus. Nature 4091007–1011. 10.1038/35059006 [DOI] [PubMed] [Google Scholar]
- Darzins A.(1993). The Pilg gene product, required for Pseudomonas aeruginosa pilus production and twitching motility, is homologous to the enteric, single-domain response regulator cheY. J Bacteriol 1755934–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Bi L., Zhou L., Guo S. J., Fleming J., Jiang H.-W., Zhou Y., Gu J., Zhong Q., et al. (2014). Mycobacterium tuberculosis proteome microarray for global studies of protein function and immunogenicity. Cell Rep 92317–2329. 10.1016/j.celrep.2014.11.023 [DOI] [PubMed] [Google Scholar]
- Déziel E., Comeau Y., Villemur R.(2001). Initiation of biofilm formation by Pseudomonas aeruginosa 57rp correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol 1831195–1204. 10.1128/JB.183.4.1195-1204.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J. M., Taly J. F., Notredame C.(2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res 39W13–W17. 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Valdez M. A., De Jesus Aceves-Sanchez M., Pedroza-Roldan C., Vega-Dominguez P. J., Prado-Montes De Oca E., Bravo-Madrigal J., Laval F., Daffe M., Koestler B., et al. (2015). The cyclic di-GMP phosphodiesterase gene Rv1357c/BCG1419c affects BCG pellicle production and in vivo maintenance. IUBMB Life 67129–138. [DOI] [PubMed] [Google Scholar]
- Geluk A., van den Eeden S. J., Dijkman K., Wilson L., Kim H. J., Franken K. L., Spencer J. S., Pessolani M. C., Pereira G. M., et al. (2011). ML1419c Peptide immunization induces Mycobacterium leprae-specific HLA-A*0201-restricted CTL Vivo with potential to kill live mycobacteria. J Immunol 1871393–1402. 10.4049/jimmunol.1100980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengenbacher M., Kaufmann S. H.(2012). Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev 36514–532. 10.1111/j.1574-6976.2012.00331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K., Kumar P., Chatterji D.(2010). Identification, activity and disulfide connectivity of c-di-GMP regulating proteins in Mycobacterium tuberculosis. PLoS One 5e15072. 10.1371/journal.pone.0015072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K. R., Kasetty S., Chatterji D.(2015). Novel functions of (p)PPGPP and cyclic di-GMP in mycobacterial physiology revealed by phenotype microarray analysis of wild-type and isogenic strains of mycobacterium smegmatis. Appl Environ Microbiol 812571–2578. 10.1128/AEM.03999-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg Z. F., Wang X. C., Wright T. A., Nan B., Ad O., Yeo J., Hammond M. C.(2016). Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3′, 3′-cGAMP). Proc Natl Acad Sci U S A 1131790–1795. 10.1073/pnas.1515287113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R.(2009). Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Henry J. T., Crosson S.(2011). Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol 65261–286. 10.1146/annurev-micro-121809-151631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman J. W., Tifrea D. F., Harwood C. S.(2005). A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci 10214422–14427. 10.1073/pnas.0507170102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman J. W., Harwood C. S.(2008). Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69376–389. 10.1111/j.1365-2958.2008.06281.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Zhou X., Fang H., Yu D., Li C., Sun B.(2013). Cyclic di-GMP mediates Mycobacterium tuberculosis dormancy and pathogenecity. Tuberculosis 93625–634. 10.1016/j.tube.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Irie Y., Borlee B. R., O'Connor J. R., Hill P. J., Harwood C. S., Wozniak D. J., Parsek M. R.(2012). Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 10920632–20636. 10.1073/pnas.1217993109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y., Parsek M. R.(2014). LC/MS/MS-based quantitative assay for the secondary messenger molecule, c-di-GMP. Methods Mol Biol 1149271–279. 10.1007/978-1-4939-0473-0_22 [DOI] [PubMed] [Google Scholar]
- Karaolis D. K., Means T. K., Yang D., Takahashi M., Yoshimura T., Muraille E., Philpott D., Schroeder J. T., Hyodo M., et al. (2007). Bacterial c-di-GMP is an immunostimulatory molecule. J Immunol 1782171–2181. 10.4049/jimmunol.178.4.2171 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L.(2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kulasakara H., Lee V., Brencic A., Liberati N., Urbach J., Miyata S., Lee D. G., Neely A. N., Hyodo M., et al. (2006). Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci 1032839–2844. 10.1073/pnas.0511090103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee V. T., Matewish J. M., Kessler J. L., Hyodo M., Hayakawa Y., Lory S.(2007). A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 651474–1484. 10.1111/j.1365-2958.2007.05879.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P.(2015). SMART: recent updates, new developments and status. Nucleic Acids Res 43D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res 43D222–D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J. H., Brothers K. M., Kuchma S. L., O'Toole G. A.(2007). SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol 1898154–8164. 10.1128/JB.00585-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E., Pultz I. S., Kulasekara H. D., Miller S. I.(2011). The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell Microbiol 131122–1129. 10.1111/j.1462-5822.2011.01619.x [DOI] [PubMed] [Google Scholar]
- Monot M., Honoré N., Garnier T., Zidane N., Sherafi D., Paniz-Mondolfi A., Matsuoka M., Taylor G. M., Donoghue H. D., et al. (2009). Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet 411282–1289. 10.1038/ng.477 [DOI] [PubMed] [Google Scholar]
- Newman J. R., Fuqua C.(1999). Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227197–203. 10.1016/S0378-1119(98)00601-5 [DOI] [PubMed] [Google Scholar]
- O'Toole G. A., Kolter R.(1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30295–304. 10.1046/j.1365-2958.1998.01062.x [DOI] [PubMed] [Google Scholar]
- O'Toole G. A.(2011). Microtiter dish biofilm formation assay. J Vis Exp, e2437 10.3791/2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr M. W., Donaldson G. P., Severin G. B., Wang J., Sintim H. O., Waters C. M., Lee V. T.(2015). Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci U S A 112E5048–E5057. 10.1073/pnas.1507245112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyrat J. M., Kahn D.(2001). Mycobacterium smegmatis: an absurd model for tuberculosis? Trends Microbiol 9472–473. 10.1016/S0966-842X(01)02168-0 [DOI] [PubMed] [Google Scholar]
- Ridley D. S., Jopling W. H.(1966). Classification of leprosy according to immunity. Int J Lepr Other Mycobact Dis 34255–273. [PubMed] [Google Scholar]
- Römling U., Galperin M. Y., Gomelsky M.(2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 771–52. 10.1128/MMBR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R. P., Fouhy Y., Lucey J. F., Crossman L. C., Spiro S., He Y. W., Zhang L. H., Heeb S., Cámara M., et al. (2006). Cell–cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A 1036712–6717. 10.1073/pnas.0600345103 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ryan R. P., Lucey J., O'Donovan K., McCarthy Y., Yang L., Tolker-Nielsen T., Dow J. M.(2009). HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ Microbiol 111126–1136. 10.1111/j.1462-2920.2008.01842.x [DOI] [PubMed] [Google Scholar]
- Ryjenkov D. A., Tarutina M., Moskvin O. V., Gomelsky M.(2005). Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 1871792–1798. 10.1128/JB.187.5.1792-1798.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A. J., Ryjenkov D. A., Gomelsky M.(2005). The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 1874774–4781. 10.1128/JB.187.14.4774-4781.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma I. M., Prakash S., Dhanaraman T., Chatterji D.(2014). Characterization of a dual-active enzyme, DcpA, involved in cyclic diguanosine monophosphate turnover in Mycobacterium smegmatis. Microbiology 1602304–2318. 10.1099/mic.0.080200-0 [DOI] [PubMed] [Google Scholar]
- Simm R., Morr M., Kader A., Nimtz M., Römling U.(2004). GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 531123–1134. 10.1111/j.1365-2958.2004.04206.x [DOI] [PubMed] [Google Scholar]
- Singh A. K., Reyrat J. M.(2009). Laboratory maintenance of Mycobacterium smegmatis. Curr Protoc Microbiol, Chapter 10, Unit10C 1. [DOI] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., et al. (1985). Measurement of protein using bicinchoninic acid. Anal Biochem 15076–85. 10.1016/0003-2697(85)90442-7 [DOI] [PubMed] [Google Scholar]
- Sompolinsky D., Lagziel A., Naveh D., Yankilevitz T.(1978). Mycobacterium haemophilum sp. nov., a new pathogen of humans. Int J Syst Evol Microbiol 2867–75. [Google Scholar]
- Sondermann H., Shikuma N. J., Yildiz F. H.(2012). You've come a long way: c-di-GMP signaling. Curr Opin Microbiol 15140–146. 10.1016/j.mib.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer J. S., Dockrell H. M., Kim H. J., Marques M. A., Williams D. L., Martins M. V., Martins M. L., Lima M. C., Sarno E. N., et al. (2005). Identification of specific proteins and peptides in Mycobacterium leprae suitable for the selective diagnosis of leprosy. J Immunol 1757930–7938. 10.4049/jimmunol.175.12.7930 [DOI] [PubMed] [Google Scholar]
- Starkey M., Hickman J. H., Ma L., Zhang N., De Long S., Hinz A., Palacios S., Manoil C., Kirisits M. J., et al. (2009). Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol 1913492–3503. 10.1128/JB.00119-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufariello J. M., Kerantzas C. A., Vilchèze C., Calder R. B., Nordberg E. K., Fischer J. A., Hartman T. E., Yang E., Driscoll T., et al. (2015). The complete genome sequence of the emerging pathogen Mycobacterium haemophilum explains its unique culture requirements. MBio 6e01313–e01315. 10.1128/mBio.01313-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A., Wood T. K.(2009). Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog 5e1000483. 10.1371/journal.ppat.1000483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2014). Weekly epidemiological record (WER).89389–400. [Google Scholar]
- Walker S. L., Lockwood D. N.(2007). Leprosy. Clin Dermatol 25165–172. 10.1016/j.clindermatol.2006.05.012 [DOI] [PubMed] [Google Scholar]
- Wassmann P., Chan C., Paul R., Beck A., Heerklotz H., Jenal U., Schirmer T.(2007). Structure of BeF3−-modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure 15915–927. 10.1016/j.str.2007.06.016 [DOI] [PubMed] [Google Scholar]
- Williams D. L., Slayden R. A., Amin A., Martinez A. N., Pittman T. L., Mira A., Mitra A., Nagaraja V., Morrison N. E., et al. (2009). Implications of high level pseudogene transcription in Mycobacterium leprae. BMC Genomics 10397. 10.1186/1471-2164-10-397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M. C., Lee V. T., Gilmore M. E., Lory S.(2003). Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell 4253–263 10.1016/S1534-5807(03)00019-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.